Study of CaDreb2c and CaDreb2h Gene Sequences and Expression in Chickpea (Cicer arietinum L.) Cultivars Growing in Northern Kazakhstan under Drought
Abstract
:1. Introduction
2. Results and Discussion
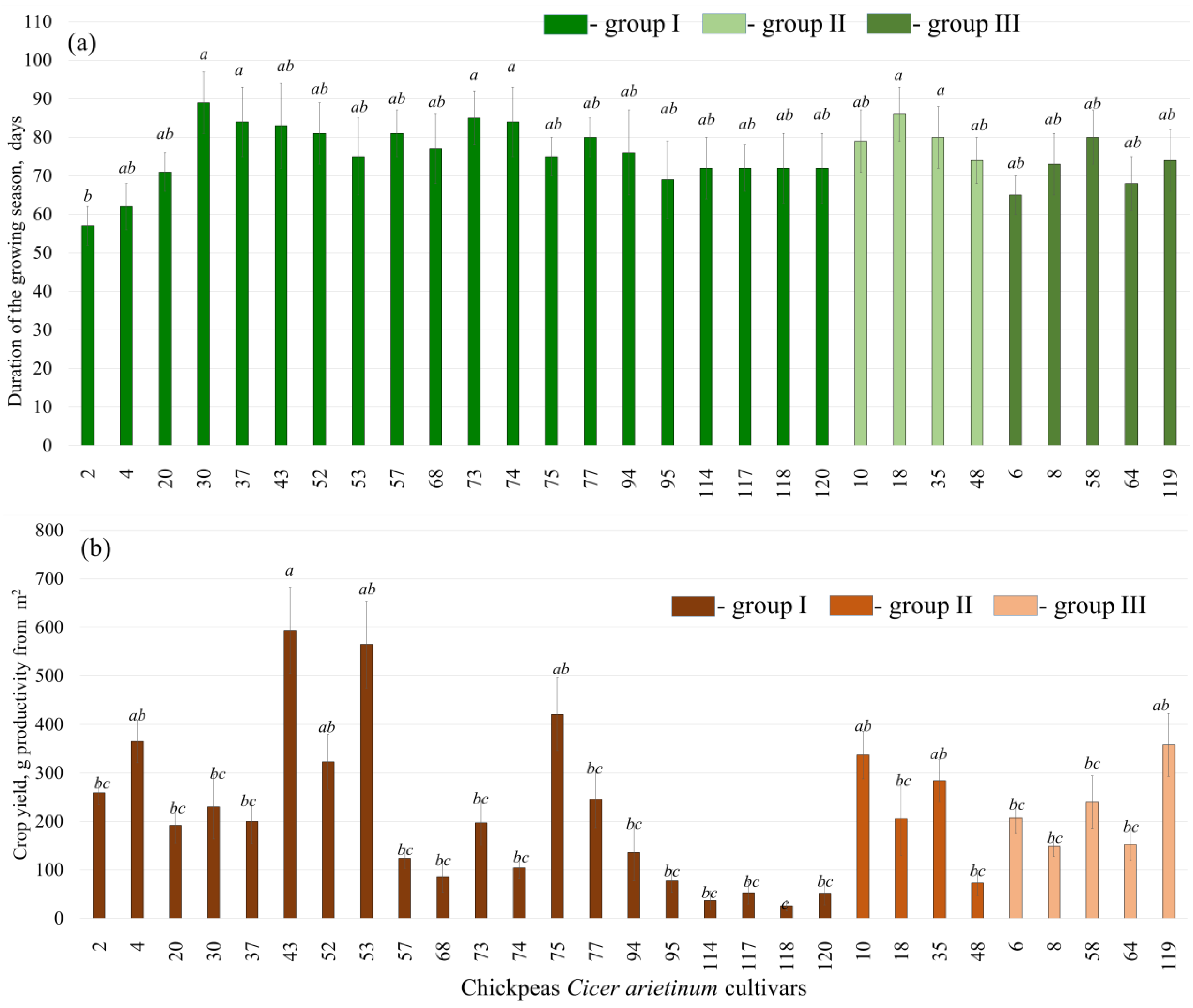
2.1. Selection of Chickpea Cultivars for Drought Experiments Based on Duration of the Growing Season and Crop Yield
2.2. CaDreb2c and CaDreb2h Gene Sequences in the Different Used Chickpea C. arietinum Cultivars
- Twenty were identical for deposited c in GenBank chickpeas CaDreb2c gene sequence (XM_012713080.2, Indian cultivar CDC Frontier) (#2, 4, 20, 30, 37, 43, 52, 53, 57, 68, 73, 74, 75, 77, 94, 95, 114, 117, 118, and 120).
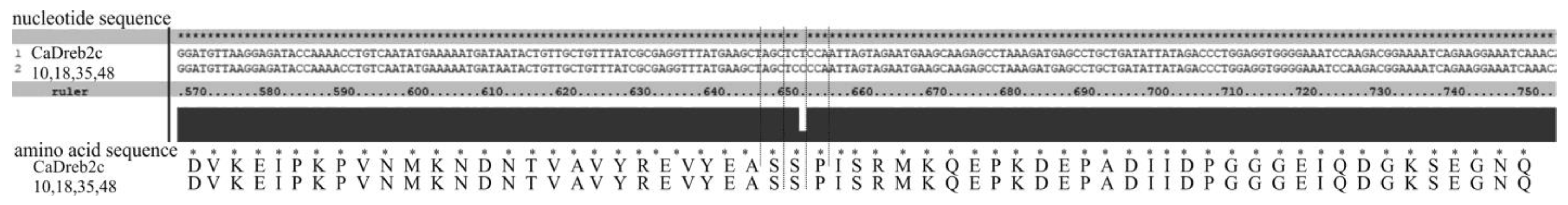
- Four differed by one synonymous substitution (T on C, 651 bp) from the deposited CaDreb2c gene sequence of cultivar CDC Frontier (#10, 18, 35, and 48) (Figure 3).
- Five differed by one synonymous substitution (C on T, 910 bp) and two non-synonymous substitutions (G on T, 790 bp; G on A, 940 bp) from the deposited in GenBank CaDreb2c gene sequence of cultivar CDC Frontier (#6, 8, 58, 64, and 119) (Figure 4). Using a search for conserved domains within a protein sequence (CD-search, National Center for Biotechnology Information, NCBI, https://www.ncbi.nlm.nih.gov, accessed on 23 July 2024), we found DNA-binding domains in plant proteins such as APETALA2 and EREBPs [19]. This domain is located closer to the beginning of the gene (interval 85–145 aa, E-value 3.74 × 10−34). Thus, the found substitutions do not change the structure of the main DNA-binding domain.


2.3. Chickpeas C. arietinum Cultivar Resistance to Drought in Experimental Conditions
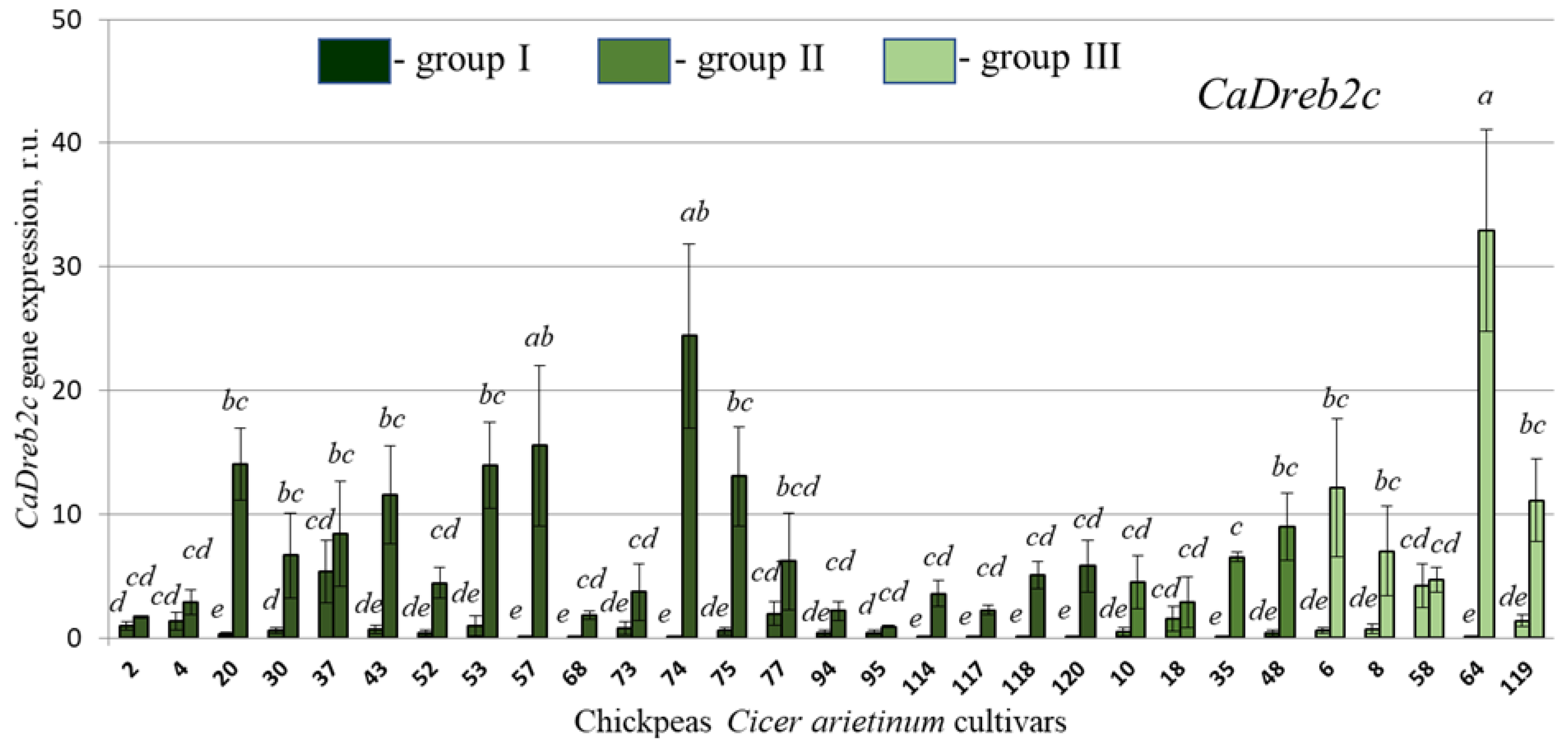
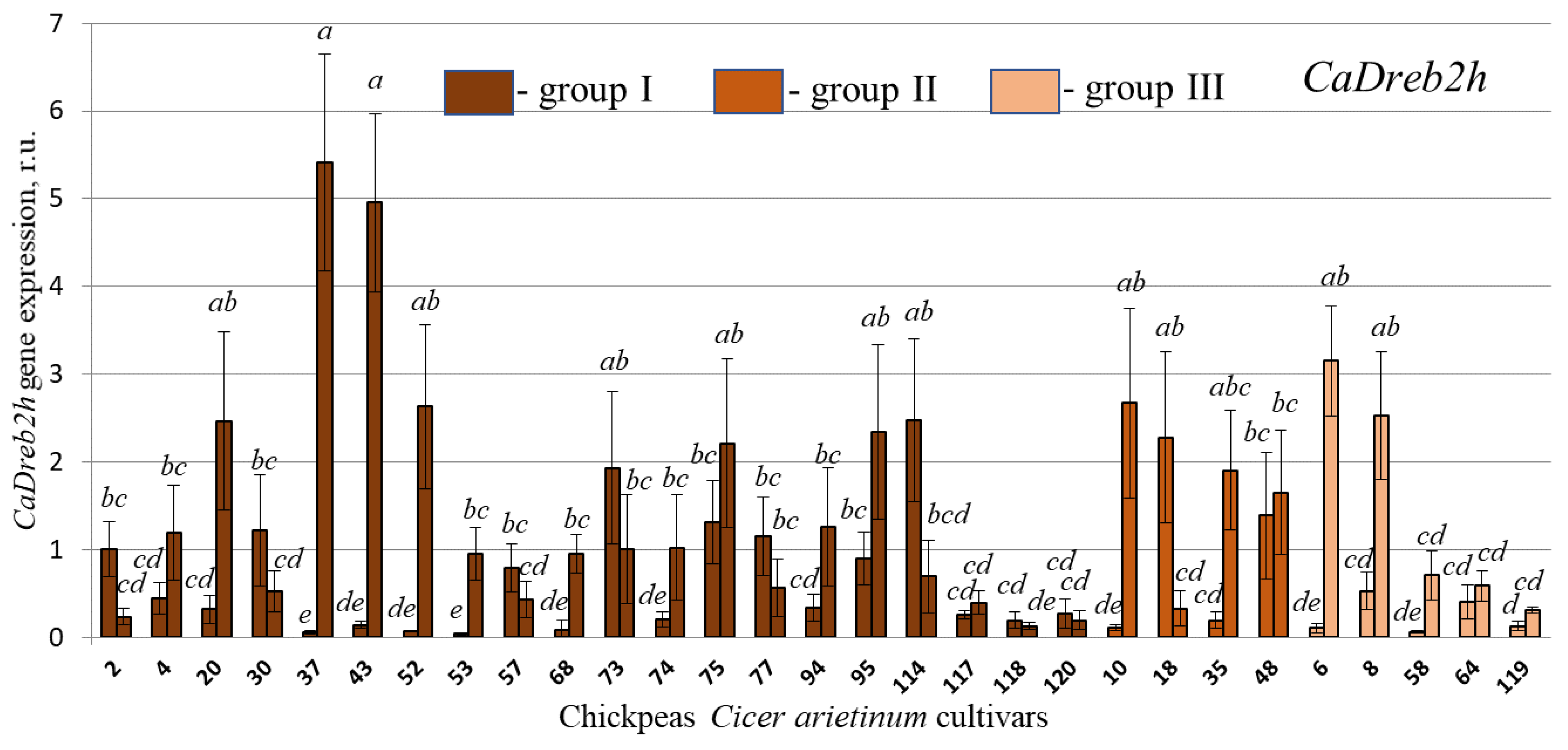
2.4. CaDreb2c and CaDreb2h Gene Expression in the Different Used Chickpea C. arietinum Cultivars in Normal and Drought Conditions
3. Materials and Methods
3.1. Plant Material
3.2. Drought Stress Experiments
3.3. Nucleic Acid Purification and Real-Time Quantitative PCR (RT-qPCR)
3.4. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kudapa, H.; Ghatak, A.; Barmukh, R.; Chaturvedi, P.; Khan, A.; Kale, S.; Fragner, L.; Chitikineni, A.; Weckwerth, W.; Varshney, R.K. Integrated multi-omics analysis reveals drought stress response mechanism in chickpea (Cicer arietinum L.). Plant Genome 2023, 17, e20337. [Google Scholar] [CrossRef]
- Arriagada, O.; Cacciuttolo, F.; Cabeza, R.A.; Carrasco, B.; Schwember, A.R. A comprehensive review on chickpea (Cicer arietinum L.) breeding for abiotic stress tolerance and climate change resilience. Int. J. Mol. Sci. 2022, 23, 6794. [Google Scholar] [CrossRef]
- Ravneet, K.; Kamlesh, P. Technological, processing and nutritional aspects of chickpea (Cicer arietinum)—A review. Trends Food Sci. Technol. 2021, 109, 448–463. [Google Scholar]
- Begum, N.; Khan, Q.U.; Liu, L.G.; Li, W.; Liu, D.; Haq, I.U. Nutritional composition, health benefits and bio-active compounds of chickpea (Cicer arietinum L.). Front. Nutr. 2023, 10, 1218468. [Google Scholar] [CrossRef] [PubMed]
- Maphosa, L.; Richards, M.F.; Norton, S.L.; Nguyen, G.N. Breeding for abiotic stress adaptation in chickpea (Cicer arietinum L.): A comprehensive review. Crop Breed. Genet. Genom. 2020, 2, e200015. [Google Scholar]
- Muhammad, A.M.; Muhammad, A.; Hina, A. Breeding for improved drought tolerance in Chickpea (Cicer arietinum L.). Plant Breed. 2017, 136, 3300–3318. [Google Scholar]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, G.; Liu, X.; Wang, Z.; Zhang, M.; Zhang, J. Genome-wide identification and expression profiling of DREB genes in Saccharum spontaneum. BMC Genom. 2021, 22, 456. [Google Scholar] [CrossRef]
- Kim, Y.H.; Yang, K.S.; Ryu, S.H.; Kim, K.Y.; Song, W.K.; Kwon, S.Y.; Lee, H.S.; Bang, J.W.; Kwak, S.S. Molecular characterization of a cDNA encoding DRE-binding transcription factor from dehydration-treated fibrous roots of sweetpotato. Plant Physiol. Biochem. 2008, 46, 196–204. [Google Scholar] [CrossRef]
- Donde, R.; Gupta, M.K.; Gouda, G.; Kumar, J.; Vadde, R.; Sahoo, K.K.; Dash, S.K.; Behera, L. Computational characterization of structural and functional roles of DREB1A, DREB1B and DREB1C in enhancing cold tolerance in rice plant. Amino Acids 2019, 51, 839–853. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Li, J.; Long, S.; Wei, S. A DREB1 gene from zoysiagrass enhances Arabidopsis tolerance to temperature stresses without growth inhibition. Plant Sci. 2019, 278, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Augustine, S.M.; Ashwin Narayan, J.; Syamaladevi, D.P.; Appunu, C.; Chakravarthi, M.; Ravichandran, V.; Tuteja, N.; Subramonian, N. Overexpression of EaDREB2 and pyramiding of EaDREB2 with the pea DNA helicase gene (PDH45) enhance drought and salinity tolerance in sugarcane (Saccharum spp. hybrid). Plant Cell Rep. 2015, 34, 247–263. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhuang, J.; Zou, Z.; Li, Q.; Xin, H.; Li, X. Overexpression of a Camellia sinensis DREB transcription factor gene (CsDREB) increases salt and drought tolerance in transgenic Arabidopsis thaliana. J. Plant Biol. 2017, 60, 452–461. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, L.; Xin, M.; Xing, H.; Zhang, Y.; Wu, J.; Xu, D.; Wang, Y.; Shang, Y.; Wei, L.; et al. Overexpression of GhABF3 increases cotton (Gossypium hirsutum L.) tolerance to salt and drought. BMC Plant Biol. 2022, 22, 313. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Akulov, A.S.; Budarina, G.A. Productivity of chick-pea depending on the elements of cultivation technology in the north of the central chernozem region. Zemledelie 2016, 4, 11–13. [Google Scholar]
- Fedyushkin, A.V.; Pasko, S.V. The productivity of chickpea, depending on seed rate and the background of mineral nutrition. Int. J. Humanit. Soc. Sci. 2019, 2-1, 69–71. [Google Scholar]
- Riechmann, J.L.; Meyerowitz, E.M. The AP2/EREBP family of plant transcription factors. Biol. Chem. 1998, 379, 633–646. [Google Scholar]
- Sazegari, S.; Niazi, A.; Ahmadi, F.S. A study on the regulatory network with promoter analysis for Arabidopsis DREB genes. Bioinformation 2015, 11, 101–106. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Kiselev, K.V.; Khristenko, V.S.; Aleynova, O.A. VaCPK20, a calcium-dependent protein kinase gene of wild grapevine Vitis amurensis Rupr., mediates cold and drought stress tolerance. J. Plant Physiol. 2015, 185, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kiselev, K.V.; Dubrovina, A.S.; Tyunin, A.P. The methylation status of plant genomic DNA influences PCR efficiency. J. Plant Physiol. 2015, 175, 59–67. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Suprun, A.R.; Aleynova, O.A.; Ogneva, Z.V.; Kalachev, A.V.; Dubrovina, A.S. External dsRNA downregulates anthocyanin biosynthesis-related genes and affects anthocyanin accumulation in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 6749. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Sahoo, A.; Tyagi, A.K.; Jain, M. Validation of internal control genes for quantitative gene expression studies in chickpea (Cicer arietinum L.). Biochem. Biophys. Res. Commun. 2010, 396, 283–288. [Google Scholar] [CrossRef]
- Aleynova, O.A.; Suprun, A.R.; Ananev, A.A.; Nityagovsky, N.N.; Ogneva, Z.V.; Dubrovina, A.S.; Kiselev, K.V. Effect of calmodulin-like gene (CML) overexpression on stilbene biosynthesis in cell cultures of Vitis amurensis Rupr. Plants 2022, 11, 171. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Aleynova, O.A.; Ogneva, Z.V.; Suprun, A.R.; Ananev, A.A.; Nityagovsky, N.N.; Dneprovskaya, A.A.; Beresh, A.A.; Dubrovina, A.S. The effect of stress hormones, ultraviolet C, and stilbene precursors on expression of calcineurin b-like protein (CBL) and CBL-interacting protein kinase (CIPK) genes in cell cultures and leaves of Vitis amurensis Rupr. Plants 2023, 12, 1562. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiselev, K.V.; Ogneva, Z.V.; Dubrovina, A.S.; Gabdola, A.Z.; Khassanova, G.Z.; Jatayev, S.A. Study of CaDreb2c and CaDreb2h Gene Sequences and Expression in Chickpea (Cicer arietinum L.) Cultivars Growing in Northern Kazakhstan under Drought. Plants 2024, 13, 2066. https://doi.org/10.3390/plants13152066
Kiselev KV, Ogneva ZV, Dubrovina AS, Gabdola AZ, Khassanova GZ, Jatayev SA. Study of CaDreb2c and CaDreb2h Gene Sequences and Expression in Chickpea (Cicer arietinum L.) Cultivars Growing in Northern Kazakhstan under Drought. Plants. 2024; 13(15):2066. https://doi.org/10.3390/plants13152066
Chicago/Turabian StyleKiselev, Konstantin V., Zlata V. Ogneva, Alexandra S. Dubrovina, Ademi Zh. Gabdola, Gulmira Zh. Khassanova, and Satyvaldy A. Jatayev. 2024. "Study of CaDreb2c and CaDreb2h Gene Sequences and Expression in Chickpea (Cicer arietinum L.) Cultivars Growing in Northern Kazakhstan under Drought" Plants 13, no. 15: 2066. https://doi.org/10.3390/plants13152066
APA StyleKiselev, K. V., Ogneva, Z. V., Dubrovina, A. S., Gabdola, A. Z., Khassanova, G. Z., & Jatayev, S. A. (2024). Study of CaDreb2c and CaDreb2h Gene Sequences and Expression in Chickpea (Cicer arietinum L.) Cultivars Growing in Northern Kazakhstan under Drought. Plants, 13(15), 2066. https://doi.org/10.3390/plants13152066









