Transcriptomic and Physiological Analyses for the Role of Hormones and Sugar in Axillary Bud Development of Wild Strawberry Stolon
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. RNA Sequencing
2.3. Measurement of Endogenous Hormones
2.4. Paraffin Section
2.5. Exogenous Hormone Treatments
2.6. Culturing Dormant Buds In Vitro
2.7. qRT-PCR Analysis
3. Results and Discussion
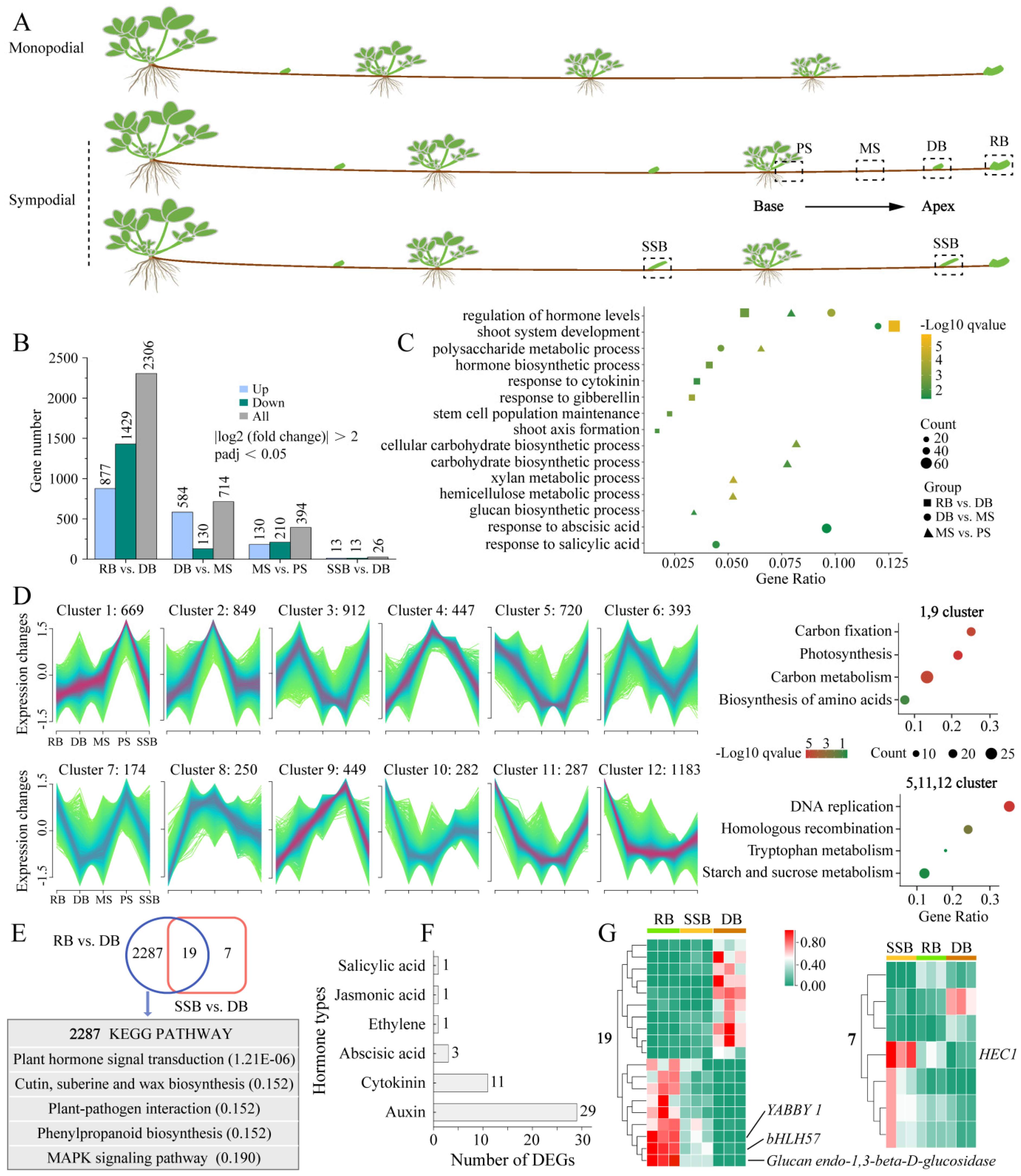
3.1. Dynamic Changes of Differentially Expressed Genes (DEGs) during Sympodial Stolon Growth in F. nilgerrensis
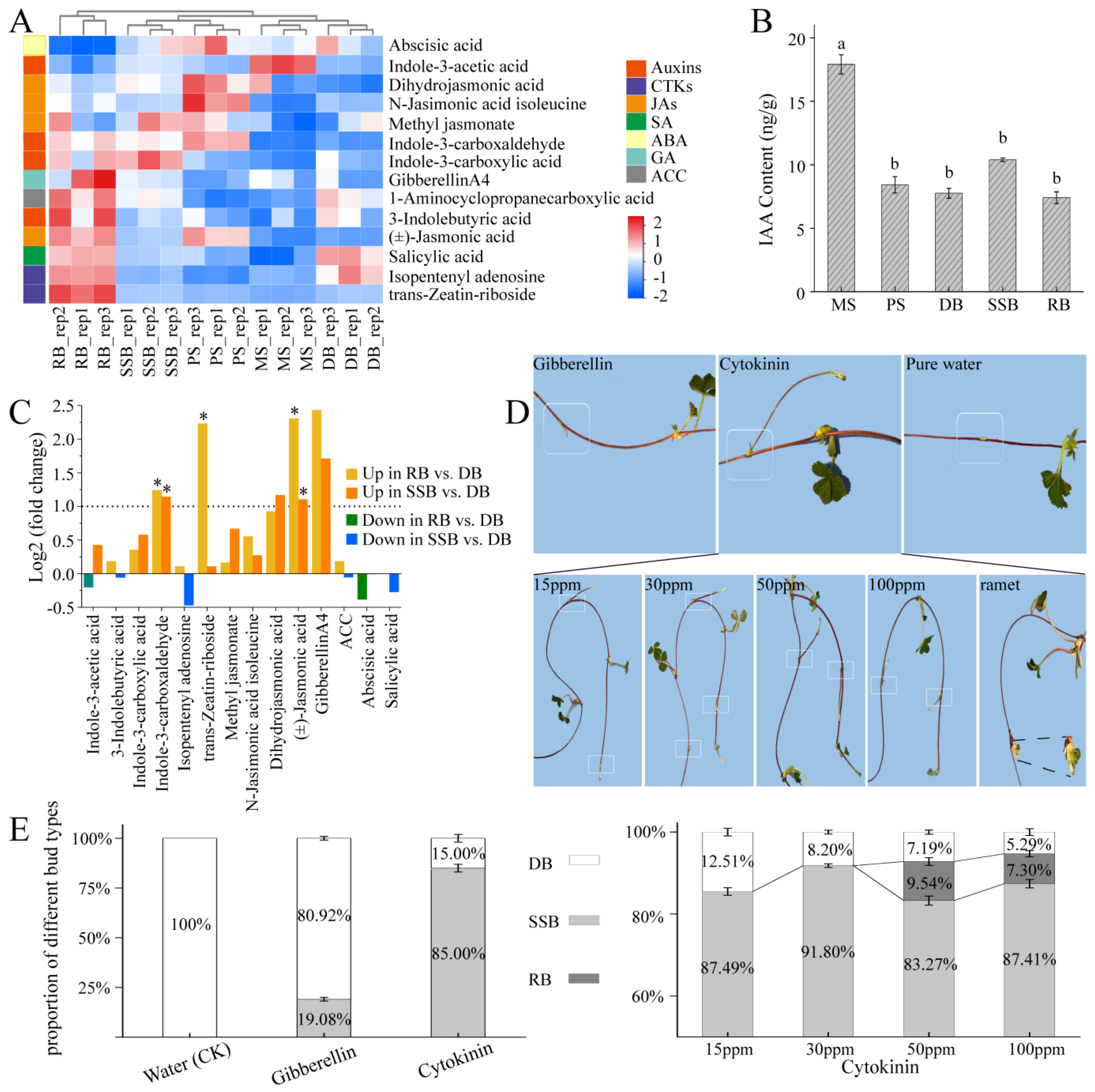
3.2. Endogenous Hormones Profile and Morphological Landscape of Sympodial Stolon in F. nilgerrensis, and Exogenous Hormone Treatment for Dormant Bud
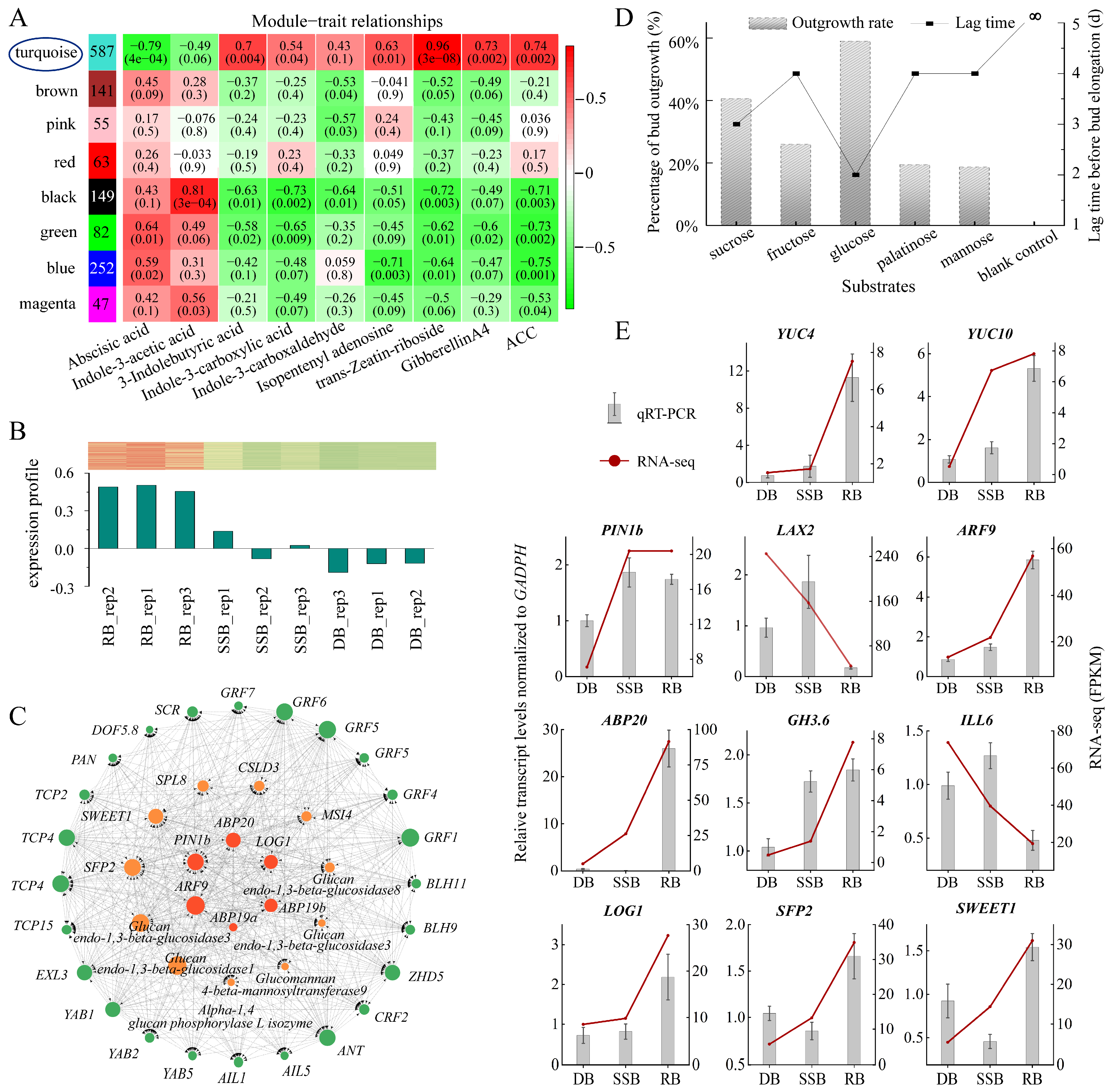
3.3. Exploring Key Genes Responsible for Ramet Formation of Sympodial Stolons in F. nilgerrensis
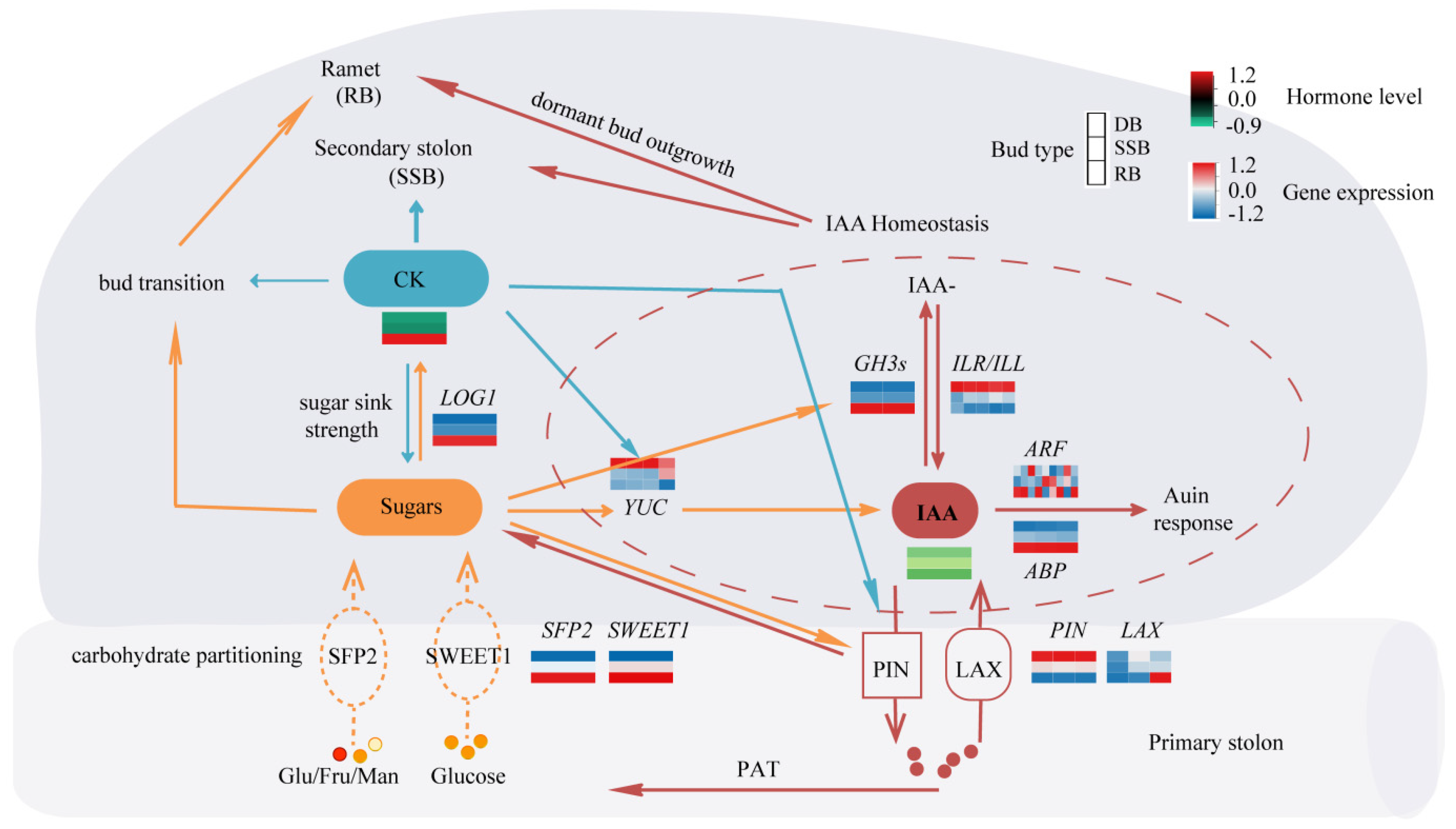
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Costes, E.; Crespel, L.; Denoyes, B.; Morel, P.; Demene, M.N.; Lauri, P.E.; Wenden, B. Bud structure, position and fate generate various branching patterns along shoots of closely related Rosaceae species: A review. Front. Plant Sci. 2014, 5, 666. [Google Scholar] [CrossRef] [PubMed]
- Caruana, J.C.; Sittmann, J.W.; Wang, W.; Liu, Z. Suppressor of Runnerless Encodes a DELLA Protein that Controls Runner Formation for Asexual Reproduction in Strawberry. Mol Plant 2018, 11, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.J.; Xue, L.; Guo, R.X.; Dai, H.P. The Fragaria Species Native to China and Their Geographical Distribution; International Society for Horticultural Science (ISHS): Leuven, Belgium, 2017. [Google Scholar]
- Sun, J.; Sun, R.; Liu, H.; Chang, L.; Li, S.; Zhao, M.; Shennan, C.; Lei, J.; Dong, J.; Zhong, C.; et al. Complete chloroplast genome sequencing of ten wild Fragaria species in China provides evidence for phylogenetic evolution of Fragaria. Genomics 2021, 113, 1170–1179. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Guan, S.C.; Wen, C.; Li, P.; Gao, Z.; Chen, X. Auxin and cytokinin coordinate the dormancy and outgrowth of axillary bud in strawberry runner. BMC Plant Biol. 2019, 19, 528. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Mo, X.; Guan, S.; Chen, X.; Xue, C. Expression Characteristics and Functional Analysis of FvYABBY5.1 in Strawberry. Acta Hortic. Sin. 2022, 49, 1458–1472. (In Chinese) [Google Scholar] [CrossRef]
- Rameau, C.; Bertheloot, J.; Leduc, N.; Andrieu, B.; Foucher, F.; Sakr, S. Multiple pathways regulate shoot branching. Front. Plant Sci. 2014, 5, 741. [Google Scholar] [CrossRef]
- Wei, J.; Yang, Q.; Ni, J.; Gao, Y.; Tang, Y.; Bai, S.; Teng, Y. Early defoliation induces auxin redistribution, promoting paradormancy release in pear buds. Plant Physiol. 2022, 190, 2739–2756. [Google Scholar] [CrossRef]
- Barbier, F.F.; Dun, E.A.; Kerr, S.C.; Chabikwa, T.G.; Beveridge, C.A. An Update on the Signals Controlling Shoot Branching. Trends Plant Sci. 2019, 24, 220–236. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, A.; Wenzl, C.; Lohmann, J.U. Beyond flexibility: Controlling stem cells in an ever changing environment. Curr. Opin. Plant Biol. 2017, 35, 117–123. [Google Scholar] [CrossRef]
- Nambara, E.; van Wees, S. Plant hormone functions and interactions in biological systems. Plant J. 2021, 105, 287–289. [Google Scholar] [CrossRef]
- Sakr, S.; Wang, M.; Dédaldéchamp, F.; Perez-Garcia, M.D.; Ogé, L.; Hamama, L.; Atanassova, R. The Sugar-Signaling Hub: Overview of Regulators and Interaction with the Hormonal and Metabolic Network. Int. J. Mol. Sci. 2018, 19, 2506. [Google Scholar] [CrossRef] [PubMed]
- Barbier, F.; Péron, T.; Lecerf, M.; Perez-Garcia, M.D.; Barrière, Q.; Rolčík, J.; Boutet-Mercey, S.; Citerne, S.; Lemoine, R.; Porcheron, B.; et al. Sucrose is an early modulator of the key hormonal mechanisms controlling bud outgrowth in Rosa hybrida. J. Exp. Bot. 2015, 66, 2569–2582. [Google Scholar] [CrossRef] [PubMed]
- Bertheloot, J.; Barbier, F.; Boudon, F.; Perez-Garcia, M.D.; Péron, T.; Citerne, S.; Dun, E.; Beveridge, C.; Godin, C.; Sakr, S. Sugar availability suppresses the auxin-induced strigolactone pathway to promote bud outgrowth. New Phytol. 2020, 225, 866–879. [Google Scholar] [CrossRef] [PubMed]
- Otori, K.; Tanabe, N.; Tamoi, M.; Shigeoka, S. Sugar Transporter Protein 1 (STP1) contributes to regulation of the genes involved in shoot branching via carbon partitioning in Arabidopsis. Biosci. Biotechnol. Biochem. 2019, 83, 472–481. [Google Scholar] [CrossRef] [PubMed]
- Tenreira, T.; Lange, M.J.P.; Lange, T.; Bres, C.; Labadie, M.; Monfort, A.; Hernould, M.; Rothan, C.; Denoyes, B. A Specific Gibberellin 20-Oxidase Dictates the Flowering-Runnering Decision in Diploid Strawberry. Plant Cell 2017, 29, 2168–2182. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Cheng, L.; Zhu, Z.; Yu, F.; Dai, C.; Liu, Z.; Guo, W.W.; Wu, X.M.; Kang, C. GRAS transcription factor loss of axillary meristems is essential for stamen and runner formation in wild strawberry. Plant Physiol. 2021, 186, 1970–1984. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wu, Z.; Zheng, J.; Koskela, E.A.; Fan, L.; Fan, G.; Gao, D.; Dong, Z.; Hou, S.; Feng, Z.; et al. The GATA factor HANABA TARANU promotes runner formation by regulating axillary bud initiation and outgrowth in cultivated strawberry. Plant J. 2022, 110, 1237–1254. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Zhao, M.; Qian, Y.; Yu, H.; Xia, J.; Wu, E. Phenotypic analysis combined with tandem mass tags (TMT) labeling reveal the heterogeneity of strawberry stolon buds. BMC Plant Biol. 2019, 19, 505. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Plunkert, M.; Luo, X.; Liu, Z. Developmental regulation of stolon and rhizome. Curr. Opin. Plant Biol. 2021, 59, 101970. [Google Scholar] [CrossRef]
- Zhang, J.; Lei, Y.; Wang, B.; Li, S.; Yu, S.; Wang, Y.; Li, H.; Liu, Y.; Ma, Y.; Dai, H.; et al. The high-quality genome of diploid strawberry (Fragaria nilgerrensis) provides new insights into anthocyanin accumulation. Plant Biotechnol. J. 2020, 18, 1908–1924. [Google Scholar] [CrossRef]
- Hu, Y.; Feng, C.; Yang, L.; Edger, P.P.; Kang, M. Genomic population structure and local adaptation of the wild strawberry Fragaria nilgerrensis. Hortic. Res. 2022, 9, uhab059. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Wang, J.; Harris, A.J.; Folta, K.M.; Zhao, M.; Kang, M. Tracing the Diploid Ancestry of the Cultivated Octoploid Strawberry. Mol. Biol. Evol. 2021, 38, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Q.; Edger, P.P.; Xue, L.; Qiong, L.; Lu, J.; Zhang, Y.; Cao, Q.; Yocca, A.E.; Platts, A.E.; Knapp, S.J.; et al. Evolutionary history and pan-genome dynamics of strawberry (Fragaria spp.). Proc. Natl. Acad. Sci. USA 2021, 118, e2105431118. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Zhang, Y.; Diao, X.; Yu, K.; Dai, X.; Qu, P.; Crabbe, M.J.C.; Zhang, T.; Qiao, Q. Evaluation of genetic diversity and population structure of Fragaria nilgerrensis using EST-SSR markers. Gene 2021, 796–797, 145791. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Feng, Y.; Dai, X.; Huang, L.; Li, J.; Tao, P.; Crabbe, M.J.C.; Zhang, T.; Qiao, Q. Dynamic Changes of DNA Methylation During Wild Strawberry (Fragaria nilgerrensis) Tissue Culture. Front. Plant Sci. 2021, 12, 765383. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Huang, L.; Li, J.; Qu, P.; Tao, P.; Crabbe, M.J.C.; Zhang, T.; Qiao, Q. Integrated transcriptome and methylome analyses reveal the molecular regulation of drought stress in wild strawberry (Fragaria nilgerrensis). BMC Plant Biol. 2022, 22, 613. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.; Futschik, M.E. Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2007, 2, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Hollender, C.A.; Geretz, A.C.; Slovin, J.P.; Liu, Z. Flower and early fruit development in a diploid strawberry, Fragaria vesca. Planta 2012, 235, 1123–1139. [Google Scholar] [CrossRef] [PubMed]
- Allen, G.C.; Flores-Vergara, M.A.; Krasynanski, S.; Kumar, S.; Thompson, W.F. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006, 1, 2320–2325. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dai, C.; Hu, C.; Liu, Z.; Kang, C. Global identification of alternative splicing via comparative analysis of SMRT- and Illumina-based RNA-seq in strawberry. Plant J. 2017, 90, 164–176. [Google Scholar] [CrossRef]
- Dong, B.-C.; Yu, G.-L.; Guo, W.; Zhang, M.-X.; Dong, M.; Yu, F.-H. How Internode Length, Position and Presence of Leaves Affect Survival and Growth of Alternanthera philoxeroides after Fragmentation? Evol. Ecol. 2010, 24, 1447–1461. [Google Scholar] [CrossRef]
- Sarojam, R.; Sappl, P.G.; Goldshmidt, A.; Efroni, I.; Floyd, S.K.; Eshed, Y.; Bowman, J.L. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 2010, 22, 2113–2130. [Google Scholar] [CrossRef]
- Ketudat Cairns, J.R.; Mahong, B.; Baiya, S.; Jeon, J.S. β-Glucosidases: Multitasking, moonlighting or simply misunderstood? Plant Sci. 2015, 241, 246–259. [Google Scholar] [CrossRef]
- Gutierrez-Gutierrez, D.A.; Fuentes-Garibay, J.A.; Viader-Salvadó, J.M.; Guerrero-Olazarán, M. Biochemical characterization of the β-glucosidase Glu1B from Coptotermes formosanus produced in Pichia pastoris. Enzym. Microb. Technol. 2023, 163, 110155. [Google Scholar] [CrossRef]
- Zhang, L.; Xiang, Z.; Li, J.; Wang, S.; Chen, Y.; Liu, Y.; Mao, D.; Luan, S.; Chen, L. bHLH57 confers chilling tolerance and grain yield improvement in rice. Plant Cell Environ. 2023, 46, 1402–1418. [Google Scholar] [CrossRef]
- Schuster, C.; Gaillochet, C.; Lohmann, J.U. Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development 2015, 142, 3343–3350. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, Z.; Wang, L.; Yan, S.; Cheng, Z.; Liu, X.; Han, L.; Chen, G.; Wang, S.; Song, W.; et al. The CsHEC1-CsOVATE module contributes to fruit neck length variation via modulating auxin biosynthesis in cucumber. Proc. Natl. Acad. Sci. USA 2022, 119, e2209717119. [Google Scholar] [CrossRef]
- Ung, K.L.; Winkler, M.; Schulz, L.; Kolb, M.; Janacek, D.P.; Dedic, E.; Stokes, D.L.; Hammes, U.Z.; Pedersen, B.P. Structures and mechanism of the plant PIN-FORMED auxin transporter. Nature 2022, 609, 605–610. [Google Scholar] [CrossRef]
- Adamowski, M.; Friml, J. PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef]
- Swarup, R.; Bhosale, R. Developmental Roles of AUX1/LAX Auxin Influx Carriers in Plants. Front. Plant Sci. 2019, 10, 1306. [Google Scholar] [CrossRef]
- Atkinson, C.J.; Else, M.A. Hydraulic conductivity and PAT determine hierarchical resource partitioning and ramet development along Fragaria stolons. J. Exp. Bot. 2012, 63, 5093–5104. [Google Scholar] [CrossRef]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef]
- Sirhindi, G.; Mir, M.A.; Abd-Allah, E.F.; Ahmad, P.; Gucel, S. Jasmonic Acid Modulates the Physio-Biochemical Attributes, Antioxidant Enzyme Activity, and Gene Expression in Glycine max under Nickel Toxicity. Front. Plant Sci. 2016, 7, 591. [Google Scholar] [CrossRef]
- Awan, S.A.; Khan, I.; Rizwan, M.; Zhang, X.; Brestic, M.; Khan, A.; El-Sheikh, M.A.; Alyemeni, M.N.; Ali, S.; Huang, L. Exogenous abscisic acid and jasmonic acid restrain polyethylene glycol-induced drought by improving the growth and antioxidative enzyme activities in pearl millet. Physiol. Plant. 2021, 172, 809–819. [Google Scholar] [CrossRef]
- Liu, R.; Finlayson, S.A. Sorghum tiller bud growth is repressed by contact with the overlying leaf. Plant Cell Environ. 2019, 42, 2120–2132. [Google Scholar] [CrossRef]
- Roman, H.; Girault, T.; Barbier, F.; Péron, T.; Brouard, N.; Pěnčík, A.; Novák, O.; Vian, A.; Sakr, S.; Lothier, J.; et al. Cytokinins Are Initial Targets of Light in the Control of Bud Outgrowth. Plant Physiol. 2016, 172, 489–509. [Google Scholar] [CrossRef] [PubMed]
- Porcher, A.; Guérin, V.; Leduc, N.; Lebrec, A.; Lothier, J.; Vian, A. Ascorbate-glutathione pathways mediated by cytokinin regulate H2O2 levels in light-controlled rose bud burst. Plant Physiol. 2021, 186, 910–928. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Cheung, L.S.; Feng, L.; Tanner, W.; Frommer, W.B. Transport of sugars. Annu. Rev. Biochem. 2015, 84, 865–894. [Google Scholar] [CrossRef]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Slawinski, L.; Israel, A.; Paillot, C.; Thibault, F.; Cordaux, R.; Atanassova, R.; Dédaldéchamp, F.; Laloi, M. Early Response to Dehydration Six-like Transporter Family: Early Origin in Streptophytes and Evolution in Land Plants. Front. Plant Sci. 2021, 12, 681929. [Google Scholar] [CrossRef]
- Fukui, K.; Arai, K.; Tanaka, Y.; Aoi, Y.; Kukshal, V.; Jez, J.M.; Kubes, M.F.; Napier, R.; Zhao, Y.; Kasahara, H.; et al. Chemical inhibition of the auxin inactivation pathway uncovers the roles of metabolic turnover in auxin homeostasis. Proc. Natl. Acad. Sci. USA 2022, 119, e2206869119. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 2012, 5, 334–338. [Google Scholar] [CrossRef]
- Aoi, Y.; Tanaka, K.; Cook, S.D.; Hayashi, K.I.; Kasahara, H. GH3 Auxin-Amido Synthetases Alter the Ratio of Indole-3-Acetic Acid and Phenylacetic Acid in Arabidopsis. Plant Cell Physiol. 2020, 61, 596–605. [Google Scholar] [CrossRef]
- Leyser, O. Auxin Signaling. Plant Physiol. 2018, 176, 465–479. [Google Scholar] [CrossRef]
- Hajný, J.; Prát, T.; Rydza, N.; Rodriguez, L.; Tan, S.; Verstraeten, I.; Domjan, D.; Mazur, E.; Smakowska-Luzan, E.; Smet, W.; et al. Receptor kinase module targets PIN-dependent auxin transport during canalization. Science 2020, 370, 550–557. [Google Scholar] [CrossRef]
- Sanchez Carranza, A.P.; Singh, A.; Steinberger, K.; Panigrahi, K.; Palme, K.; Dovzhenko, A.; Dal Bosco, C. Hydrolases of the ILR1-like family of Arabidopsis thaliana modulate auxin response by regulating auxin homeostasis in the endoplasmic reticulum. Sci. Rep. 2016, 6, 24212. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Pérez-Garcia, M.D.; Davière, J.M.; Barbier, F.; Ogé, L.; Gentilhomme, J.; Voisine, L.; Péron, T.; Launay-Avon, A.; Clément, G.; et al. Outgrowth of the axillary bud in rose is controlled by sugar metabolism and signalling. J. Exp. Bot. 2021, 72, 3044–3060. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ogé, L.; Pérez Garcia, M.D.; Launay-Avon, A.; Clément, G.; Le Gourrierec, J.; Hamama, L.; Sakr, S. Antagonistic Effect of Sucrose Availability and Auxin on Rosa Axillary Bud Metabolism and Signaling, Based on the Transcriptomics and Metabolomics Analysis. Front. Plant Sci. 2022, 13, 830840. [Google Scholar] [CrossRef]
- Kurakawa, T.; Ueda, N.; Maekawa, M.; Kobayashi, K.; Kojima, M.; Nagato, Y.; Sakakibara, H.; Kyozuka, J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 2007, 445, 652–655. [Google Scholar] [CrossRef]
- Jones, B.; Gunnerås, S.A.; Petersson, S.V.; Tarkowski, P.; Graham, N.; May, S.; Dolezal, K.; Sandberg, G.; Ljung, K. Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell 2010, 22, 2956–2969. [Google Scholar] [CrossRef]
- Peleg, Z.; Reguera, M.; Tumimbang, E.; Walia, H.; Blumwald, E. Cytokinin-mediated source/sink modifications improve drought tolerance and increase grain yield in rice under water-stress. Plant Biotechnol. J. 2011, 9, 747–758. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, C.; Yu, X.; Tian, Y.; Wang, W.; Zhang, Y.; Bai, W.; Yang, N.; Zhang, T.; Zheng, H.; et al. Auxin regulates source-sink carbohydrate partitioning and reproductive organ development in rice. Proc. Natl. Acad. Sci. USA 2022, 119, e2121671119. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lan, G.; Wu, M.; Zhang, Q.; Yuan, B.; Shi, G.; Zhu, N.; Zheng, Y.; Cao, Q.; Qiao, Q.; Zhang, T. Transcriptomic and Physiological Analyses for the Role of Hormones and Sugar in Axillary Bud Development of Wild Strawberry Stolon. Plants 2024, 13, 2241. https://doi.org/10.3390/plants13162241
Lan G, Wu M, Zhang Q, Yuan B, Shi G, Zhu N, Zheng Y, Cao Q, Qiao Q, Zhang T. Transcriptomic and Physiological Analyses for the Role of Hormones and Sugar in Axillary Bud Development of Wild Strawberry Stolon. Plants. 2024; 13(16):2241. https://doi.org/10.3390/plants13162241
Chicago/Turabian StyleLan, Genqian, Mingzhao Wu, Qihang Zhang, Bo Yuan, Guangxin Shi, Ni Zhu, Yibingyue Zheng, Qiang Cao, Qin Qiao, and Ticao Zhang. 2024. "Transcriptomic and Physiological Analyses for the Role of Hormones and Sugar in Axillary Bud Development of Wild Strawberry Stolon" Plants 13, no. 16: 2241. https://doi.org/10.3390/plants13162241





