A First Approach for the In Vitro Cultivation, Storage, and DNA Barcoding of the Endangered Endemic Species Euonymus koopmannii
Abstract
1. Introduction
2. Results
2.1. Explant Sterilization
2.2. Direct Shoot Regeneration from Internodes and Nodal Explants
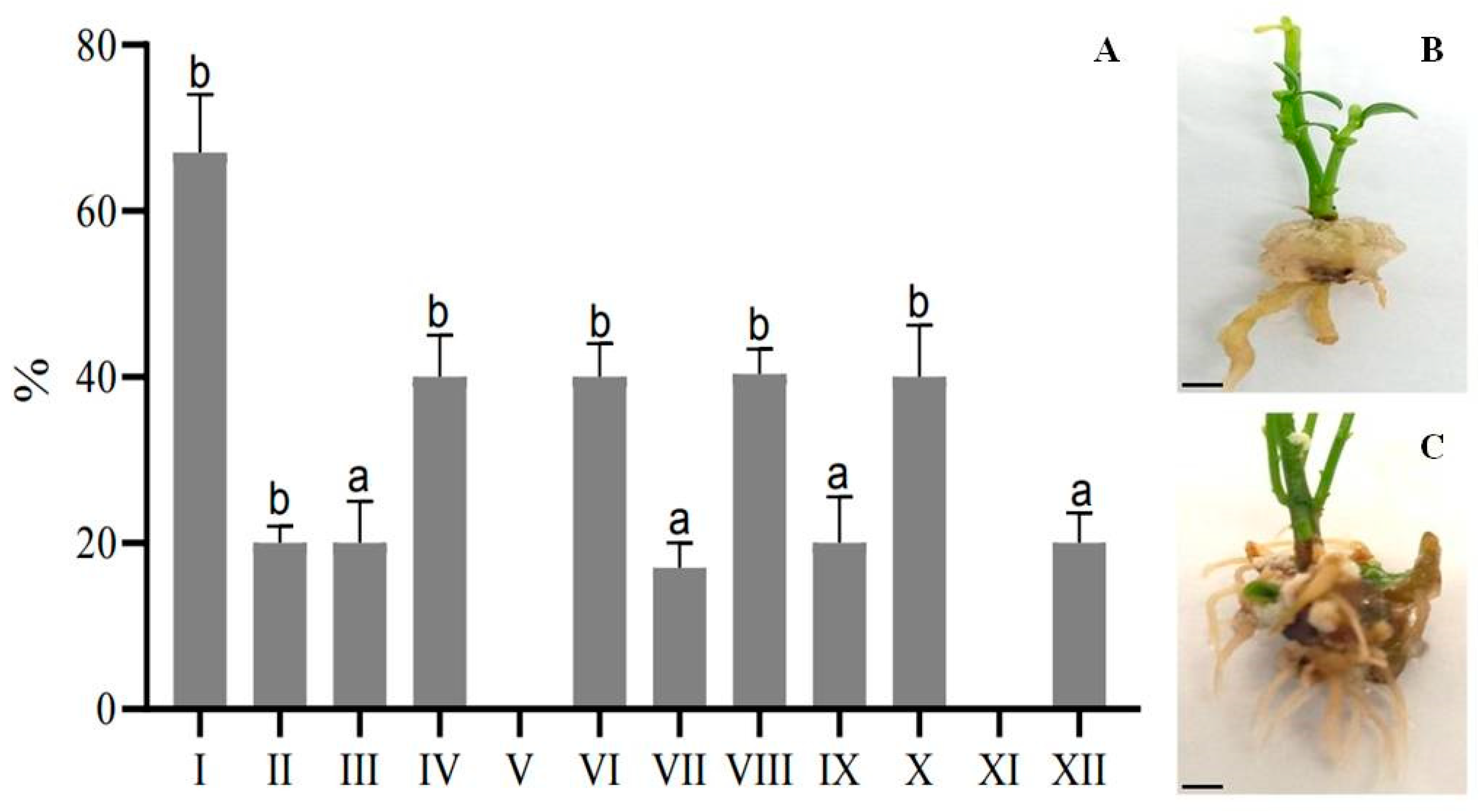
2.3. Root Formation
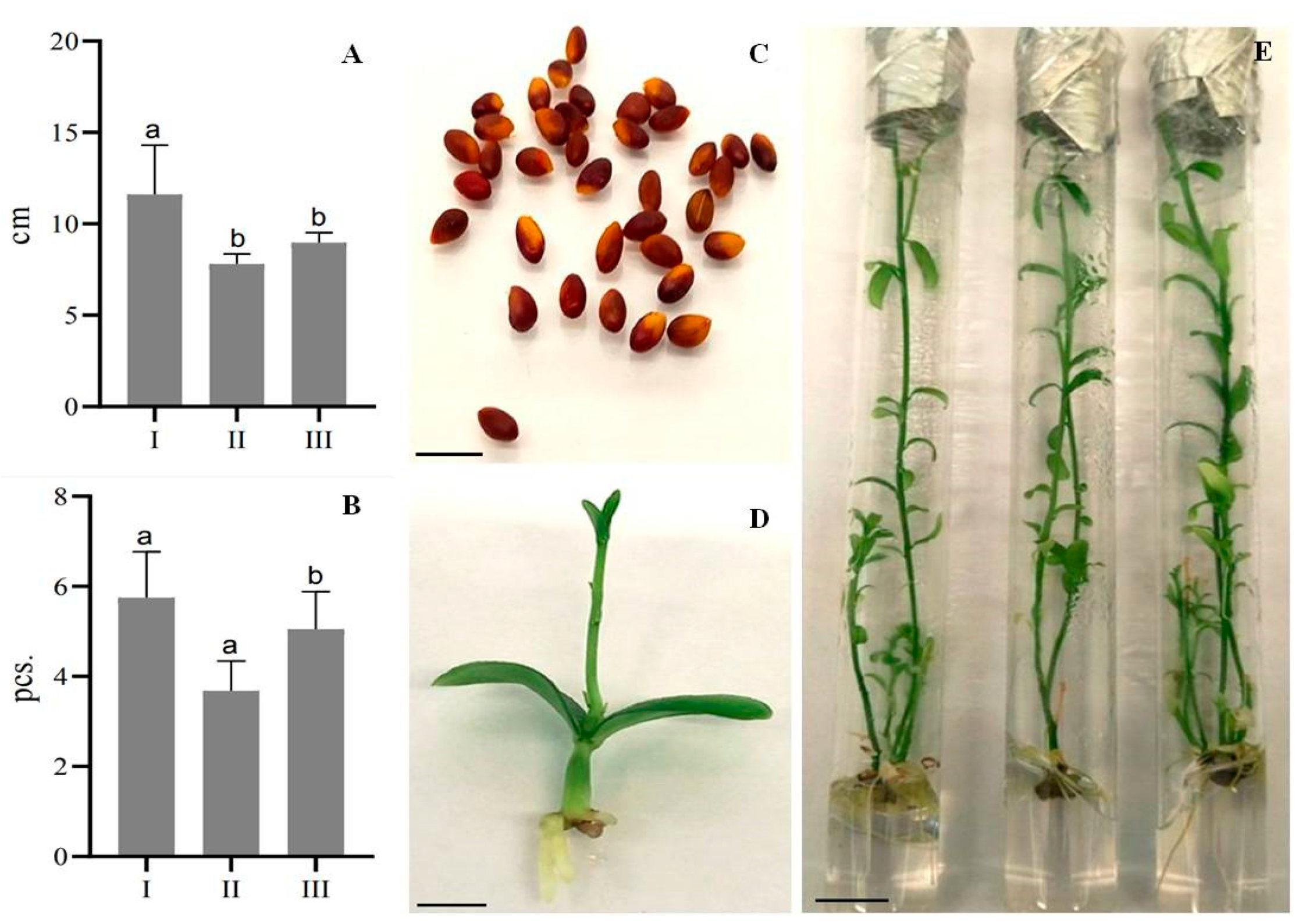
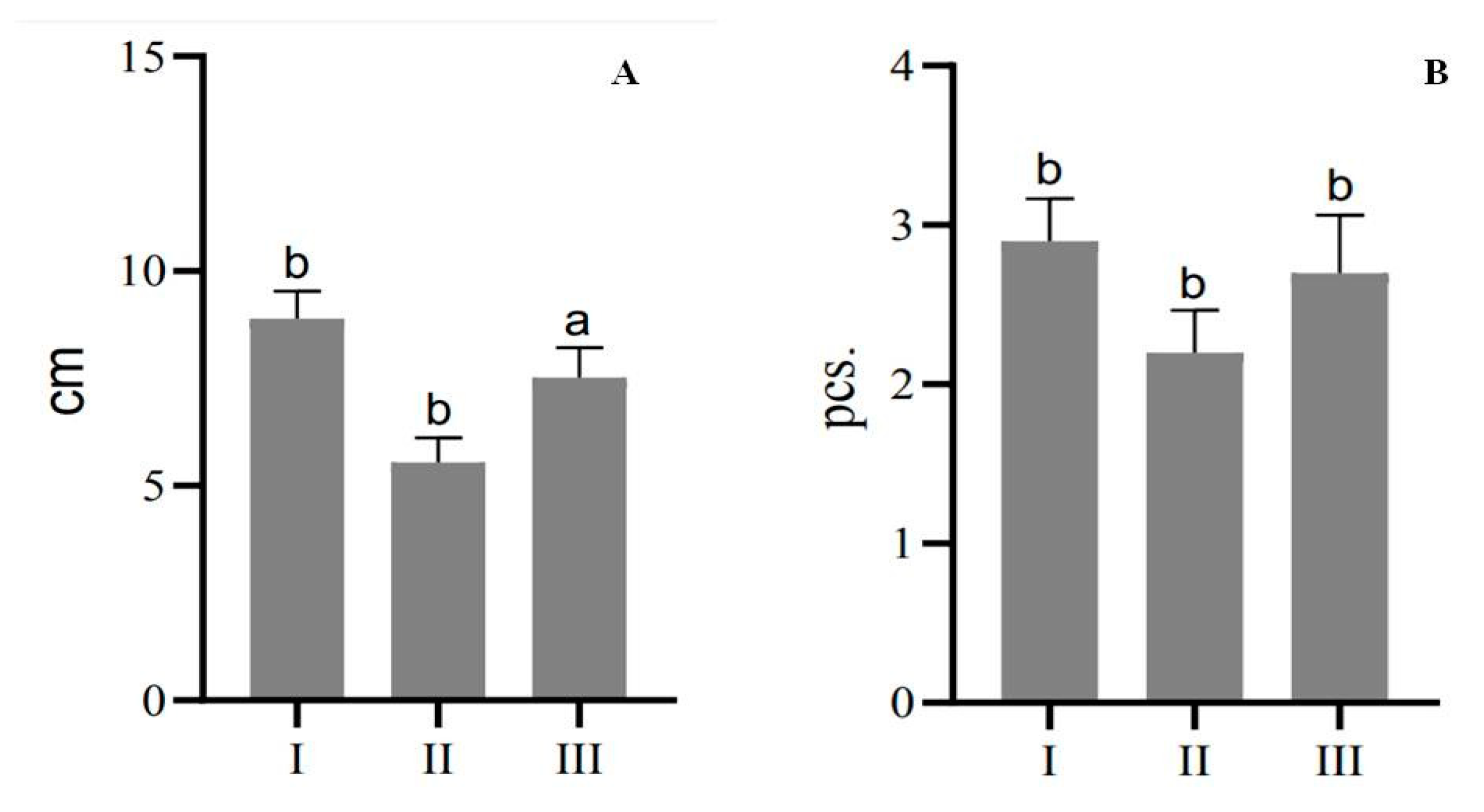
2.4. Micropropagation of Plants from Internode Explants and Seedlings
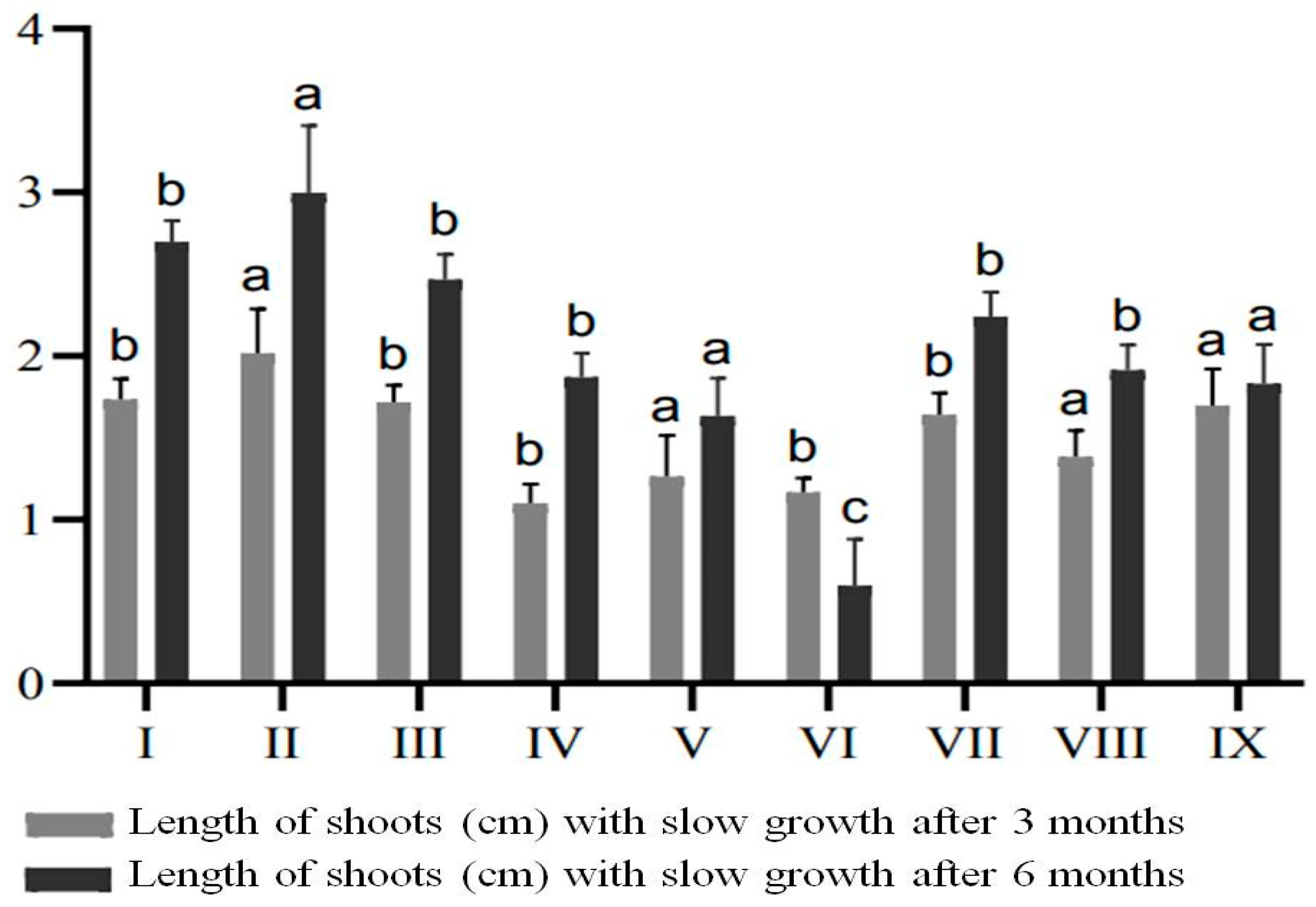
2.5. In vitro Preservation of Plants by Slow Growth Storage
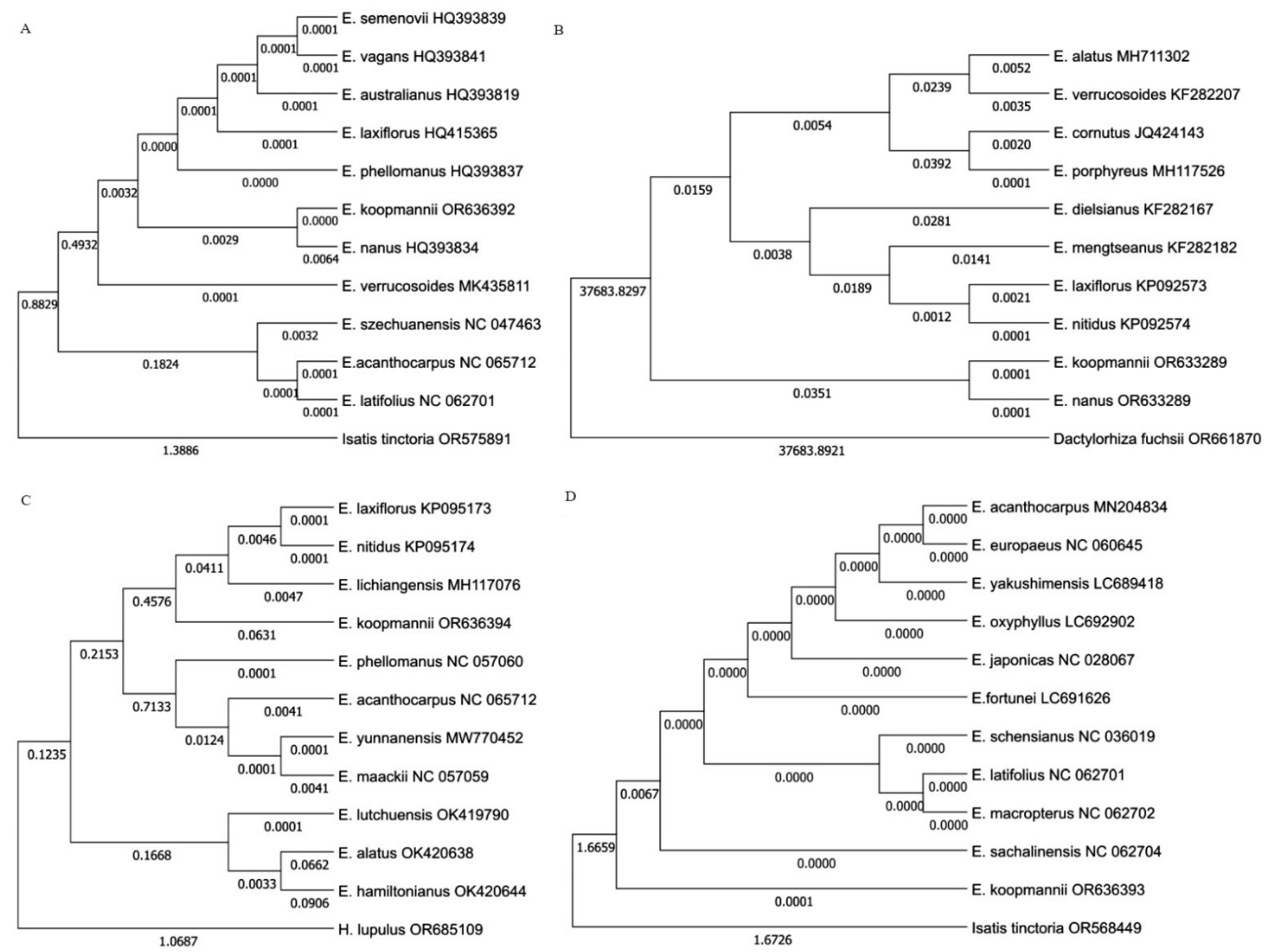
2.6. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Material and Explant Preparation and Disinfection
4.2. Direct Regeneration, Rooting and Micropropagation
4.3. Slow Growth Storage
4.4. DNA Barcoding
4.5. Statistical Analyses
- a—number of shoots after the first passage
- b—number of shoots after the third passage
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Y.; Dong, Y.; Liu, Y.; Yu, X.; Yang, M.; Huang, Y. Comparative Analyses of Euonymus Chloroplast Genomes: Genetic Structure, Screening for Loci with Suitable Polymorphism, Positive Selection Genes, and Phylogenetic Relationships within Celastrineae. Front. Plant Sci. 2021, 11, 593984. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-N.; Xie, L.; Li, J.; Zhang, Z. Phylogeny of Euonymus L. inferred from molecular and morphological data. J. Syst. Evol. 2013, 52, 1–12. [Google Scholar] [CrossRef]
- Wittmack, L. Garten-Zeitung; P. Parey: Berlin, Germany, 1883; Volume 2. [Google Scholar]
- Shukurov, E.D. Central Asian transboundary project for the conservation of biodiversity of the Western Tien Shan. In Atlas of Biological Diversity of the Western Tien Shan Book; Bismarck LLC: Bishkek, Kyrgyzstan, 2005. (In Russian) [Google Scholar]
- Ivashchenko, A.A. Reserves and National Parks of Kazakhstan; Almatykitap: Almaty, Kazakhstan, 2006; p. 284. (In Russian) [Google Scholar]
- Available online: https://e-history.kz/ru/kazakhstanika/show/11175 (accessed on 22 February 2024).
- Ruiz-Riaguas, A.; Fernandez-de Cordova, M.L.; Llorent-Martinez, E.J. Phenolic profile and antioxidant activity of Euonymus japonicus Thunb. Nat. Prod. Res. 2022, 36, 3445–3449. [Google Scholar] [CrossRef] [PubMed]
- Kukula-Koch, W.; Widelski, J.; Koch, W.; Głowniak, K. HPLC, Two-Dimensional TLC Determination of Phenolic Content, and an In Vitro Perspective to Antioxidant Potential of Euonymus verrucosus Scop. Extracts. Acta Chromatogr. 2015, 27, 743–754. [Google Scholar] [CrossRef]
- Shin, D.Y.; Kim, B.S.; Lee, H.Y.; Park, Y.M.; Kim, Y.W.; Kim, M.J.; Yang, H.J.; Kim, M.S.; Bae, J.S. Euonymus alatus (Thunb.) Siebold leaf extract enhanced immunostimulatory effects in a cyclophosphamide-induced immunosuppressed rat model. Food Nutr. Res. 2023, 67, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.L.; Li, K.; Yang, Z.; Chen, B.; Zhang, T. Dulcitol suppresses proliferation and migration of hepatocellular carcinoma via regulating SIRT1/p53 pathway. Phytomedicine 2020, 66, 153112. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Qin, J.; Chang, R.; Zeng, Q.; Cheng, X.; Zhang, F.; Jin, H.; Zhang, W. Chemical Constituents of Plants from the Genus Euonymus. Chem. Biodivers. 2012, 9, 1055–1076. [Google Scholar] [CrossRef] [PubMed]
- Popova, E.V.; Nosov, A.V.; Titova, M.V.; Kochkina, D.V.; Fomenkov, A.A.; Kulichenko, I.E.; Nosov, A.M. Promising biotechnologies: Collections of cell cultures of higher plants as the basis for the development and production of drugs. J. Plant Physiol. 2021, 68, 227–244. (In Russian) [Google Scholar]
- Fomenkov, A.A.; Sidorov, R.A.; Nosov, A.V. Suspension cell culture of Euonymus maximoviczianus is a new object in studies of the biosynthesis of lower alkyl esters of fatty acids. Cell Biol. Plant Biotechnol. 2013, 61. (In Russian) [Google Scholar]
- Smith, C.C.; Jernstedt, J.A. In vitro development of adventitious shoots in Euonymus alatus (Celastraceae). Sci. Hortic. 1989, 41, 161–169. [Google Scholar] [CrossRef]
- Chen, Y.; Lu, L.; Deng, W.; Yang, X.; McAvoy, R.; Zhao, D.; Pei, Y.; Luo, K.; Duan, H.; Smith, W.; et al. In vitro regeneration and Agrobacterium-mediated genetic transformation of Euonymus alatus. Plant Cell Rep. 2006, 25, 1043–1051. [Google Scholar] [CrossRef]
- Woo, H.-A.; Ku, S.S.; Jie, E.Y.; Kim, H.; Kim, H.-S.; Cho, H.S.; Jeong, W.-J.; Park, S.U.; Min, S.R.; Kim, S.W. Efficient plant regeneration from embryogenic cell suspension cultures of Euonymus alatus. Sci. Rep. 2021, 11, 15120. [Google Scholar] [CrossRef]
- Ning, K.; Zhou, T.; Jiang, C.Z.; Wu, H.M.; Jiang, J.L.; Chen, J.; El-kassaby, Y.A.; Ma, Y. Rapid and efficient leaf regeneration propagation system for Euonymus bungeanus. Biol. Plant. 2021, 65, 118–125. [Google Scholar] [CrossRef]
- Shang, A.-Q.; Cai, H.; Yan, X.-J.; Hu, H.-Z.; Zhao, L.-J. Plant Regeneration from In Vitro Cultured Hypocotyl Explants of Euonymus japonicus Cu Zhi. Agric. Sci. China 2006, 5, 196–201. [Google Scholar] [CrossRef]
- Levy, L.; Damsteegt, V.; Welliver, R. First Report of Plum pox virus (Sharka Disease) in Prunus persica in the United States. Plant Dis. 2000, 84, 202. [Google Scholar] [CrossRef]
- Benelli, C.; Tarraf, W.; Izgu, T.; De Carlo, A. In Vitro Conservation through Slow Growth Storage Technique of Fruit Species: An Overview of the Last 10 Years. Plants 2022, 11, 3188. [Google Scholar] [CrossRef]
- Bautista-Aguilar, J.R.; Iglesias-Andreu, L.G.; Martínez-Castillo, J.; Ramírez-Mosqueda, M.A.; Ortiz-García, M.M. In Vitro Conservation and Genetic Stability in Vanilla planifolia Jacks. HortScience 2021, 56, 1494–1498. [Google Scholar] [CrossRef]
- Choi, K.S.; Park, S. The complete chloroplast genome sequence of Euonymus japonicus (Celastraceae). Mitochondrial DNA Part A 2016, 27, 3577–3578. [Google Scholar] [CrossRef]
- Park, Y.S.; Kang, J.-S.; Park, J.Y.; Shim, H.; Yang, H.O.; Kang, J.H.; Yang, T.-J. Analysis of the complete plastomes and nuclear ribosomal DNAs from Euonymus hamiltonianus and its relatives sheds light on their diversity and evolution. PLoS ONE 2022, 17, e0275590. [Google Scholar] [CrossRef]
- Cai, H.; Gu, X.; Li, Y.; Ren, Y.; Yan, S.; Yang, M. Cold Resistance of Euonymus japonicus Beihaidao Leaves and Its Chloroplast Genome Structure and Comparison with Celastraceae Species. Plants 2022, 11, 2449. [Google Scholar] [CrossRef]
- Wang, W.-C.; Chen, S.-Y.; Zhang, X.-Z. Characterization of the complete chloroplast genome of the golden crane butterfly, Euonymus schensianus (Celastraceae). Conserv. Genet. Resour. 2017, 9, 545–547. [Google Scholar] [CrossRef]
- Lee, J.; Kang, S.J.; Shim, H.; Lee, S.C.; Kim, N.H.; Jang, W.; Park, J.Y.; Kang, J.H.; Lee, W.H.; Lee, T.J.; et al. Characterization of Chloroplast Genomes, Nuclear Ribosomal DNAs, and Polymorphic SSR Markers Using Whole Genome Sequences of Two Euonymus hamiltonianus Phenotypes. Plant Breed. Biotechnol. 2019, 7, 50–61. [Google Scholar] [CrossRef]
- Hua, W.; Chen, C. The complete chloroplast genome sequence of the plant Euonymus fortunei (Celastraceae). Mitochondrial DNA Part B 2018, 3, 637–639. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, H.; Zheng, M.; Jiang, J. The complete chloroplast genome of Euonymus szechuanensis. Mitochondrial DNA Part B 2020, 5, 1130–1131. [Google Scholar] [CrossRef]
- Li, H.; Chen, M.; Wang, Z.; Hao, Z.; Zhao, X.; Zhu, W.; Liu, L.; Guo, W. Characterization of the Complete Chloroplast Genome and Phylogenetic Implications of Euonymus microcarpus (Oliv.) Sprague. Genes 2022, 13, 2352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.-T.; Sun, J.; Xu, L.; Yang, Y.Y.; Zhan, Z.L.; Xing, Y.P.; Zhao, R.; Li, S.N.; Zhang, D.C.; Kang, T.G. The complete mitochondrial genome of Euonymus alatus (celastraceae, Euonymus L.). Mitochondrial DNA Part B 2021, 6, 182–184. [Google Scholar] [CrossRef] [PubMed]
- Gubaidullin, N.; Kali, B.; Tussipkan, D.; Manabayeva, S.A. A promising strategy for conservation of Central Asian endemic plant Euonymus koopmannii. Biodivers. J. Biol. Divers. 2024, 25, 3114–3120. [Google Scholar]
- Linh, N.N.; Hang, P.L.B.; Hue, H.T.T.; Ha, N.H.; Hanh, H.H.; Ton, N.D.; Hien, L.T.T. Species discrimination of novel chloroplast DNA barcodes and their application for identification of Panax (Aralioideae, Araliaceae). PhytoKeys 2022, 188, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Veldman, S.; Ju, Y.; Otieno, J.N.; Abihudi, S.; Posthouwer, C.; Gravendeel, B.; van Andel, T.R.; de Boer, H.J. DNA barcoding augments conventional methods for identification of medicinal plant species traded at Tanzanian markets. J. Ethnopharmacol. 2020, 250, 112495. [Google Scholar] [CrossRef]
- Ma, H.-L.; Zhu, Z.-B.; Zhang, X.-M.; Miao, Y.-Y.; Guo, Q.-S. Species identification of the medicinal plant Tulipa edulis (Liliaceae) by DNA barcode marker. Biochem. Syst. Ecol. 2014, 55, 362–368. [Google Scholar] [CrossRef]
- Jones, L.; Twyford, A.D.; Ford, C.R.; Rich, T.C.G.; Davies, H.; Forrest, L.L.; Hart, M.L.; McHaffie, H.; Brown, M.R.; Hollingsworth, P.M.; et al. Barcode UK: A complete DNA barcoding resource for the flowering plants and conifers of the United Kingdom. Mol. Ecol. Resour. 2021, 21, 2050–2062. [Google Scholar] [CrossRef]
- Singh, V.; Deen, B.; Singh, S. Micropropagation of minor fruit crops of India: A review. Agric. Rev. 2023, 44, 259–263. [Google Scholar] [CrossRef]
- Chokheli, V.A.; Dmitriev, P.A.; Rajput, V.D. Recent Development in Micropropagation Techniques for Rare Plant Species. Plants 2020, 9, 1733. [Google Scholar] [CrossRef] [PubMed]
- Abeuova, L.S.; Kali, B.R.; Rakhimzhanova, A.O.; Bekkuzhina, S.S.; Manabayeva, S.A. High frequency direct shoot regeneration from Kazakh commercial potato cultivars. PeerJ 2020, 8, e9447. [Google Scholar] [CrossRef]
- Zhumabek, A.T.; Rakhimzhanova, A.O.; Bekkuzhina, S.S.; Ramankulov, Y.M.; Manabayeva, S.A. Somatic embryogenesis and plant regeneration from upland switchgrass cultivars. Res. Crops 2020, 21, 349–354. [Google Scholar]
- Pasternak, T.P.; Steinmacher, D. Plant Growth Regulation in Cell and Tissue Culture In Vitro. Plants 2024, 13, 327. [Google Scholar] [CrossRef]
- Beasley, R.; Pijut, P. Regeneration of Plants from Fraxinus nigra Marsh. Hypocotyls. HortSci. A Publ. Am. Soc. Hortic. Sci. 2013, 48, 887–890. [Google Scholar] [CrossRef]
- Hesami, M.; Daneshvar, M.H. In Vitro Adventitious Shoot Regeneration through Direct and Indirect Organogenesis from Seedling-derived Hypocotyl Segments of Ficus religiosa L.: An Important Medicinal Plant. HortScience 2018, 53, 55–61. [Google Scholar] [CrossRef]
- Li, P.; Shang, Y.; Zhou, W.; Hu, X.; Mao, W.; Li, J.; Li, J.; Chen, X. Development of an efficient regeneration system for the precious and fast-growing timber tree Toona ciliata. Plant Biotechnol. 2018, 35, 51–58. [Google Scholar] [CrossRef]
- Verma, S.K.; Gantait, S.; Mukherjee, E.; Gurel, E. Enhanced somatic embryogenesis, plant regeneration and total phenolic content estimation in Lycium barbarum L.: A highly nutritive and medicinal plant. J. Crop. Sci. Biotechnol. 2022, 25, 547–555. [Google Scholar] [CrossRef]
- Ning, X.; Liu, Y.; Jia, M.; Wang, Q.; Sun, Z.; Ji, L.; Mayo, K.H.; Zhou, Y.; Sun, L. Pectic polysaccharides from Radix Sophorae Tonkinensis exhibit significant antioxidant effects. Carbohydr. Polym. 2021, 262, 117925. [Google Scholar] [CrossRef] [PubMed]
- Shang, A.; Sun, Z.; Zhao, L. High Efficient Regeneration In Vitro from Hypocotyl of Euonymus fortunei var. radicans. Sci. Silvae Sin. 2009, 45, 136–141. [Google Scholar]
- Chen, Y.; Lu, L.; Duan, H.; Deng, W.; McAvoy, R.; Smith, W.; Thammina, C.; von Bodman, S.; Li, Y.; Ye, D.; et al. Biotech approach to neutralize the invasiveness of burning bush (Euonymus alatus), a progress report on development of its genetic transformation system and functional analysis of sterile genes. Acta Hortic. 2008, 769, 21–29. [Google Scholar] [CrossRef]
- Mitrofanova, O.V.; Lesnikova-Sedoshenko, N.P.; Ivanova, N.N.; Smykova, N.V.; Mitrofanova, I.V. In vitro propagation and preservation of promising chrysanthemum cultivars and hybrid forms. In Proceedings of the XXX International Horticultural Congress IHC2018: II International Symposium on Micropropagation and In Vitro Techniques 1285, Istanbul, Turkey, 12–16 August 2018. [Google Scholar]
- Pan, X.; Zhang, W.; Li, X. In vitro conservation of native Chinese wild grape (Vitis heyneana Roem. & Schult) by slow growth culture. VITIS J. Grapevine Res. 2014, 53, 207–214. [Google Scholar]
- Turdiyev, T.T.; Kovalchuk, I.Y.; Kabylbekova, B.Z.; Chukanova, N.I.; Frolov, S.N. In vitro germplasm cold storage of fruit and berry plants of Kazakhstan. Eurasian J. Biosci. 2020, 14, 1213–1219. [Google Scholar]
- Li, H.; Xiao, W.; Tong, T.; Li, Y.; Zhang, M.; Lin, X.; Zou, X.; Wu, Q.; Guo, X. The specific DNA barcodes based on chloroplast genes for species identification of Orchidaceae plants. Sci. Rep. 2021, 11, 1424. [Google Scholar] [CrossRef] [PubMed]
- Mishra, M.; Jingade, P.; Huded, A.K.C. DNA barcoding analysis and phylogenetic relationships of Indian wild coffee species. Turk. J. Bot. 2022, 46, 109–122. [Google Scholar] [CrossRef]
- Hajdari, A.; Pulaj, B.; Schmiderer, C.; Mala, X.; Wilson, B.; Lluga-Rizani, K.; Mustafa, B. A phylogenetic analysis of the wild Tulipa species (Liliaceae) of Kosovo based on plastid and nuclear DNA sequence. Adv. Genet. 2021, 2, e202100016. [Google Scholar] [CrossRef] [PubMed]
- Jamdade, R.; Mosa, K.A.; El-Keblawy, A.; Al Shaer, K.; Al Harthi, E.; Al Sallani, M.; Al Jasmi, M.; Gairola, S.; Shabana, H.; Mahmoud, T. DNA barcodes for accurate identification of selected medicinal plants (Caryophyllales): Toward barcoding flowering plants of the United Arab Emirates. Diversity 2022, 14, 262. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, M.; Chen, X.; Chen, X.; Hu, Y.; Gao, J.; Pan, W.; Xin, Y.; Wu, J.; Du, Y.; et al. Developing an efficient DNA barcoding system to differentiate between Lilium species. BMC Plant Biol. 2021, 21, 465. [Google Scholar] [CrossRef]
- Chinnkar, M.; Jadhav, P. Evaluating DNA barcoding using five loci (matK, ITS, trnH-psbA, rpoB, and rbcL) for species identification and phylogenetic analysis of Capsicum frutescens. J. Appl. Biol. Biotechnol. 2023, 11, 97–103. [Google Scholar] [CrossRef]
- Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Sutula, M.; Kakanay, A.; Tussipkan, D.; Dzhumanov, S.; Manabayeva, S. Phylogenetic Analysis of Rare and Endangered Tulipa Species (Liliaceae) of Kazakhstan Based on Universal Barcoding Markers. Biology 2024, 13, 365. [Google Scholar] [CrossRef]
- Lu, G.; Moriyama, E.N. Vector NTI, a balanced all-in-one sequence analysis suite. Brief. Bioinform. 2005, 5, 378–388. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Zhang, C.-Y.; Wang, F.-Y.; Yan, H.-F.; Hao, G.; Hu, C.-M.; Ge, X.-J. Testing DNA barcoding in closely related groups of Lysimachia L. (Myrsinaceae). Mol. Ecol. Resour. 2011, 12, 98–108. [Google Scholar] [CrossRef]
- Kuzmina, M.L.; Johnson, K.L.; Barron, H.R.; Hebert, P.D. Identification of the vascular plants of Churchill, Manitoba, using a DNA barcode library. BMC Ecol. 2012, 12, 25. [Google Scholar] [CrossRef]
- Parmentier, I.; Duminil, J.; Kuzmina, M.; Philippe, M.; Thomas, D.W.; Kenfack, D.; Chuyong, G.B.; Cruaud, C.; Hardy, O.J. How effective are DNA barcodes in the identification of African rainforest trees? PLoS ONE 2013, 8, e54921. [Google Scholar] [CrossRef]
- Khan, A.M.; Bhadauria, S. Molecular characterization of keratin degrading fungi isolated from semi-arid soil by PCR using ITS4 and ITS5 primers. J. King Saud Univ.-Sci. 2019, 31, 1418–1423. [Google Scholar] [CrossRef]

| No of Variants | Sterilization Options | Type of Explant | Shoot Formation In Vitro (%) | |||
|---|---|---|---|---|---|---|
| Step 1 | Exposure Time | Step 2 | Exposure Time | |||
| I | 1% sodium hypochlorite | 30 min one time | 5% sodium hypochlorite + Tween 20 | 10 min | axillary buds | 67.5 ± 0.12 |
| seeds | 63.3 ± 0.10 | |||||
| II | 5 min | axillary buds | 85.2 ± 0.09 | |||
| seeds | 87.5 ± 0.07 | |||||
| III | 1% KMnO4 solution | 3% sodium hypochlorite + Tween 20 | 10 min | axillary buds | 49.4 ± 0.14 | |
| seeds | 43.3 ± 0.51 | |||||
| Explant Type | Number of Shoots after the First Passage, Pcs | Number of Shoots after the Second Passage, Pcs | Number of Shoots after the Third Passage, Pcs | Multiplication Rate |
|---|---|---|---|---|
| Seedlings | 4 | 31 | 114 | 28.5 |
| Internodes | 7 | 20 | 43 | 6.1 |
| matK Gene | ITS Region | psbA-trnH Region | rbcL Gene | |
|---|---|---|---|---|
| Total aligned length (bp) | 334 | 501 | 242 | 444 |
| GC content (%) | 32.38 | 59.6 | 26.4 | 45.7 |
| Codon count | 101 | - | 70 | 148 |
| Number of monomorphic sites | 165 | 403 | 38 | 441 |
| Number of polymorphic sites | 132 | 91 | 139 | 3 |
| Total number of InDels sites | 37 | 7 | 65 | 0 |
| Overlapping InDels sites | 0 | 2 | 38 | 0 |
| Number of singleton variables sites | 2 | 41 | 5 | 3 |
| Total number of mutations (Eta) | 134 | 107 | 196 | 3 |
| Parsimony informative sites (PIC) | 130 | 50 | 83 | 0 |
| Nucleotide diversity (Pi) | 0.1931 | 0.0625 | 0.4282 | 0.0012 |
| Tajima’s neutrality test () | 0.1723 | 0.0494 | 0.3833 | 0.0012 |
| Mean nucleotide difference (k) | 57.345 | 30.889 | 75.782 | 0.545 |
| Number of Haplotypes (h) | 5 | 9 | 10 | 2 |
| Haplotype diversity (Hd) | 0.709 | 0.978 | 0.982 | 0.182 |
| Variance of Haplotype diversity | 0.01865 | 0.00292 | 0.00215 | 0.02061 |
| Sequence conservation (C) | 0.581 | 0.814 | 0.199 | 0.993 |
| Conservation threshold (CT) | 0.68 | 0.91 | 0.29 | 1 |
| PGRs, mg/L | MSDR I | MSDR II | MSDR III |
|---|---|---|---|
| BAP | 0.5 | 1.0 | 2.0 |
| NAA | 0.1 | 0.5 | 1.0 |
| Kinetin | − | 0.2 | − |
| Options | MS | IBA, mg/L | IAA, mg/L | NAA, mg/L |
|---|---|---|---|---|
| MSR I | ½ | 0.5 | 0.05 | − |
| MSR II | full | |||
| MSR III | ½ | 1.0 | 0.1 | − |
| MSR IV | full | |||
| MSR V | ½ | 2.0 | 0.5 | − |
| MSR VI | full | |||
| MSR VII | ½ | 0.5 | − | 0.1 |
| MSR VIII | full | |||
| MSR IX | ½ | 1.0 | − | 0.5 |
| MSR X | full | |||
| MSR XI | ½ | 2.0 | − | 0.05 |
| MSR XII | full |
| Variations of MS Medium | ABA, mg/L | Mannitol, mg/L | CCC, mg/L |
|---|---|---|---|
| MSP I | 2.0 | − | − |
| MSP II | 5.0 | − | − |
| MSP III | 10.0 | − | − |
| MSP IV | − | 5.0 | − |
| MSP V | − | 8.0 | − |
| MSP VI | − | 10.0 | − |
| MSP VII | − | − | 0.2 |
| MSP VIII | − | − | 0.5 |
| MSP IX | − | − | 1.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kali, B.; Bekkuzhina, S.; Tussipkan, D.; Manabayeva, S. A First Approach for the In Vitro Cultivation, Storage, and DNA Barcoding of the Endangered Endemic Species Euonymus koopmannii. Plants 2024, 13, 2174. https://doi.org/10.3390/plants13162174
Kali B, Bekkuzhina S, Tussipkan D, Manabayeva S. A First Approach for the In Vitro Cultivation, Storage, and DNA Barcoding of the Endangered Endemic Species Euonymus koopmannii. Plants. 2024; 13(16):2174. https://doi.org/10.3390/plants13162174
Chicago/Turabian StyleKali, Balnur, Sara Bekkuzhina, Dilnur Tussipkan, and Shuga Manabayeva. 2024. "A First Approach for the In Vitro Cultivation, Storage, and DNA Barcoding of the Endangered Endemic Species Euonymus koopmannii" Plants 13, no. 16: 2174. https://doi.org/10.3390/plants13162174
APA StyleKali, B., Bekkuzhina, S., Tussipkan, D., & Manabayeva, S. (2024). A First Approach for the In Vitro Cultivation, Storage, and DNA Barcoding of the Endangered Endemic Species Euonymus koopmannii. Plants, 13(16), 2174. https://doi.org/10.3390/plants13162174






