‘Organ’ising Floral Organ Development
Abstract
:1. Introduction
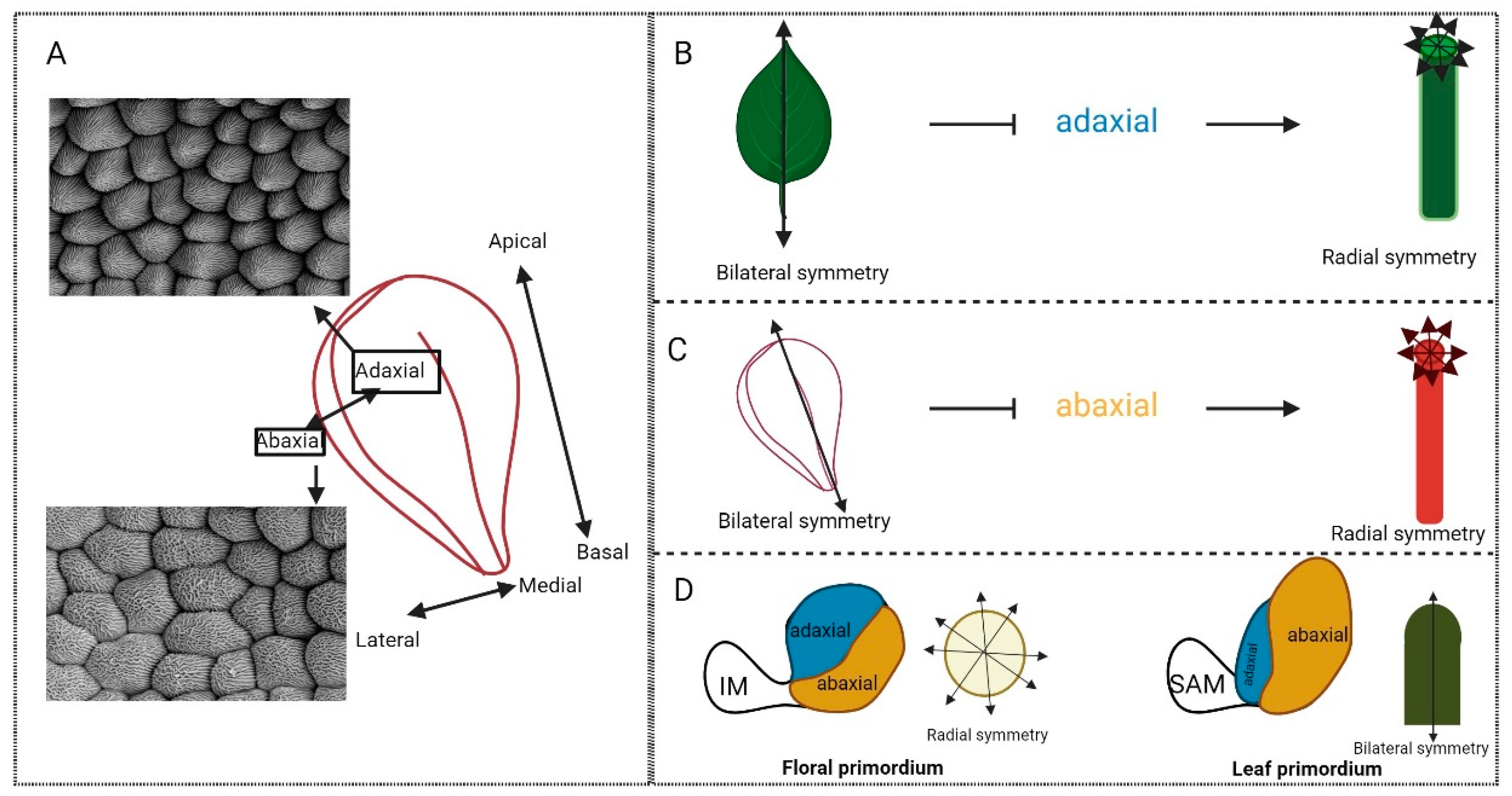
2. Symmetry
2.1. The Beauty of Biological Symmetry
2.2. Symmetry Establishment in Plants
2.3. Morphogens and Phytohormones in Plant Symmetry Establishment
2.4. Transitions in Flowers and Floral Organ Symmetry
3. Determining Organ Identity Establishment
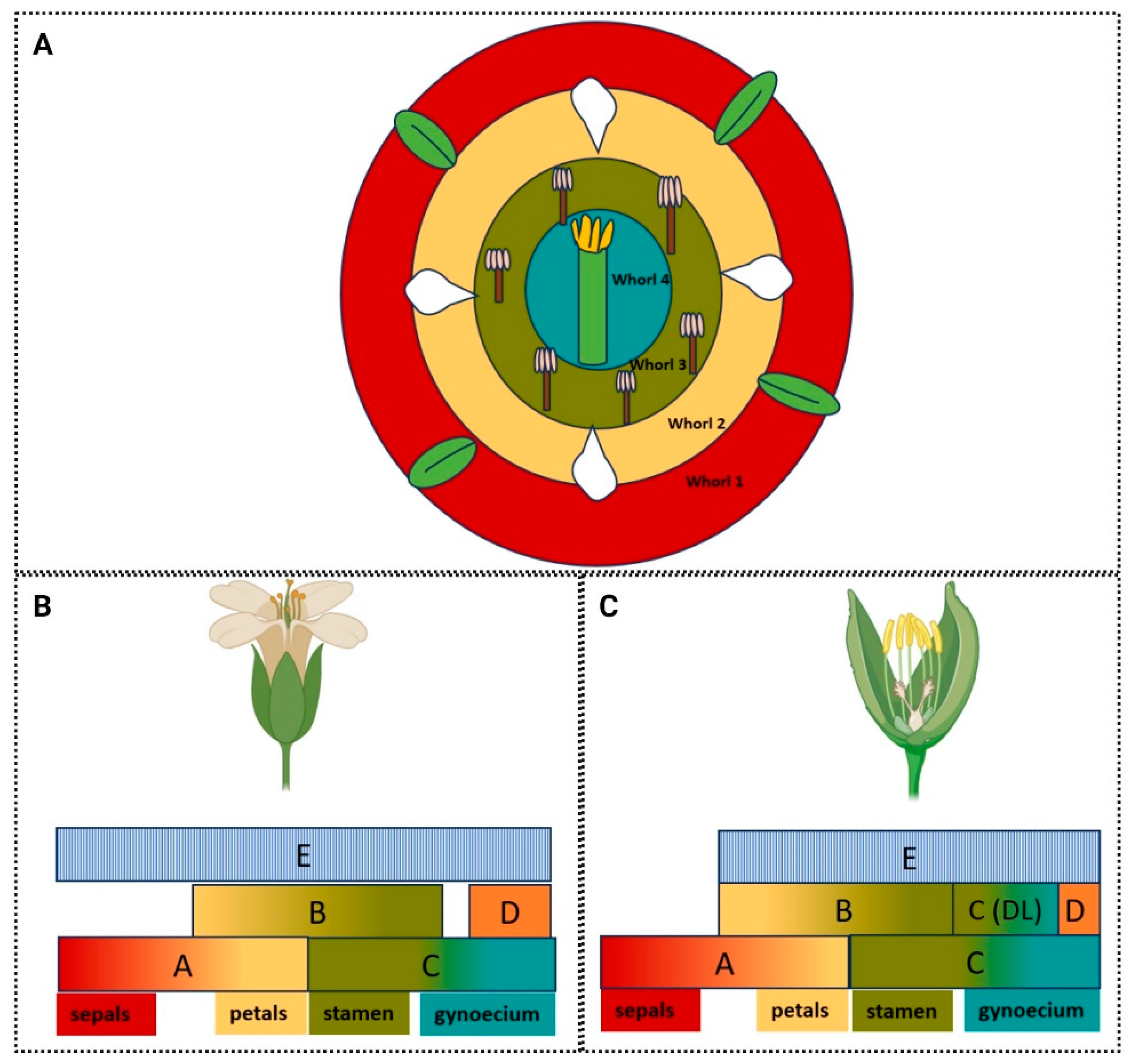
3.1. Flower Identity and Distinction from Leaf-Like Lateral Organs Is Conferred by ABC Genes
3.2. The ABCDE Model: An Extension of the ABC Model Describes the Designation of Floral Organ Identity
3.3. Diversity of the ABCDE Model to Suit Environment and Function
3.4. Interactions with the ABCDE Model Confer Specificity at a Tissue and Organ Level
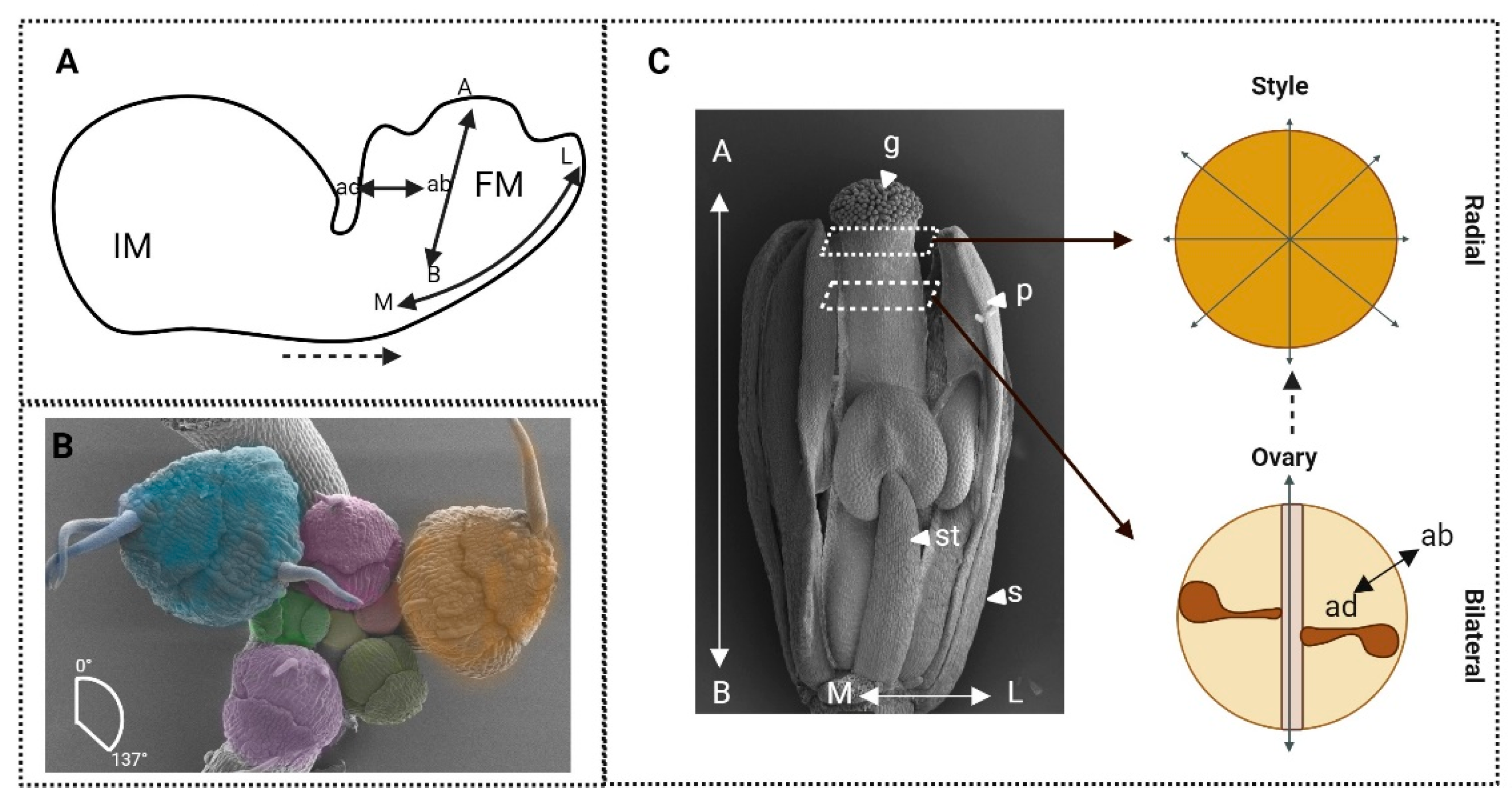
4. ‘Organ’isation across the Axes
4.1. Shaping Identity: How Regulation of Axis Identity Informs the Identity of Floral Organs
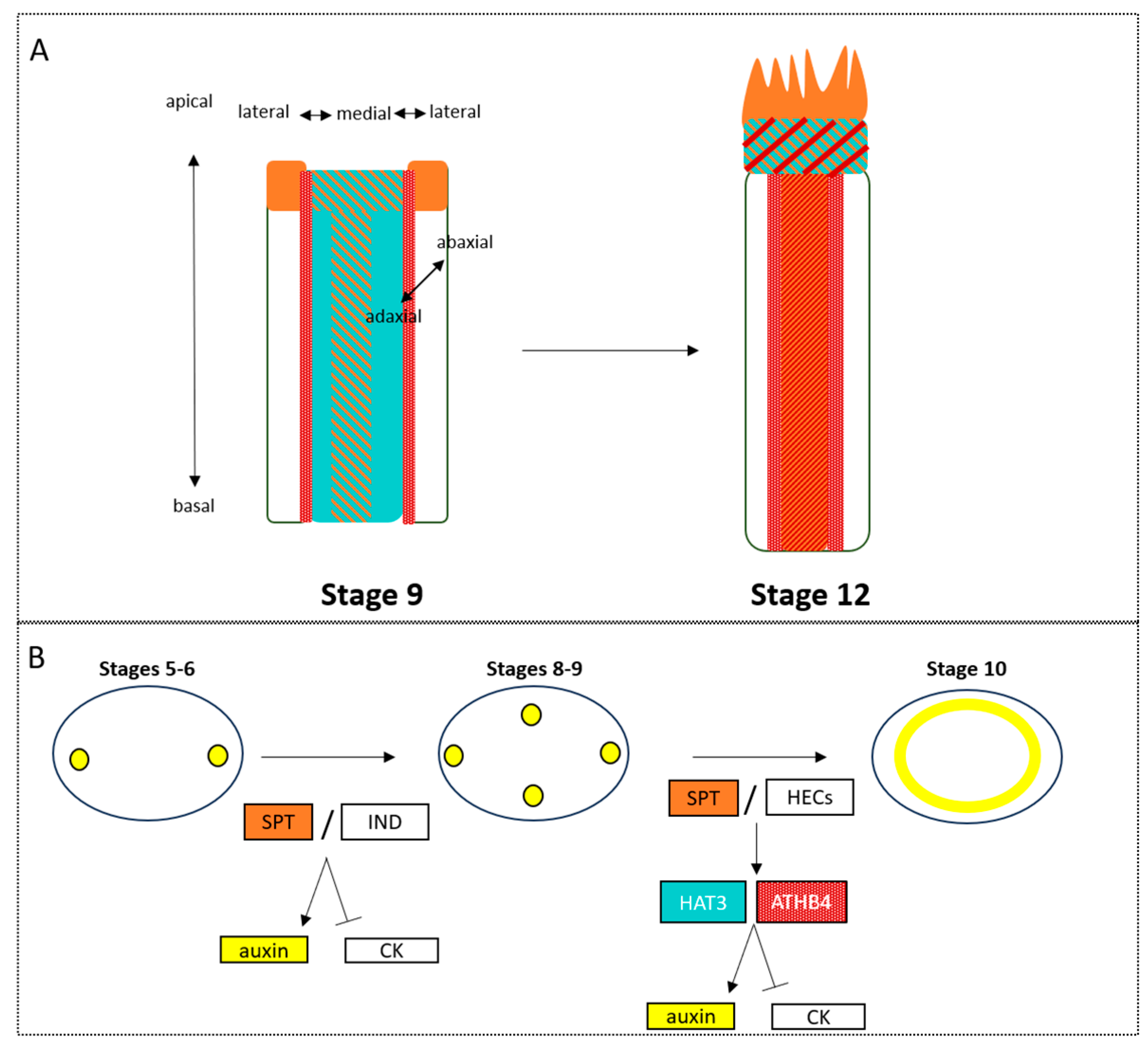
4.2. The Role of Phytohormones in Establishing Axes
4.3. The Functional Importance of Axis Specification
4.4. The Role of Homeobox Genes in Specifying Axis and Plant Organ Shape
4.5. How Axiality Informs Identity and Symmetry in the Development of Floral Organ Structure
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ollerton, J.; Winfree, R.; Tarrant, S. How many flowering plants are pollinated by animals? Oikos 2011, 120, 321–326. [Google Scholar] [CrossRef]
- Smyth, D.R. Morphogenesis of flowers—Our evolving view. Plant Cell 2005, 17, 330–341. [Google Scholar] [CrossRef] [PubMed]
- Vijayraghavan, U.; Prasad, K.; Meyerowitz, E. Specification and maintenance of the floral meristem: Interactions between positively-acting promoters of flowering and negative regulators. Curr. Sci. 2005, 89, 1835–1843. [Google Scholar]
- Saedler, H.; Huijser, P. Molecular biology of flower development in Antirrhinum majus (snapdragon). Gene 1993, 135, 239–243. [Google Scholar] [CrossRef]
- Steeves, T.A.; Sussex, I.M. Patterns in Plant Development; Cambridge University Press: Cambridge, UK, 1989. [Google Scholar]
- Mandel, M.A.; Yanofsky, M.F. A gene triggering flower formation in Arabidopsis. Nature 1995, 377, 522–524. [Google Scholar] [CrossRef] [PubMed]
- Weigel, D.; Alvarez, J.; Smyth, D.R.; Yanofsky, M.F.; Meyerowitz, E.M. LEAFY controls floral meristem identity in Arabidopsis. Cell 1992, 69, 843–859. [Google Scholar] [CrossRef] [PubMed]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, W.; Krumlauf, R. Homeobox genes and axial patterning. Cell 1992, 68, 283–302. [Google Scholar] [CrossRef]
- Kmita, M.; Duboule, D. Organizing axes in time and space; 25 years of colinear tinkering. Science 2003, 301, 331–333. [Google Scholar] [CrossRef]
- Lewis, J.; Slack, J.; Wolpert, L. Thresholds in development. J. Theor. Biol. 1977, 65, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Gross, D.J. The role of symmetry in fundamental physics. Proc. Natl. Acad. Sci. USA 1996, 93, 14256–14259. [Google Scholar] [CrossRef] [PubMed]
- McManus, I.C. Symmetry and asymmetry in aesthetics and the arts. Eur. Rev. 2005, 13, 157–180. [Google Scholar] [CrossRef]
- Edelstein-Keshet, L. Mathematical Models in Biology; SIAM: Philadelphia, PA, USA, 2005. [Google Scholar]
- Perrett, D.I.; Burt, D.M.; Penton-Voak, I.S.; Lee, K.J.; Rowland, D.A.; Edwards, R. Symmetry and human facial attractiveness. Evol. Hum. Behav. 1999, 20, 295–307. [Google Scholar] [CrossRef]
- Møller, A.P.; Eriksson, M. Pollinator preference for symmetrical flowers and sexual selection in plants. Oikos 1995, 73, 15–22. [Google Scholar] [CrossRef]
- Little, A.C.; Jones, B.C.; DeBruine, L.M. Facial attractiveness: Evolutionary based research. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 1638–1659. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, I.; Gumbert, A.; Hempel de Ibarra, N.; Kunze, J.; Giurfa, M. Symmetry is in the eye of the ‘beeholder’: Innate preference for bilateral symmetry in flower-naïve bumblebees. Naturwissenschaften 2004, 91, 374–377. [Google Scholar] [CrossRef] [PubMed]
- Helversen, D.v.; Holderied, M.W.; Helversen, O.v. Echoes of bat-pollinated bell-shaped flowers: Conspicuous for nectar-feeding bats? J. Exp. Biol. 2003, 206, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Hůla, M.; Flegr, J. What flowers do we like? The influence of shape and color on the rating of flower beauty. PeerJ 2016, 4, e2106. [Google Scholar] [CrossRef]
- Endress, P.K.; Doyle, J.A. Reconstructing the ancestral angiosperm flower and its initial specializations. Am. J. Bot. 2009, 96, 22–66. [Google Scholar] [CrossRef]
- Luo, D.; Carpenter, R.; Vincent, C.; Copsey, L.; Coen, E. Origin of floral asymmetry in Antirrhinum. Nature 1996, 383, 794–799. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, R.; Coen, E.S. Floral homeotic mutations produced by transposon-mutagenesis in Antirrhinum majus. Genes Dev. 1990, 4, 1483–1493. [Google Scholar] [CrossRef] [PubMed]
- Remizowa, M.V.; Shipunov, A.B.; Sokoloff, D.D. When asymmetry mimics zygomorphy: Flower development in Chamaelirium japonicum (Melanthiaceae, Liliales). Bot. Pac. 2023, 12, 3–14. [Google Scholar] [CrossRef]
- Luo, D.; Carpenter, R.; Copsey, L.; Vincent, C.; Clark, J.; Coen, E. Control of organ asymmetry in flowers of Antirrhinum. Cell 1999, 99, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.; Rocheta, M.; Galego, L. Genetic control of flower shape in Antirrhinum majus. Development 1997, 124, 1387–1392. [Google Scholar] [CrossRef] [PubMed]
- Galego, L.; Almeida, J. Role of DIVARICATA in the control of dorsoventral asymmetry in Antirrhinum flowers. Genes Dev. 2002, 16, 880–891. [Google Scholar] [CrossRef]
- Kramer, E.M. Floral architecture: Regulation and diversity of floral shape and pattern. In Annual Plant Reviews Online; Wiley: Hoboken, NJ, USA, 2018; pp. 121–148. [Google Scholar]
- Endress, P.K. Antirrhinum and Asteridae–evolutionary changes of floral symmetry. Symp. Soc. Exp. Biol. 1998, 51, 133–140. [Google Scholar] [PubMed]
- Hileman, L.C.; Kramer, E.M.; Baum, D.A. Differential regulation of symmetry genes and the evolution of floral morphologies. Proc. Natl. Acad. Sci. USA 2003, 100, 12814–12819. [Google Scholar] [CrossRef]
- Cubas, P.; Vincent, C.; Coen, E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature 1999, 401, 157–161. [Google Scholar] [CrossRef]
- Citerne, H.L.; Reyes, E.; Le Guilloux, M.; Delannoy, E.; Simonnet, F.; Sauquet, H.; Weston, P.H.; Nadot, S.; Damerval, C. Characterization of CYCLOIDEA-like genes in Proteaceae, a basal eudicot family with multiple shifts in floral symmetry. Ann. Bot. 2017, 119, 367–378. [Google Scholar] [CrossRef]
- Running, M.P.; Meyerowitz, E.M. Mutations in the PERIANTHIA gene of Arabidopsis specifically alter floral organ number and initiation pattern. Development 1996, 122, 1261–1269. [Google Scholar] [CrossRef] [PubMed]
- Endress, P.K. Evolution of floral symmetry. Curr. Opin. Plant Biol. 2001, 4, 86–91. [Google Scholar] [CrossRef]
- Jabbour, F.; Damerval, C.; Nadot, S. Evolutionary trends in the flowers of Asteridae: Is polyandry an alternative to zygomorphy? Ann. Bot. 2008, 102, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Ronse Decraene, L.; Smets, E. Merosity in flowers: Definition, origin, and taxonomic significance. Plant Syst. Evol. 1994, 191, 83–104. [Google Scholar] [CrossRef]
- Chandler, J.W.; Werr, W. Arabidopsis floral phytomer development: Auxin response relative to biphasic modes of organ initiation. J. Exp. Bot. 2014, 65, 3097–3110. [Google Scholar] [CrossRef]
- Bossinger, G.; Smyth, D.R. Initiation patterns of flower and floral organ development in Arabidopsis thaliana. Development 1996, 122, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, A.; Kitazawa, M.S.; Fujimoto, K. A design principle for floral organ number and arrangement in flowers with bilateral symmetry. Development 2020, 147, dev182907. [Google Scholar] [CrossRef]
- Driever, W.; Nüsslein-Volhard, C. A gradient of bicoid protein in Drosophila embryos. Cell 1988, 54, 83–93. [Google Scholar] [CrossRef]
- Chan, S.-K.; Struhl, G. Sequence-specific RNA binding by bicoid. Nature 1997, 388, 634. [Google Scholar] [CrossRef]
- Briscoe, J.; Small, S. Morphogen rules: Design principles of gradient-mediated embryo patterning. Development 2015, 142, 3996–4009. [Google Scholar] [CrossRef]
- Turing, A. A reaction-diffusion model for development, The chemical basis of morphogenesis. Philos. Trans. R. Soc. Lond. Ser. B 1952, 237, 37–72. [Google Scholar]
- Sabatini, S.; Beis, D.; Wolkenfelt, H.; Murfett, J.; Guilfoyle, T.; Malamy, J.; Benfey, P.; Leyser, O.; Bechtold, N.; Weisbeek, P.; et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 1999, 99, 463–472. [Google Scholar] [CrossRef]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef]
- Grieneisen, V.A.; Xu, J.; Marée, A.F.; Hogeweg, P.; Scheres, B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 2007, 449, 1008–1013. [Google Scholar] [CrossRef]
- Petersson, S.V.; Johansson, A.I.; Kowalczyk, M.; Makoveychuk, A.; Wang, J.Y.; Moritz, T.; Grebe, M.; Benfey, P.N.; Sandberg, G.; Ljung, K. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 2009, 21, 1659–1668. [Google Scholar] [CrossRef]
- Bhalerao, R.P.; Bennett, M.J. The case for morphogens in plants. Nat. Cell Biol. 2003, 5, 939–943. [Google Scholar] [CrossRef] [PubMed]
- BERGBUSCH, V.L. A note on the manipulation of flower symmetry in Antirrhinum majus. Ann. Bot. 1999, 83, 483–488. [Google Scholar] [CrossRef]
- Chandler, J.W. Founder cell specification. Trends Plant Sci. 2011, 16, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Lampugnani, E.R.; Kilinc, A.; Smyth, D.R. Auxin controls petal initiation in Arabidopsis. Development 2013, 140, 185–194. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, W.; Mirabet, V.; Hong, L.; Bovio, S.; Strauss, S.; Schwarz, E.M.; Tsugawa, S.; Wang, Z.; Smith, R.S. Robust organ size requires robust timing of initiation orchestrated by focused auxin and cytokinin signalling. Nat. Plants 2020, 6, 686–698. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Wu, M.-F.; Winter, C.M.; Wagner, D. LEAFY and polar auxin transport coordinately regulate Arabidopsis flower development. Plants 2014, 3, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Moubayidin, L.; Østergaard, L. Dynamic control of auxin distribution imposes a bilateral-to-radial symmetry switch during gynoecium development. Curr. Biol. 2014, 24, 2743–2748. [Google Scholar] [CrossRef] [PubMed]
- Reyes, E.; Sauquet, H.; Nadot, S. Perianth symmetry changed at least 199 times in angiosperm evolution. Taxon 2016, 65, 945–964. [Google Scholar] [CrossRef]
- Mair, O. Zur Entwicklungsgeschichte Monosymmetrischer Dicotylen-Blüten. Botany Thesis, Universität Augsburg, Augsburg, Germany, 1977. [Google Scholar]
- Jabbour, F.; Ronse De Craene, L.P.; Nadot, S.; Damerval, C. Establishment of zygomorphy on an ontogenic spiral and evolution of perianth in the tribe Delphinieae (Ranunculaceae). Ann. Bot. 2009, 104, 809–822. [Google Scholar] [CrossRef]
- Gómez-Felipe, A.; Kierzkowski, D.; de Folter, S. The relationship between AGAMOUS and cytokinin signaling in the establishment of carpeloid features. Plants 2021, 10, 827. [Google Scholar] [CrossRef]
- Damerval, C.; Citerne, H.; Le Guilloux, M.; Domenichini, S.; Dutheil, J.; Ronse de Craene, L.; Nadot, S. Asymmetric morphogenetic cues along the transverse plane: Shift from disymmetry to zygomorphy in the flower of Fumarioideae. Am. J. Bot. 2013, 100, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Moubayidin, L. Floral symmetry: The geometry of plant reproduction. Emerg. Top. Life Sci. 2022, 6, 259–269. [Google Scholar] [CrossRef]
- Moubayidin, L.; Østergaard, L. Symmetry matters. New Phytol. 2015, 207, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Carabelli, M.; Turchi, L.; Morelli, G.; Østergaard, L.; Ruberti, I.; Moubayidin, L. Coordination of biradial-to-radial symmetry and tissue polarity by HD-ZIP II proteins. Nat. Commun. 2021, 12, 4321. [Google Scholar] [CrossRef]
- Ballester, P.; Martínez-Godoy, M.A.; Ezquerro, M.; Navarrete-Gómez, M.; Trigueros, M.; Rodríguez-Concepción, M.; Ferrándiz, C. A transcriptional complex of NGATHA and bHLH transcription factors directs stigma development in Arabidopsis. Plant Cell 2021, 33, 3645–3657. [Google Scholar] [CrossRef]
- Larsson, E.; Roberts, C.J.; Claes, A.R.; Franks, R.G.; Sundberg, E. Polar auxin transport is essential for medial versus lateral tissue specification and vascular-mediated valve outgrowth in Arabidopsis gynoecia. Plant Physiol. 2014, 166, 1998–2012. [Google Scholar] [CrossRef]
- Alvarez, J.; Smyth, D.R. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 1999, 126, 2377–2386. [Google Scholar] [CrossRef]
- Bowman, J.L.; Moyroud, E. Reflections on the ABC model of flower development. Plant Cell 2024, 36, 1334–1357. [Google Scholar] [CrossRef] [PubMed]
- Goethe, J.v. Versuch die Metamorphose der Pflanzen zu Erklären. Ettinger, Gotha. Translated by A. Arber 1946. Goethe’s botany. Chron. Bot. 1790, 10, 63–126. [Google Scholar]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genetic interactions among floral homeotic genes of Arabidopsis. Development 1991, 112, 1–20. [Google Scholar] [CrossRef]
- Sablowski, R. Control of patterning, growth, and differentiation by floral organ identity genes. J. Exp. Bot. 2015, 66, 1065–1073. [Google Scholar] [CrossRef]
- Fukuda, Y.; Suzuki, K.; Murata, J. The function of each sepal in pollinator behavior and effective pollination in Aconitum japonicum var. montanum. Plant Species Biol. 2001, 16, 151–157. [Google Scholar] [CrossRef]
- Dohzono, I.; Suzuki, K. Bumblebee-pollination and temporal change of the calyx tube length in Clematis stans (Ranunculaceae). J. Plant Res. 2002, 115, 355–359. [Google Scholar] [CrossRef]
- Endress, P.K.; Matthews, M.L. Elaborate petals and staminodes in eudicots: Diversity, function, and evolution. Org. Divers. Evol. 2006, 6, 257–293. [Google Scholar] [CrossRef]
- Huang, J.; Yang, L.; Yang, L.; Wu, X.; Cui, X.; Zhang, L.; Hui, J.; Zhao, Y.; Yang, H.; Liu, S. Stigma receptors control intraspecies and interspecies barriers in Brassicaceae. Nature 2023, 614, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Edlund, A.F.; Swanson, R.; Preuss, D. Pollen and stigma structure and function: The role of diversity in pollination. Plant Cell 2004, 16, S84–S97. [Google Scholar] [CrossRef] [PubMed]
- Jofuku, K.D.; Den Boer, B.; Van Montagu, M.; Okamuro, J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar] [PubMed]
- Favaro, R.; Pinyopich, A.; Battaglia, R.; Kooiker, M.; Borghi, L.; Ditta, G.; Yanofsky, M.F.; Kater, M.M.; Colombo, L. MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 2003, 15, 2603–2611. [Google Scholar] [CrossRef] [PubMed]
- Pinyopich, A.; Ditta, G.S.; Savidge, B.; Liljegren, S.J.; Baumann, E.; Wisman, E.; Yanofsky, M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003, 424, 85–88. [Google Scholar] [CrossRef]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef]
- Jetha, K.; Theißen, G.; Melzer, R. Arabidopsis SEPALLATA proteins differ in cooperative DNA-binding during the formation of floral quartet-like complexes. Nucleic Acids Res. 2014, 42, 10927–10942. [Google Scholar] [CrossRef]
- Litt, A.; Kramer, E.M. The ABC model and the diversification of floral organ identity. Semin. Cell Dev. Biol. 2010, 21, 129–137. [Google Scholar] [CrossRef]
- Huijser, P.; Klein, J.; Lönnig, W.; Meijer, H.; Saedler, H.; Sommer, H. Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J. 1992, 11, 1239–1249. [Google Scholar] [CrossRef]
- Sommer, H.; Beltran, J.-P.; Huijser, P.; Pape, H.; Lönnig, W.-E.; Saedler, H.; Schwarz-Sommer, Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 1990, 9, 605–613. [Google Scholar] [CrossRef]
- Tröbner, W.; Ramirez, L.; Motte, P.; Hue, I.; Huijser, P.; Lönnig, W.; Saedler, H.; Sommer, H.; Schwarz-Sommer, Z. GLOBOSA: A homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 1992, 11, 4693–4704. [Google Scholar] [CrossRef] [PubMed]
- Davies, B.; Motte, P.; Keck, E.; Saedler, H.; Sommer, H.; Schwarz-Sommer, Z. PLENA and FARINELLI: Redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J. 1999, 18, 4023–4034. [Google Scholar] [CrossRef] [PubMed]
- Kramer, E.M.; Jaramillo, M.A.; Di Stilio, V.S. Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 2004, 166, 1011–1023. [Google Scholar] [CrossRef] [PubMed]
- Motte, P.; Saedler, H.; Schwarz-Sommer, Z. STYLOSA and FISTULATA: Regulatory components of the homeotic control of Antirrhinum floral organogenesis. Development 1998, 125, 71–84. [Google Scholar] [CrossRef]
- Wilkinson, M.; de Andrade Silva, E.; Zachgo, S.; Saedler, H.; Schwarz-Sommer, Z. CHORIPETALA and DESPENTEADO: General regulators during plant development and potential floral targets of FIMBRIATA-mediated degradation. Development 2000, 127, 3725–3734. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Gouy, M.; Yang, Y.-W.; Sharp, P.M.; Li, W.-H. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 1989, 86, 6201–6205. [Google Scholar] [CrossRef] [PubMed]
- Salse, J.; Piégu, B.; Cooke, R.; Delseny, M. Synteny between Arabidopsis thaliana and rice at the genome level: A tool to identify conservation in the ongoing rice genome sequencing project. Nucleic Acids Res. 2002, 30, 2316–2328. [Google Scholar] [CrossRef]
- Rudall, P.J.; Bateman, R.M. Evolution of zygomorphy in monocot flowers: Iterative patterns and developmental constraints. New Phytol. 2004, 162, 25–44. [Google Scholar] [CrossRef]
- Kater, M.M.; Dreni, L.; Colombo, L. Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J. Exp. Bot. 2006, 57, 3433–3444. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Lee, D.Y.; Miyao, A.; Hirochika, H.; An, G.; Hirano, H.-Y. Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 2006, 18, 15–28. [Google Scholar] [CrossRef]
- Nagasawa, N.; Miyoshi, M.; Sano, Y.; Satoh, H.; Hirano, H.; Sakai, H.; Nagato, Y. SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 2003, 130, 705–718. [Google Scholar] [CrossRef] [PubMed]
- Pelaz, S.; Tapia-López, R.; Alvarez-Buylla, E.R.; Yanofsky, M.F. Conversion of leaves into petals in Arabidopsis. Curr. Biol. 2001, 11, 182–184. [Google Scholar] [CrossRef] [PubMed]
- Theißen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Theißen, G.; Melzer, R.; Rümpler, F. MADS-domain transcription factors and the floral quartet model of flower development: Linking plant development and evolution. Development 2016, 143, 3259–3271. [Google Scholar] [CrossRef] [PubMed]
- Goslin, K.; Finocchio, A.; Wellmer, F. Floral Homeotic Factors: A Question of Specificity. Plants 2023, 12, 1128. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.S.; Lelli, K.M.; Joshi, R. Hox specificity: Unique roles for cofactors and collaborators. Curr. Top. Dev. Biol. 2009, 88, 63–101. [Google Scholar] [PubMed]
- Slattery, M.; Riley, T.; Liu, P.; Abe, N.; Gomez-Alcala, P.; Dror, I.; Zhou, T.; Rohs, R.; Honig, B.; Bussemaker, H.J. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 2011, 147, 1270–1282. [Google Scholar] [CrossRef] [PubMed]
- Brambilla, V.; Battaglia, R.; Colombo, M.; Masiero, S.; Bencivenga, S.; Kater, M.M.; Colombo, L. Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. Plant Cell 2007, 19, 2544–2556. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Ubaldo, H.; Campos, S.E.; López-Gómez, P.; Luna-García, V.; Zúñiga-Mayo, V.M.; Armas-Caballero, G.E.; González-Aguilera, K.L.; DeLuna, A.; Marsch-Martínez, N.; Espinosa-Soto, C.; et al. The protein–protein interaction landscape of transcription factors during gynoecium development in Arabidopsis. Mol. Plant 2023, 16, 260–278. [Google Scholar] [CrossRef]
- Heisler, M.G.; Atkinson, A.; Bylstra, Y.H.; Walsh, R.; Smyth, D.R. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 2001, 128, 1089–1098. [Google Scholar] [CrossRef]
- Gremski, K.; Ditta, G.; Yanofsky, M.F. The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 2007, 134, 3593–3601. [Google Scholar] [CrossRef] [PubMed]
- Crawford, B.C.; Yanofsky, M.F. HALF FILLED promotes reproductive tract development and fertilization efficiency in Arabidopsis thaliana. Development 2011, 138, 2999–3009. [Google Scholar] [CrossRef] [PubMed]
- Hackbusch, J.; Richter, K.; Müller, J.; Salamini, F.; Uhrig, J.F. A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 4908–4912. [Google Scholar] [CrossRef] [PubMed]
- Smaczniak, C.; Immink, R.G.; Muiño, J.M.; Blanvillain, R.; Busscher, M.; Busscher-Lange, J.; Dinh, Q.; Liu, S.; Westphal, A.H.; Boeren, S. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 2012, 109, 1560–1565. [Google Scholar] [CrossRef] [PubMed]
- Żyła, N.; Babula-Skowrońska, D. Evolutionary Consequences of Functional and Regulatory Divergence of HD-Zip I Transcription Factors as a Source of Diversity in Protein Interaction Networks in Plants. J. Mol. Evol. 2023, 91, 581–597. [Google Scholar] [CrossRef]
- Pramila, T.; Miles, S.; GuhaThakurta, D.; Jemiolo, D.; Breeden, L.L. Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes Dev. 2002, 16, 3034–3045. [Google Scholar] [CrossRef] [PubMed]
- Bou-Torrent, J.; Salla-Martret, M.; Brandt, R.; Musielak, T.; Palauqui, J.-C.; Martínez-García, J.F.; Wenkel, S. ATHB4 and HAT3, two class II HD-ZIP transcription factors, control leaf development in Arabidopsis. Plant Signal. Behav. 2012, 7, 1382–1387. [Google Scholar] [CrossRef]
- Díaz, J.; Álvarez-Buylla, E.R. Spatio-temporal dynamics of the patterning of Arabidopsis flower meristem. Front. Plant Sci. 2021, 12, 585139. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jürgens, G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef]
- Hill, J.P.; Lord, E.M. Floral development in Arabidopsis thaliana: A comparison of the wild type and the homeotic pistillata mutant. Can. J. Bot. 1989, 67, 2922–2936. [Google Scholar] [CrossRef]
- Chandler, J. Floral meristem initiation and emergence in plants. Cell. Mol. Life Sci. 2012, 69, 3807–3818. [Google Scholar] [CrossRef] [PubMed]
- Tucker, S.C. Unidirectional organ initiation in leguminous flowers. Am. J. Bot. 1984, 71, 1139–1148. [Google Scholar] [CrossRef]
- Kwiatkowska, D. Flower primordium formation at the Arabidopsis shoot apex: Quantitative analysis of surface geometry and growth. J. Exp. Bot. 2006, 57, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xu, Z.; Chua, N.-h. Auxin polar transport is essential for the establishment of bilateral symmetry during early plant embryogenesis. Plant Cell 1993, 5, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Robert, H.S.; Grones, P.; Stepanova, A.N.; Robles, L.M.; Lokerse, A.S.; Alonso, J.M.; Weijers, D.; Friml, J. Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr. Biol. 2013, 23, 2506–2512. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Zhang, Z.J.; Laux, T. Transcriptional Activation of Arabidopsis Axis Patterning Genes WOX8/9 Links Zygote Polarity to Embryo Development. Dev. Cell 2011, 20, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Sussex, I. Experiments on the cause of dorsiventrality in leaves. Nature 1951, 167, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Wang, Y.; Yu, T.; Cunha, A.; Wu, B.; Vernoux, T.; Meyerowitz, E.; Jiao, Y. Auxin depletion from leaf primordia contributes to organ patterning. Proc. Natl. Acad. Sci. USA 2014, 111, 18769–18774. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.M.; Dong, J.Q.; Xue, J.S.; Wang, H.; Yang, Z.N.; Jiao, Y.L.; Xu, L.; Huang, H. Model for the role of auxin polar transport in patterning of the leaf adaxial-abaxial axis. Plant J. 2017, 92, 469–480. [Google Scholar] [CrossRef]
- Nakata, M.; Matsumoto, N.; Tsugeki, R.; Rikirsch, E.; Laux, T.; Okada, K. Roles of the middle domain–specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell 2012, 24, 519–535. [Google Scholar] [CrossRef]
- Nakata, M.; Okada, K. The three-domain model: A new model for the early development of leaves in Arabidopsis thaliana. Plant Signal. Behav. 2012, 7, 1423–1427. [Google Scholar] [CrossRef]
- Pyke, K.A.; Page, A.M. Plastid ontogeny during petal development in Arabidopsis. Plant Physiol. 1998, 116, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Vignolini, S.; Moyroud, E.; Hingant, T.; Banks, H.; Rudall, P.J.; Steiner, U.; Glover, B.J. The flower of H ibiscus trionum is both visibly and measurably iridescent. New Phytol. 2015, 205, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Moyroud, E.; Wenzel, T.; Middleton, R.; Rudall, P.J.; Banks, H.; Reed, A.; Mellers, G.; Killoran, P.; Westwood, M.M.; Steiner, U. Disorder in convergent floral nanostructures enhances signalling to bees. Nature 2017, 550, 469–474. [Google Scholar] [CrossRef]
- Matsumoto, N.; Okada, K. A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev. 2001, 15, 3355–3364. [Google Scholar] [CrossRef]
- Waites, R.; Hudson, A. phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 1995, 121, 2143–2154. [Google Scholar] [CrossRef]
- Cavallini-Speisser, Q.; Morel, P.; Monniaux, M. Petal cellular identities. Front. Plant Sci. 2021, 12, 745507. [Google Scholar] [CrossRef]
- Gorton, H.L.; Vogelmann, T.C. Effects of epidermal cell shape and pigmentation on optical properties of Antirrhinum petals at visible and ultraviolet wavelengths. Plant Physiol. 1996, 112, 879–888. [Google Scholar] [CrossRef]
- Kay, Q.; Daoud, H.; Stirton, C. Pigment distribution, light reflection and cell structure in petals. Bot. J. Linn. Soc. 1981, 83, 57–83. [Google Scholar] [CrossRef]
- Kevan, P.G.; Lane, M.A. Flower petal microtexture is a tactile cue for bees. Proc. Natl. Acad. Sci. USA 1985, 82, 4750–4752. [Google Scholar] [CrossRef]
- Baudino, S.; Caissard, J.-C.; Bergougnoux, V.; Jullien, F.; Magnard, J.-L.; Scalliet, G.; Cock, J.M.; Hugueney, P. Production and emission of volatile compounds by petal cells. Plant Signal. Behav. 2007, 2, 525–526. [Google Scholar] [CrossRef] [PubMed]
- Roeder, A.H.; Chickarmane, V.; Cunha, A.; Obara, B.; Manjunath, B.; Meyerowitz, E.M. Variability in the control of cell division underlies sepal epidermal patterning in Arabidopsis thaliana. PLoS Biol. 2010, 8, e1000367. [Google Scholar] [CrossRef] [PubMed]
- Urbanus, S.L.; de Folter, S.; Shchennikova, A.V.; Kaufmann, K.; Immink, R.G.; Angenent, G.C. In planta localisation patterns of MADS domain proteins during floral development in Arabidopsis thaliana. BMC Plant Biol. 2009, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Emery, J.F.; Floyd, S.K.; Alvarez, J.; Eshed, Y.; Hawker, N.P.; Izhaki, A.; Baum, S.F.; Bowman, J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003, 13, 1768–1774. [Google Scholar] [CrossRef] [PubMed]
- Izhaki, A.; Bowman, J.L. KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 2007, 19, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Ruberti, I.; Sessa, G.; Ciolfi, A.; Possenti, M.; Carabelli, M.; Morelli, G. Plant adaptation to dynamically changing environment: The shade avoidance response. Biotechnol. Adv. 2012, 30, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Reymond, M.C.; Brunoud, G.; Chauvet, A.; Martínez-Garcia, J.F.; Martin-Magniette, M.-L.; Monéger, F.; Scutt, C.P. A light-regulated genetic module was recruited to carpel development in Arabidopsis following a structural change to SPATULA. Plant Cell 2012, 24, 2812–2825. [Google Scholar] [CrossRef] [PubMed]
- Wuest, S.E.; O’Maoileidigh, D.S.; Rae, L.; Kwasniewska, K.; Raganelli, A.; Hanczaryk, K.; Lohan, A.J.; Loftus, B.; Graciet, E.; Wellmer, F. Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc. Natl. Acad. Sci. USA 2012, 109, 13452–13457. [Google Scholar] [CrossRef] [PubMed]
- Endress, P.K. Symmetry in flowers: Diversity and evolution. Int. J. Plant Sci. 1999, 160, S3–S23. [Google Scholar] [CrossRef]
- Ciarbelli, A.R.; Ciolfi, A.; Salvucci, S.; Ruzza, V.; Possenti, M.; Carabelli, M.; Fruscalzo, A.; Sessa, G.; Morelli, G.; Ruberti, I. The Arabidopsis homeodomain-leucine zipper II gene family: Diversity and redundancy. Plant Mol. Biol. 2008, 68, 465–478. [Google Scholar] [CrossRef]
- Turchi, L.; Carabelli, M.; Ruzza, V.; Possenti, M.; Sassi, M.; Peñalosa, A.; Sessa, G.; Salvi, S.; Forte, V.; Morelli, G. Arabidopsis HD-Zip II transcription factors control apical embryo development and meristem function. Development 2013, 140, 2118–2129. [Google Scholar] [CrossRef] [PubMed]
- Stahle, M.I.; Kuehlich, J.; Staron, L.; von Arnim, A.G.; Golz, J.F. YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 2009, 21, 3105–3118. [Google Scholar] [CrossRef] [PubMed]
- McConnell, J.R.; Barton, M.K. Leaf polarity and meristem formation in Arabidopsis. Development 1998, 125, 2935–2942. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Alique, D.; Xiong, Y.; Hu, J.; Cao, X.; Lü, S.; Long, M.; Wang, Y.; Wabnik, K.; Jiao, Y. Differential growth dynamics control aerial organ geometry. Curr. Biol. 2022, 32, 4854–4868.e4855. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maio, K.A.; Moubayidin, L. ‘Organ’ising Floral Organ Development. Plants 2024, 13, 1595. https://doi.org/10.3390/plants13121595
Maio KA, Moubayidin L. ‘Organ’ising Floral Organ Development. Plants. 2024; 13(12):1595. https://doi.org/10.3390/plants13121595
Chicago/Turabian StyleMaio, Kestrel A., and Laila Moubayidin. 2024. "‘Organ’ising Floral Organ Development" Plants 13, no. 12: 1595. https://doi.org/10.3390/plants13121595
APA StyleMaio, K. A., & Moubayidin, L. (2024). ‘Organ’ising Floral Organ Development. Plants, 13(12), 1595. https://doi.org/10.3390/plants13121595




