Automatic Disease Detection from Strawberry Leaf Based on Improved YOLOv8
Abstract
:1. Introduction
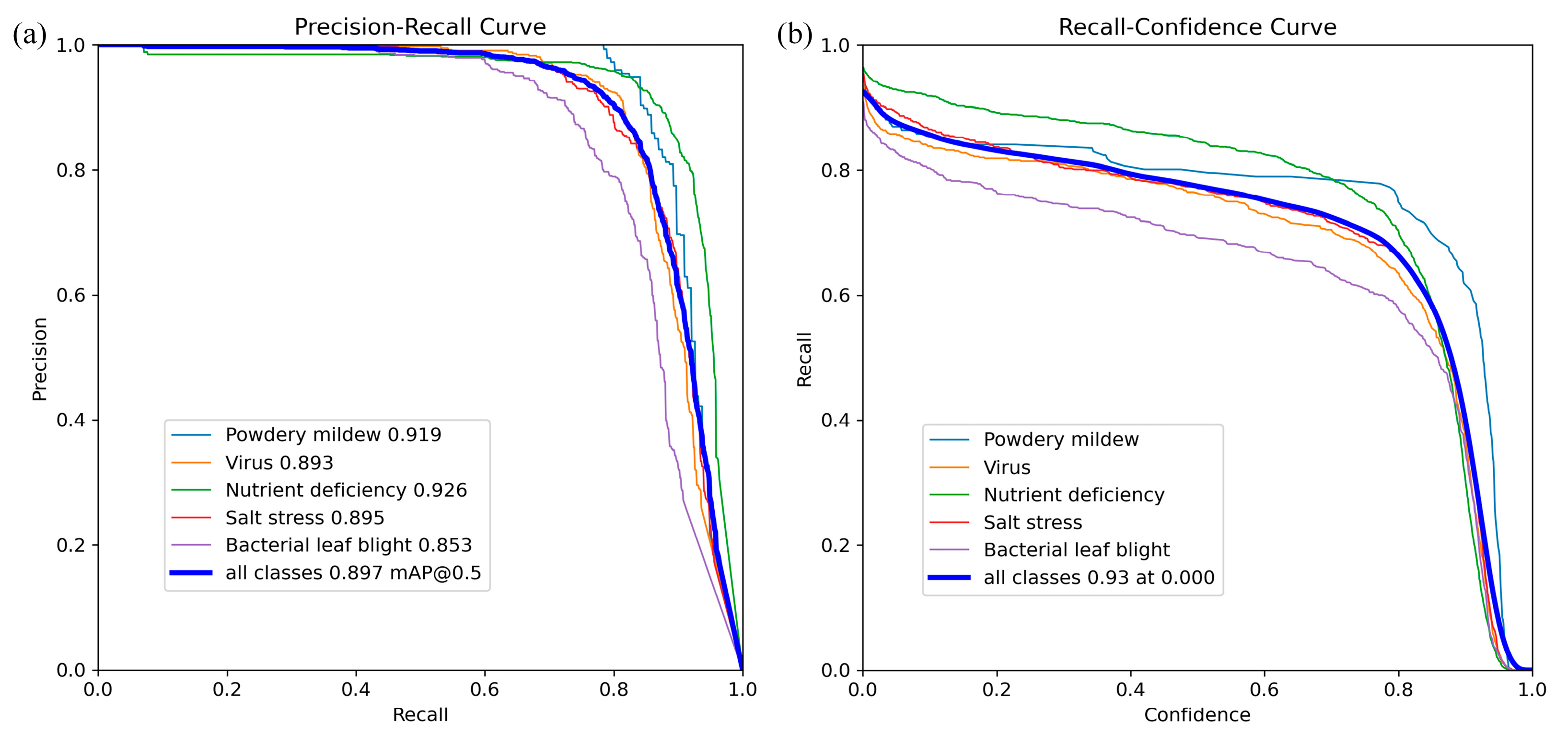
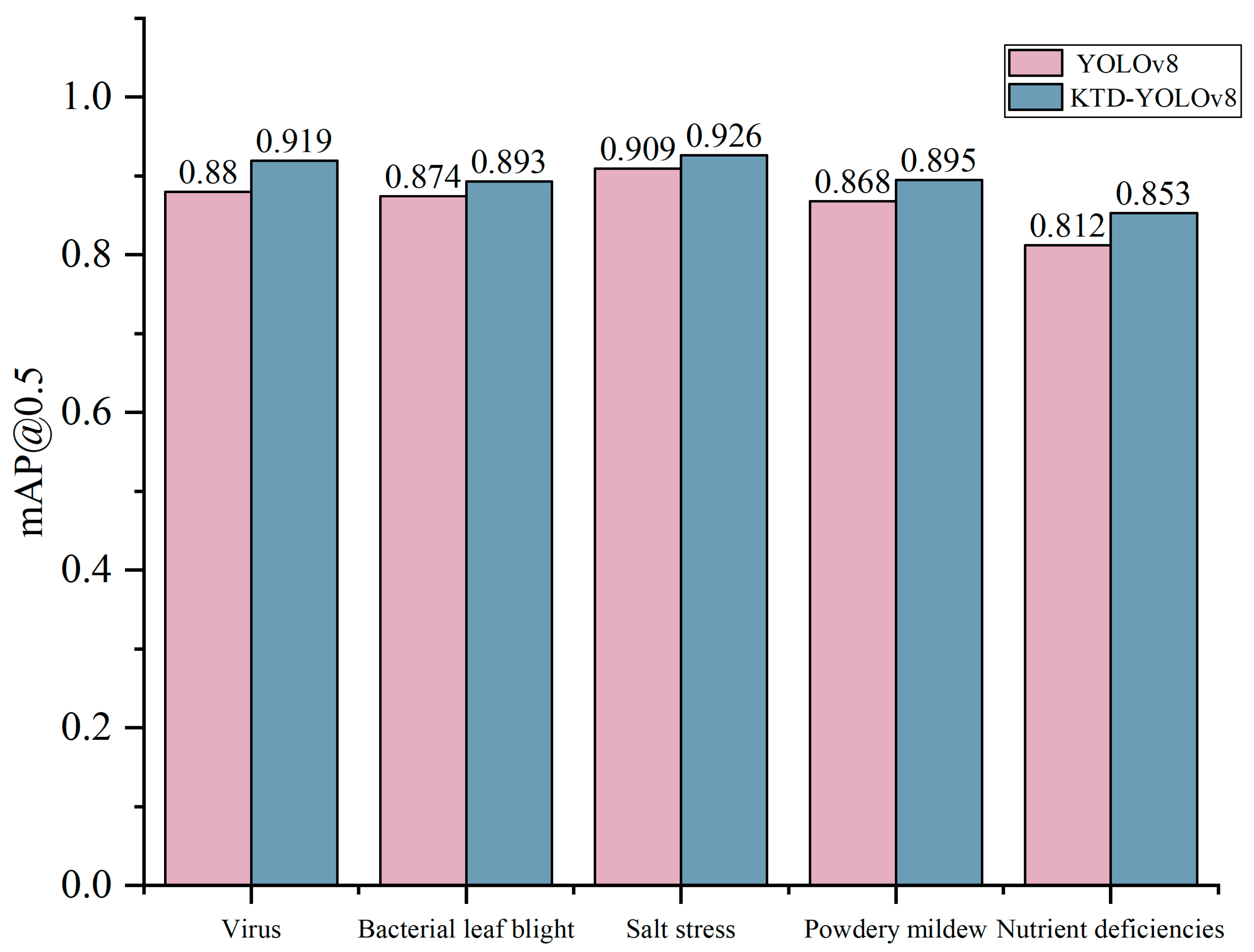
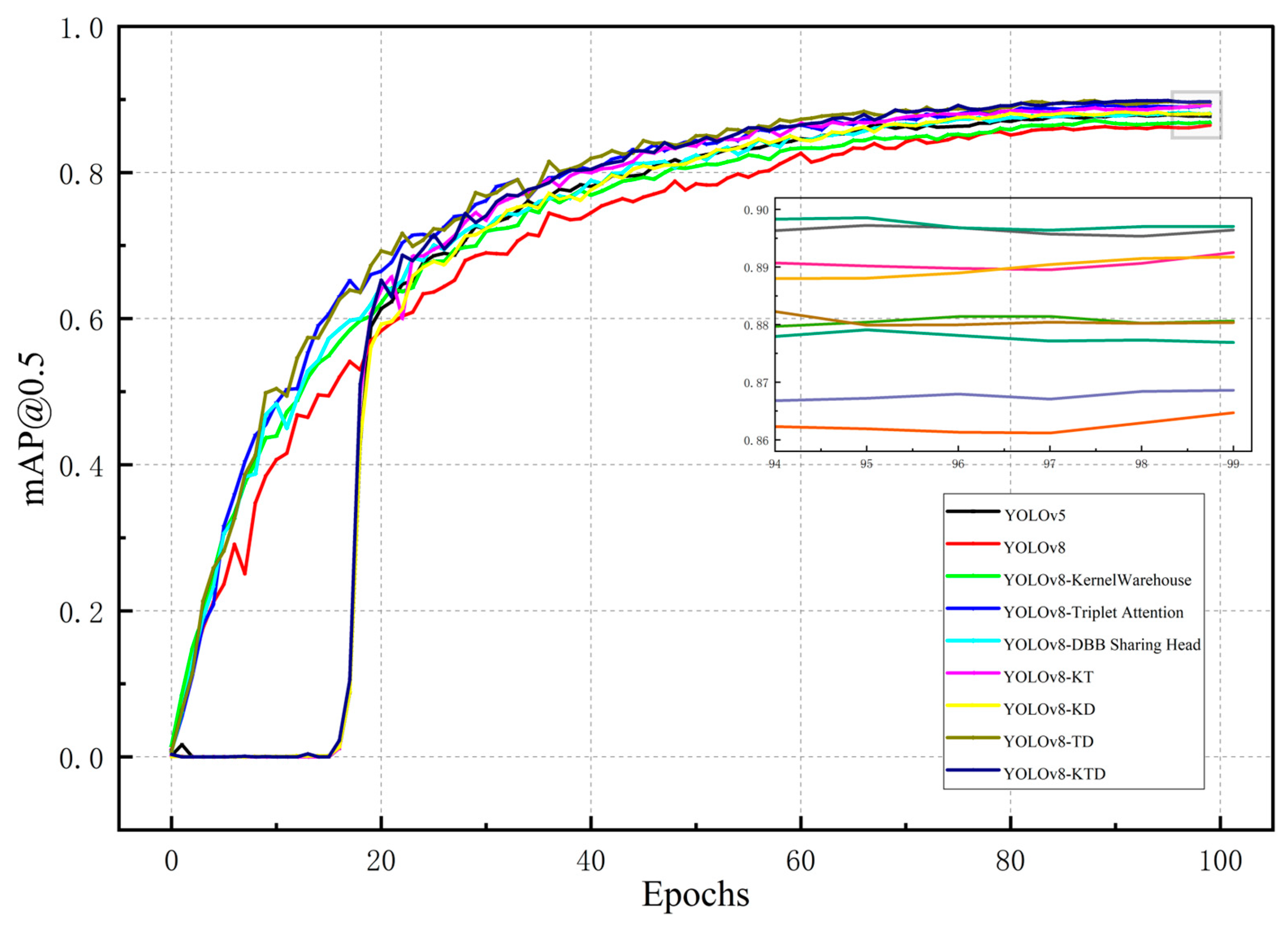
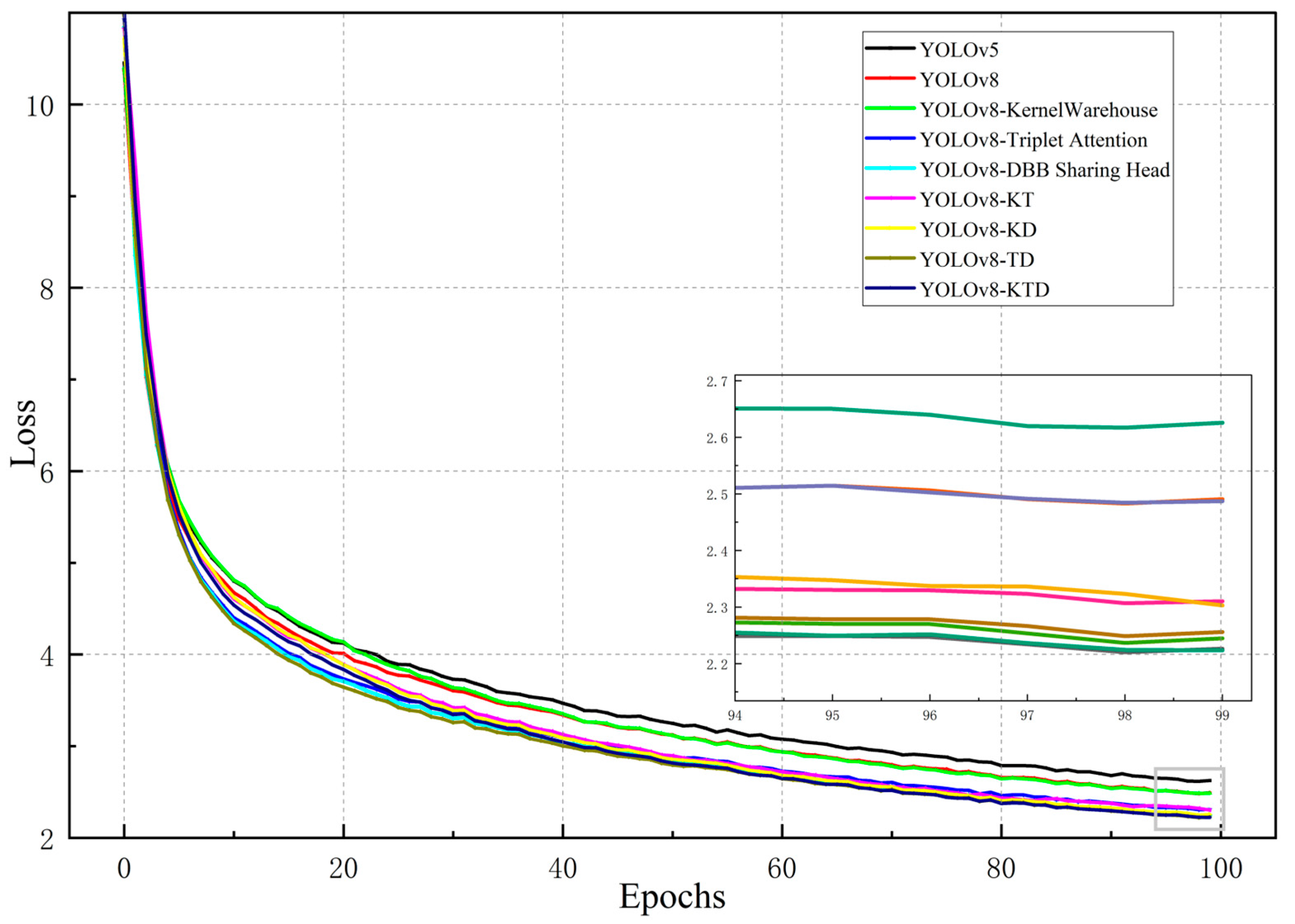
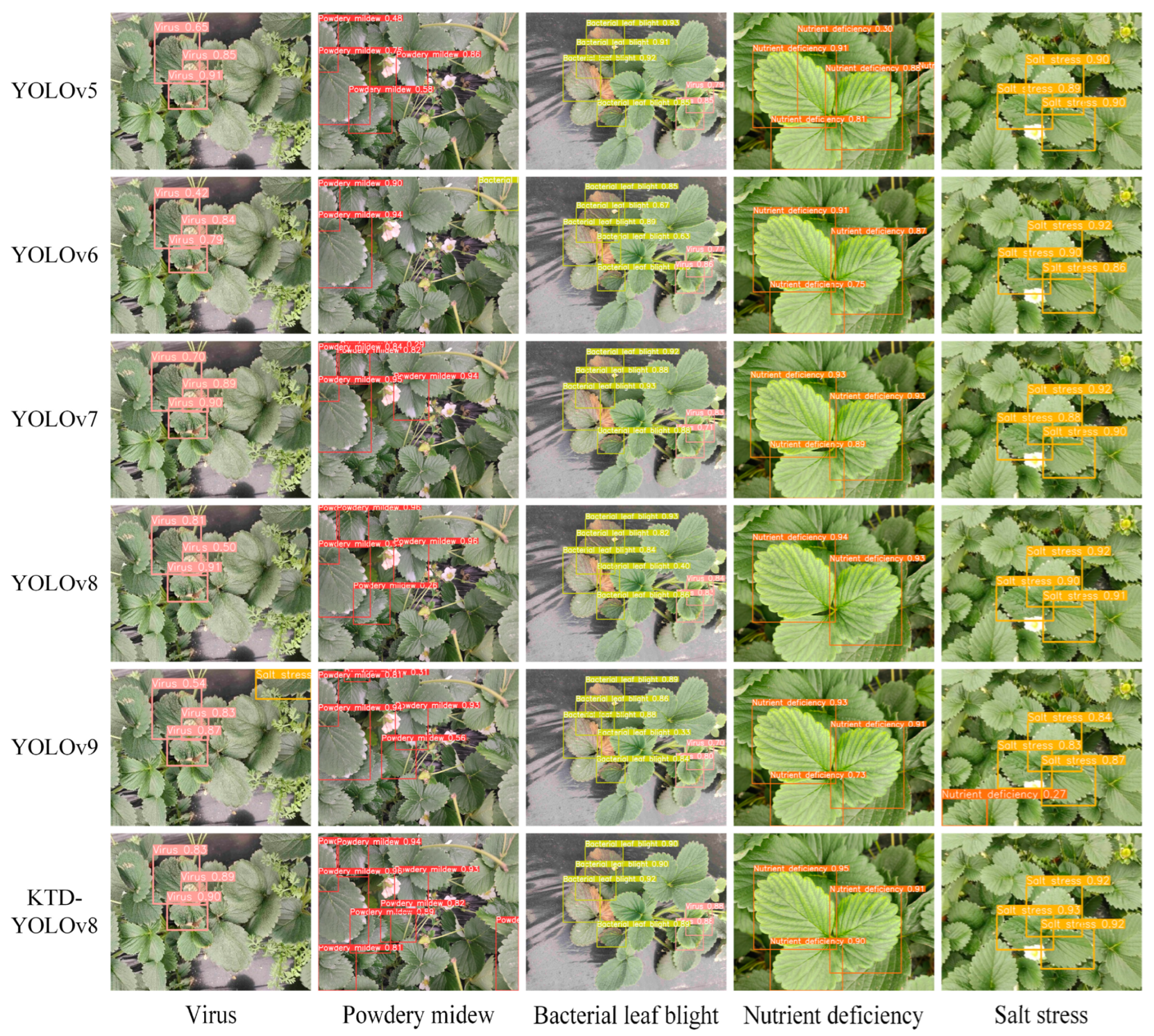
2. Results and Discussion
3. Materials and Methods
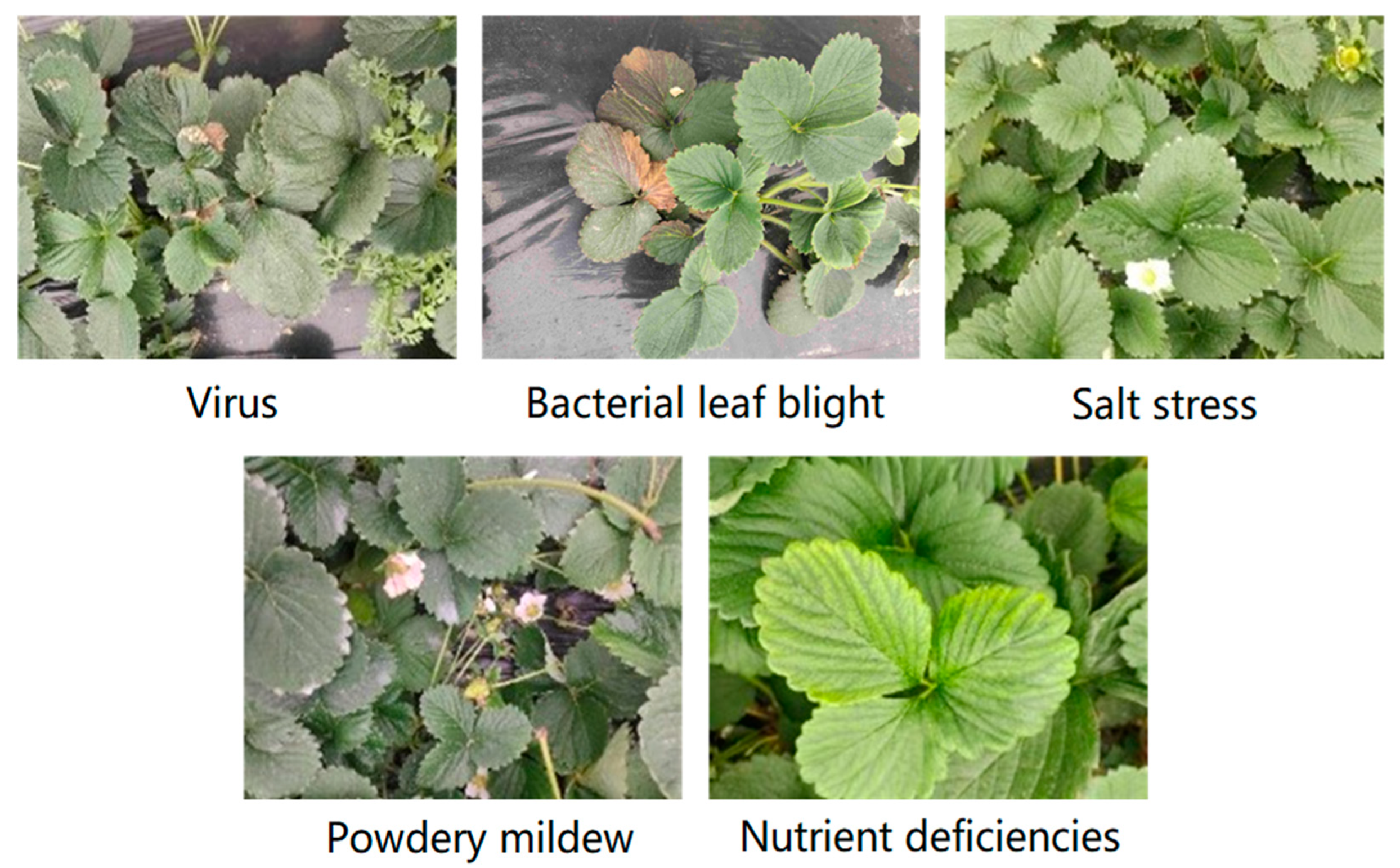
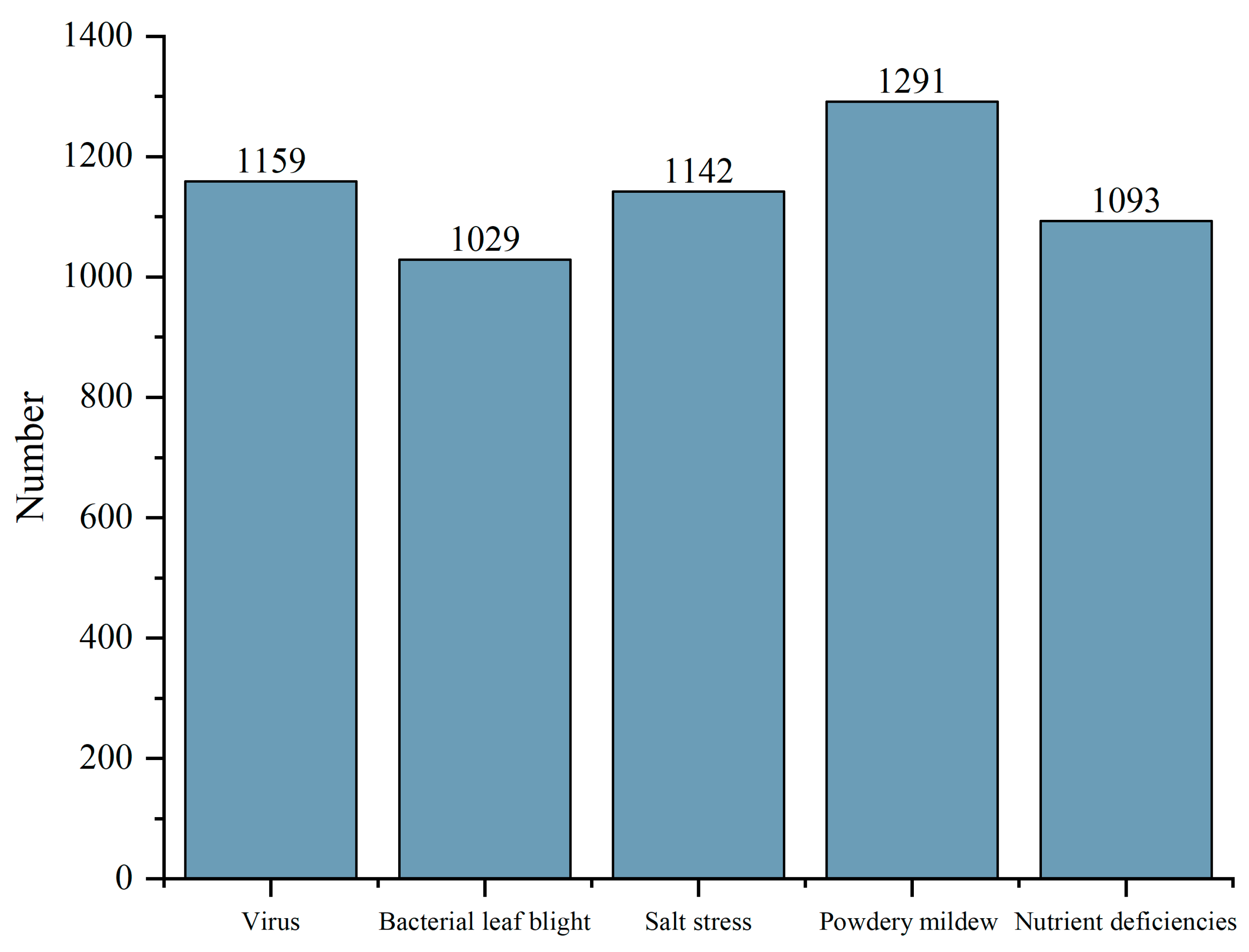
3.1. Image Dataset
3.2. Image Enhancement
3.3. Experimental Platform
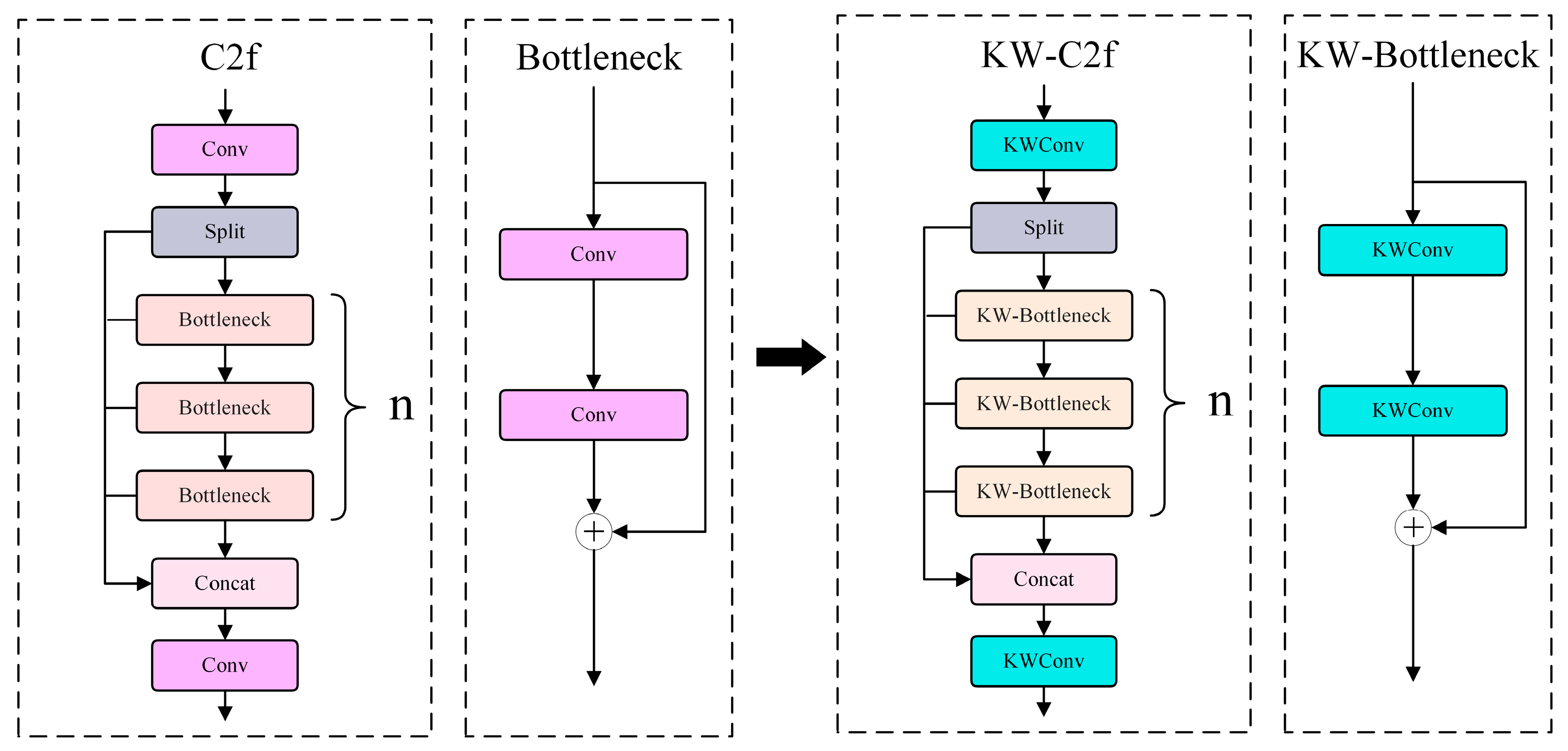
3.4. KTD-YOLOv8 Model
- KernelWarehouse convolution (KWConv)
- 2.
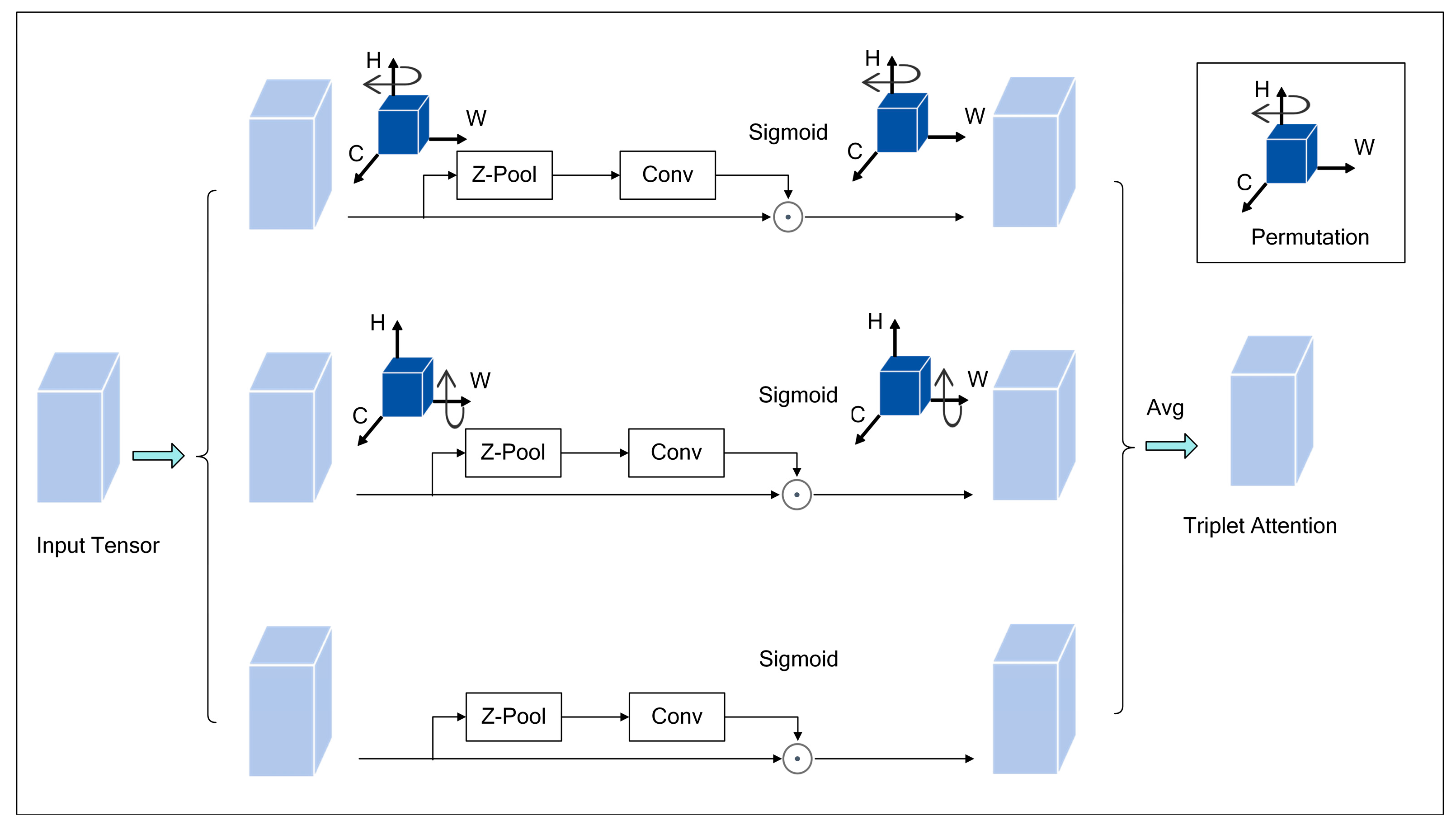
- Triplet Attention mechanism
- 3.
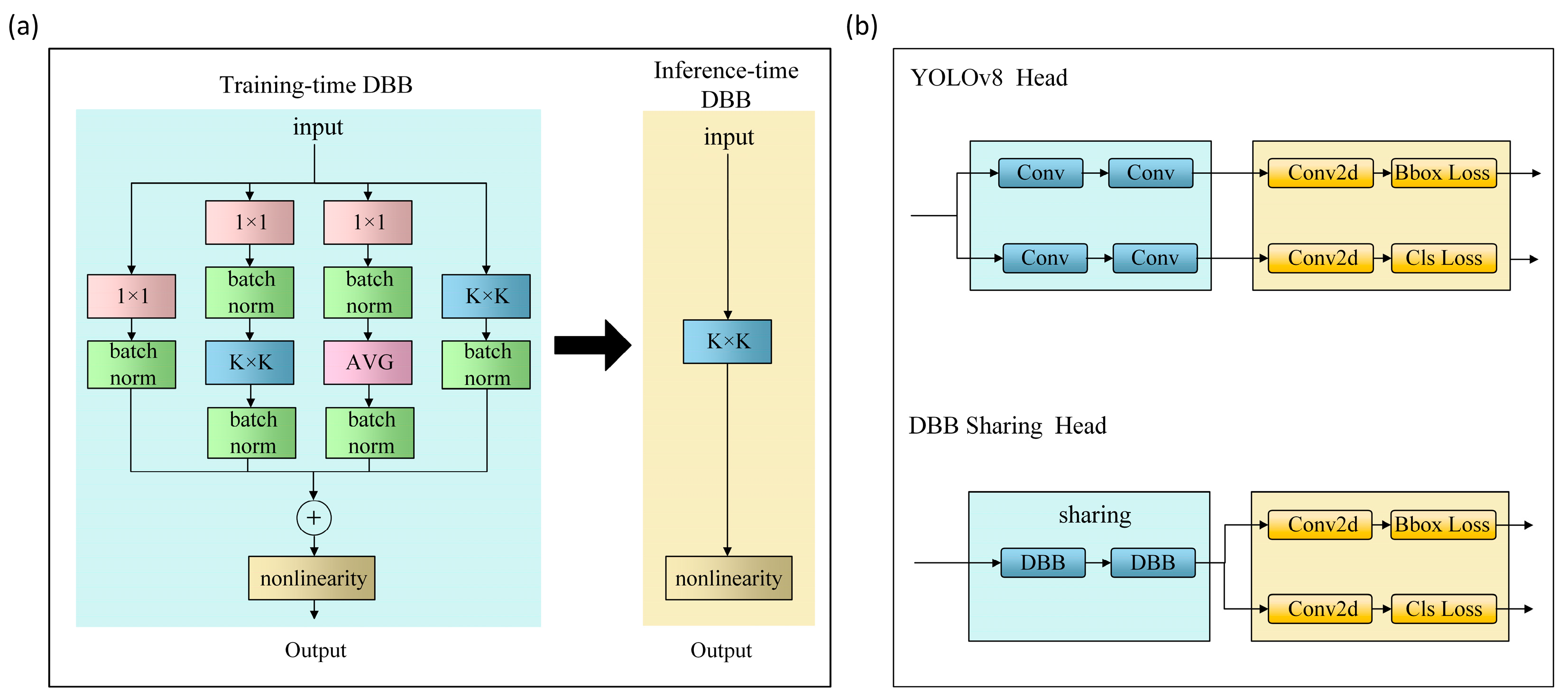
- DBB Sharing Head
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Giampieri, F.; Tulipani, S.; Alvarez-Suarez, J.M.; Quiles, J.L.; Mezzetti, B.; Battino, M. The strawberry: Composition, nutritional quality, and impact on human health. Nutrition 2012, 28, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, C.; Gupta, D.; Gupta, U.; Hazarika, B.B. Leaf disease detection using machine learning and deep learning: Review and challenges. Appl. Soft Comput. 2023, 145, 110534. [Google Scholar] [CrossRef]
- Li, G.; Jiao, L.; Chen, P.; Liu, K.; Wang, R.; Dong, S.; Kang, C. Spatial convolutional self-attention-based transformer module for strawberry disease identification under complex background. Comput. Electron. Agric. 2023, 212, 108121. [Google Scholar] [CrossRef]
- Camargo, A.; Smith, J. Image pattern classification for the identification of disease causing agents in plants. Comput. Electron. Agric. 2009, 66, 121–125. [Google Scholar] [CrossRef]
- Maas, J.L. Strawberry disease management. In Diseases of Fruits and Vegetables: Volume II: Diagnosis and Management; Springer: Dordrecht, The Netherland, 2004; pp. 441–483. [Google Scholar]
- You, J.; Jiang, K.; Lee, J. Deep metric learning-based strawberry disease detection with unknowns. Front. Plant Sci. 2022, 13, 891785. [Google Scholar] [CrossRef]
- Li, Z.; Li, B.-x.; Li, Z.-h.; Zhan, Y.-f.; Wang, L.-h.; Gong, Q. Research progress in crop disease and pest identification based on deep learning. Hubei Agric. Sci. 2023, 62, 165. [Google Scholar]
- Bharate, A.A.; Shirdhonkar, M. A review on plant disease detection using image processing. In Proceedings of the 2017 International Conference on Intelligent Sustainable Systems (ICISS), Palladam, India, 7–8 December 2017; pp. 103–109. [Google Scholar]
- Radhakrishnan, S. An improved machine learning algorithm for predicting blast disease in paddy crop. Mater. Today Proc. 2020, 33, 682–686. [Google Scholar] [CrossRef]
- Thakur, P.S.; Khanna, P.; Sheorey, T.; Ojha, A. Trends in vision-based machine learning techniques for plant disease identification: A systematic review. Expert Syst. Appl. 2022, 208, 118117. [Google Scholar] [CrossRef]
- Liu, J.; Abbas, I.; Noor, R.S. Development of deep learning-based variable rate agrochemical spraying system for targeted weeds control in strawberry crop. Agronomy 2021, 11, 1480. [Google Scholar] [CrossRef]
- Meshram, A.T.; Vanalkar, A.V.; Kalambe, K.B.; Badar, A.M. Pesticide spraying robot for precision agriculture: A categorical literature review and future trends. J. Field Robot. 2022, 39, 153–171. [Google Scholar] [CrossRef]
- Wu, G.; Fang, Y.; Jiang, Q.; Cui, M.; Li, N.; Ou, Y.; Diao, Z.; Zhang, B. Early identification of strawberry leaves disease utilizing hyperspectral imaging combing with spectral features, multiple vegetation indices and textural features. Comput. Electron. Agric. 2023, 204, 107553. [Google Scholar]
- Aggarwal, M.; Khullar, V.; Goyal, N. Exploring classification of rice leaf diseases using machine learning and deep learning. In Proceedings of the 2023 3rd International Conference on Innovative Practices in Technology and Management (ICIPTM), Uttar Pradesh, India, 22–24 February 2023; pp. 1–6. [Google Scholar]
- Javidan, S.M.; Banakar, A.; Vakilian, K.A.; Ampatzidis, Y. Diagnosis of grape leaf diseases using automatic K-means clustering and machine learning. Smart Agric. Technol. 2023, 3, 100081. [Google Scholar] [CrossRef]
- Zhang, S.; Shang, Y.; Wang, L. Plant disease recognition based on plant leaf image. J. Anim. Plant Sci. 2015, 25, 42–45. [Google Scholar]
- Dargan, S.; Kumar, M.; Ayyagari, M.R.; Kumar, G. A survey of deep learning and its applications: A new paradigm to machine learning. Arch. Comput. Methods Eng. 2020, 27, 1071–1092. [Google Scholar] [CrossRef]
- Dash, A.; Sethy, P.K.; Behera, S.K. Maize disease identification based on optimized support vector machine using deep feature of DenseNet201. J. Agric. Food Res. 2023, 14, 100824. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, S.; Li, E.; Yang, G.; Liang, Z.; Tan, M. MD-YOLO: Multi-scale Dense YOLO for small target pest detection. Comput. Electron. Agric. 2023, 213, 108233. [Google Scholar] [CrossRef]
- Ozguven, M.M.; Adem, K. Automatic detection and classification of leaf spot disease in sugar beet using deep learning algorithms. Phys. A Stat. Mech. Its Appl. 2019, 535, 122537. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, J.; Wu, S. Multiple disease detection method for greenhouse-cultivated strawberry based on multiscale feature fusion Faster R_CNN. Comput. Electron. Agric. 2022, 199, 107176. [Google Scholar] [CrossRef]
- Abbas, I.; Liu, J.; Amin, M.; Tariq, A.; Tunio, M.H. Strawberry fungal leaf scorch disease identification in real-time strawberry field using deep learning architectures. Plants 2021, 10, 2643. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Wu, H.; Yu, Y.; Sun, H.; Zhang, H. Detection of powdery mildew on strawberry leaves based on DAC-YOLOv4 model. Comput. Electron. Agric. 2022, 202, 107418. [Google Scholar] [CrossRef]
- Li, C.; Yao, A. KernelWarehouse: Towards Parameter-Efficient Dynamic Convolution. arXiv 2023, arXiv:2308.08361. [Google Scholar]
- Misra, D.; Nalamada, T.; Arasanipalai, A.U.; Hou, Q. Rotate to attend: Convolutional triplet attention module. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Virtual, 5–9 January 2021; pp. 3139–3148. [Google Scholar]
- Ding, X.; Zhang, X.; Han, J.; Ding, G. Diverse branch block: Building a convolution as an inception-like unit. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 10886–10895. [Google Scholar]
- Qi, Y.; He, Y.; Qi, X.; Zhang, Y.; Yang, G. Dynamic snake convolution based on topological geometric constraints for tubular structure segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 2–6 October 2023; pp. 6070–6079. [Google Scholar]
- Sunkara, R.; Luo, T. No more strided convolutions or pooling: A new CNN building block for low-resolution images and small objects. In Proceedings of the Joint European Conference on Machine Learning and Knowledge Discovery in Databases, Grenoble, France, 19–23 September 2022; pp. 443–459. [Google Scholar]
- Jin, G.; Taniguchi, R.-I.; Qu, F. Auxiliary detection head for one-stage object detection. IEEE Access 2020, 8, 85740–85749. [Google Scholar] [CrossRef]
- Maji, D.; Nagori, S.; Mathew, M.; Poddar, D. Yolo-pose: Enhancing yolo for multi person pose estimation using object keypoint similarity loss. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 2637–2646. [Google Scholar]
- Yang, L.; Zhang, R.-Y.; Li, L.; Xie, X. Simam: A simple, parameter-free attention module for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 11863–11874. [Google Scholar]
- Huang, H.; Chen, Z.; Zou, Y.; Lu, M.; Chen, C. Channel prior convolutional attention for medical image segmentation. arXiv 2023, arXiv:2306.05196. [Google Scholar] [CrossRef]
- Tong, K.; Wu, Y. Rethinking PASCAL-VOC and MS-COCO dataset for small object detection. J. Vis. Commun. Image Represent. 2023, 93, 103830. [Google Scholar] [CrossRef]


| Baseline | Convolution | Attention | Head | Accuracy/ % | Recalls/ % | mAP@0.5/% | GFLOPS | Parameters | Inference Time/ms |
|---|---|---|---|---|---|---|---|---|---|
| √ | 89.1 | 77.6 | 86.9 | 28.8 | 1.113 × 107 | 13.1 | |||
| √ | √ | 91.3 | 77.2 | 87.9 | 14.2 | 1.123 × 107 | 17.1 | ||
| √ | √ | 90.0 | 81.0 | 89.3 | 28.5 | 1.114 × 107 | 14.2 | ||
| √ | √ | 89.2 | 79.4 | 88.1 | 31.8 | 1.327 × 107 | 11.1 | ||
| √ | √ | √ | 89.5 | 80.3 | 88.2 | 17.6 | 1.338 × 107 | 12.3 | |
| √ | √ | √ | 92.1 | 79.0 | 89.2 | 14.3 | 1.124 × 107 | 18.6 | |
| √ | √ | √ | 91.9 | 80.2 | 89.6 | 31.9 | 1.327 × 107 | 11.8 | |
| √ | √ | √ | √ | 90.0 | 81.3 | 89.7 | 17.7 | 1.343 × 107 | 12.1 |
| Convolution | Accuracy/% | Recall/% | mAP@0.5/% | GFLOPS |
|---|---|---|---|---|
| YOLOv8s | 89.1 | 77.6 | 86.9 | 28.8 |
| DySnakeConv | 88.8 | 79.2 | 87.9 | 31.6 |
| SPDConv | 90.3 | 78.6 | 88.1 | 43.0 |
| KWConv | 91.3 | 77.2 | 87.9 | 14.2 |
| Head | Accuracy (%) | Recall (%) | mAP@0.5 (%) | GFLOPS |
|---|---|---|---|---|
| YOLOv8s | 89.1 | 77.6 | 86.9 | 28.8 |
| Aux Head | 89.8 | 80.0 | 88.2 | 36.8 |
| Pose Head | 89.9 | 79.0 | 88.2 | 39.7 |
| DBB Sharing Head | 89.2 | 79.4 | 88.1 | 31.8 |
| Attention | Accuracy/% | Recall/% | mAP@0.5/% | GFLOPS |
|---|---|---|---|---|
| YOLOv8s | 89.1 | 77.6 | 86.9 | 28.8 |
| SimAM | 88.1 | 79.9 | 88.7 | 28.4 |
| CPCA | 86.5 | 80.1 | 87.6 | 29.4 |
| Triplet Attention | 90.0 | 81.0 | 89.3 | 28.5 |
| Arithmetic | Accuracy (%) | Recall (%) | mAP@0.5 (%) | GFLOPS | Parameters | Inference Time (ms) |
|---|---|---|---|---|---|---|
| YOLOv5 | 86.9 | 77.8 | 86.5 | 14.2 | 0.711 × 107 | 12.5 |
| YOLOv6 | 86.5 | 73.6 | 83.2 | 44.0 | 1.629 × 107 | 13.2 |
| YOLOv7 | 89.8 | 78.7 | 88.0 | 103.2 | 3.650 × 107 | 21.0 |
| YOLOv8 | 89.1 | 77.6 | 86.9 | 28.8 | 1.113 × 107 | 13.1 |
| YOLOv9 | 89.2 | 80.0 | 89.4 | 237.7 | 5.097 × 107 | 30.2 |
| KTD-YOLOv8 | 90.0 | 81.3 | 89.7 | 17.7 | 1.343 × 107 | 12.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Peng, Y.; Wei, C.; Zheng, Y.; Yang, C.; Zou, T. Automatic Disease Detection from Strawberry Leaf Based on Improved YOLOv8. Plants 2024, 13, 2556. https://doi.org/10.3390/plants13182556
He Y, Peng Y, Wei C, Zheng Y, Yang C, Zou T. Automatic Disease Detection from Strawberry Leaf Based on Improved YOLOv8. Plants. 2024; 13(18):2556. https://doi.org/10.3390/plants13182556
Chicago/Turabian StyleHe, Yuelong, Yunfeng Peng, Chuyong Wei, Yuda Zheng, Changcai Yang, and Tengyue Zou. 2024. "Automatic Disease Detection from Strawberry Leaf Based on Improved YOLOv8" Plants 13, no. 18: 2556. https://doi.org/10.3390/plants13182556
APA StyleHe, Y., Peng, Y., Wei, C., Zheng, Y., Yang, C., & Zou, T. (2024). Automatic Disease Detection from Strawberry Leaf Based on Improved YOLOv8. Plants, 13(18), 2556. https://doi.org/10.3390/plants13182556






