Agro-Physiological and DNA Methylation Responses to Salinity Stress in Wheat (Triticum aestivum L.), Aegilops cylindrica Host, and Their Introgressed Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Field Experiment
2.3. Agronomic Traits
2.4. Salinity Tolerance Indices
- Stress tolerance index:
- 2.
- Tolerance index:
- 3.
- Stress susceptibility index:
- 4.
- Yield stability index:
- 5.
- Yield index:
- 6.
- Geometric mean productivity:
- 7.
- Mean productivity:
- 8.
- Harmonic mean:
- 9.
- Mean Relative Performance:
- 10.
- Yield loss index
2.5. Physiological Traits
2.6. Proline Content
2.7. Total Soluble Sugars (TSS)
2.8. Malondialdehyde (MDA)
2.9. DPPH Radical-Scavenging Activity
2.10. Chlorophyll and Carotenoid Content
2.11. Leaf and Root Na and K Concentrations
2.12. Epigenetic and Gene Expression Analyses
2.13. DNA Methylation Analysis
2.14. RNA Extraction and cDNA Synthesis
2.15. Quantitative Real-Time PCR
2.16. Statistical Analyses
3. Results
3.1. Grain Yield and Related Traits
3.2. Physiological Responses of Introgressed Lines to Salinity
3.2.1. Proline Content in Leaf and Root
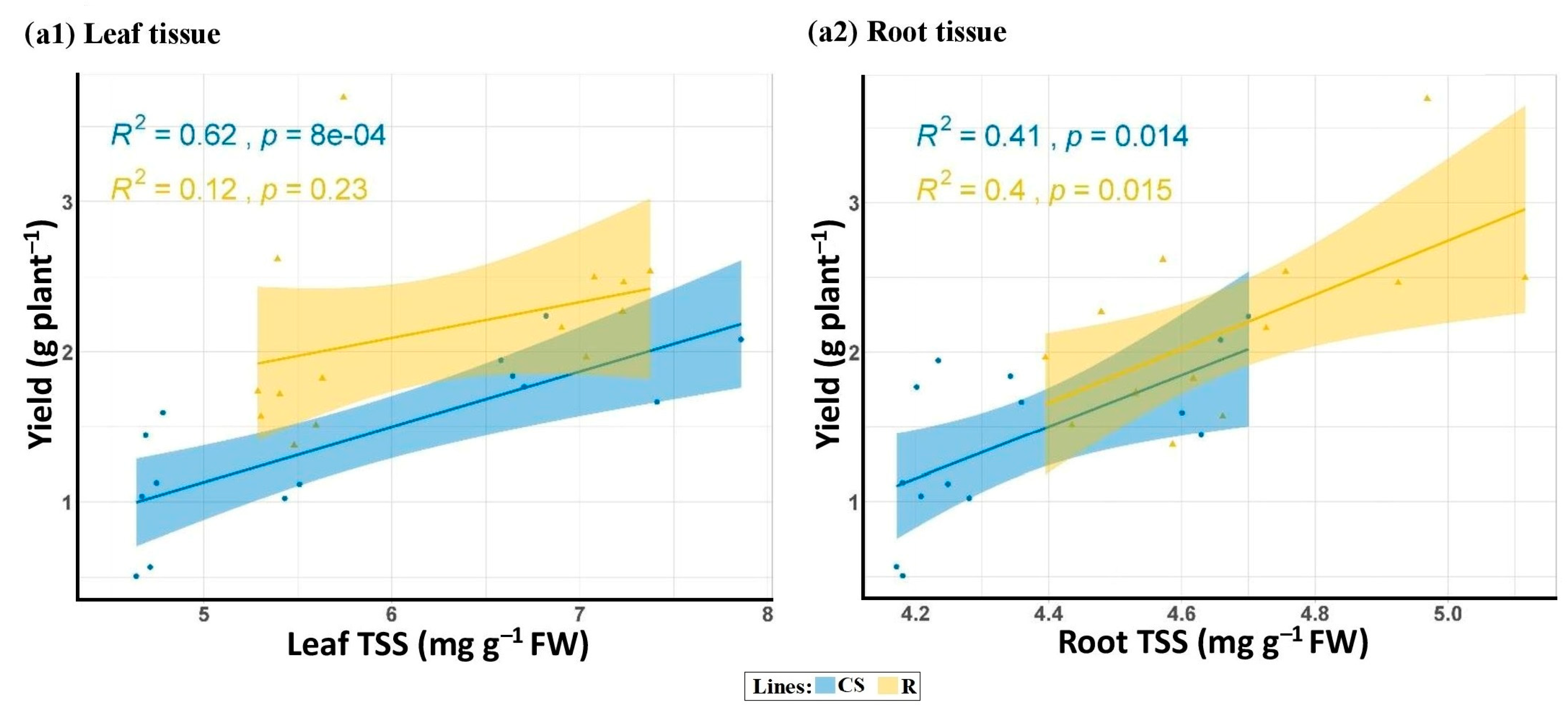
3.2.2. Total Soluble Sugar (TSS) in Leaf and Root
3.2.3. Leaf Relative Water Content (RWC)
3.2.4. Malondialdehyde (MDA)
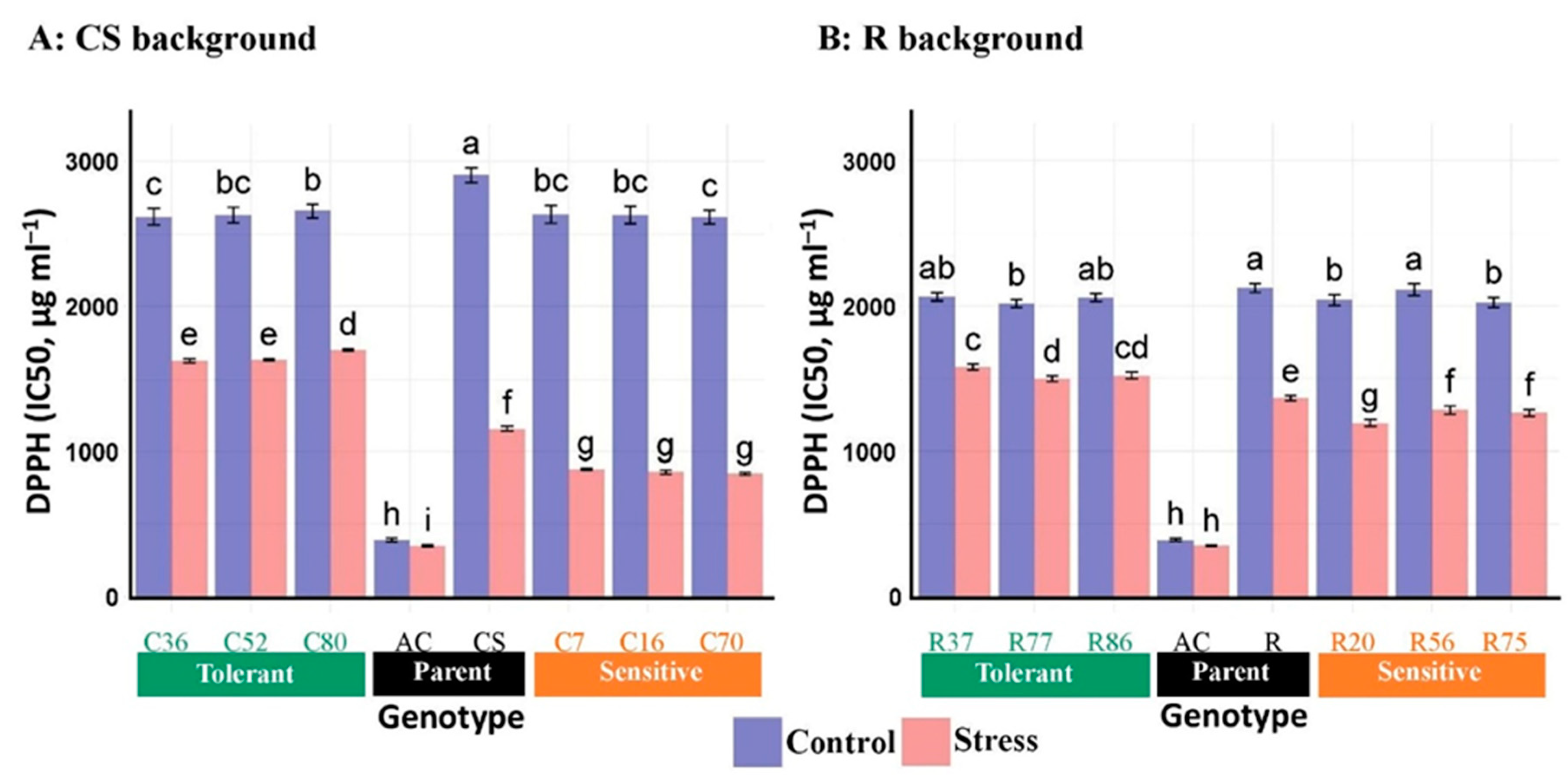
3.2.5. DPPH Radical Scavenging Activity
3.2.6. Chlorophyll and Carotenoid Content
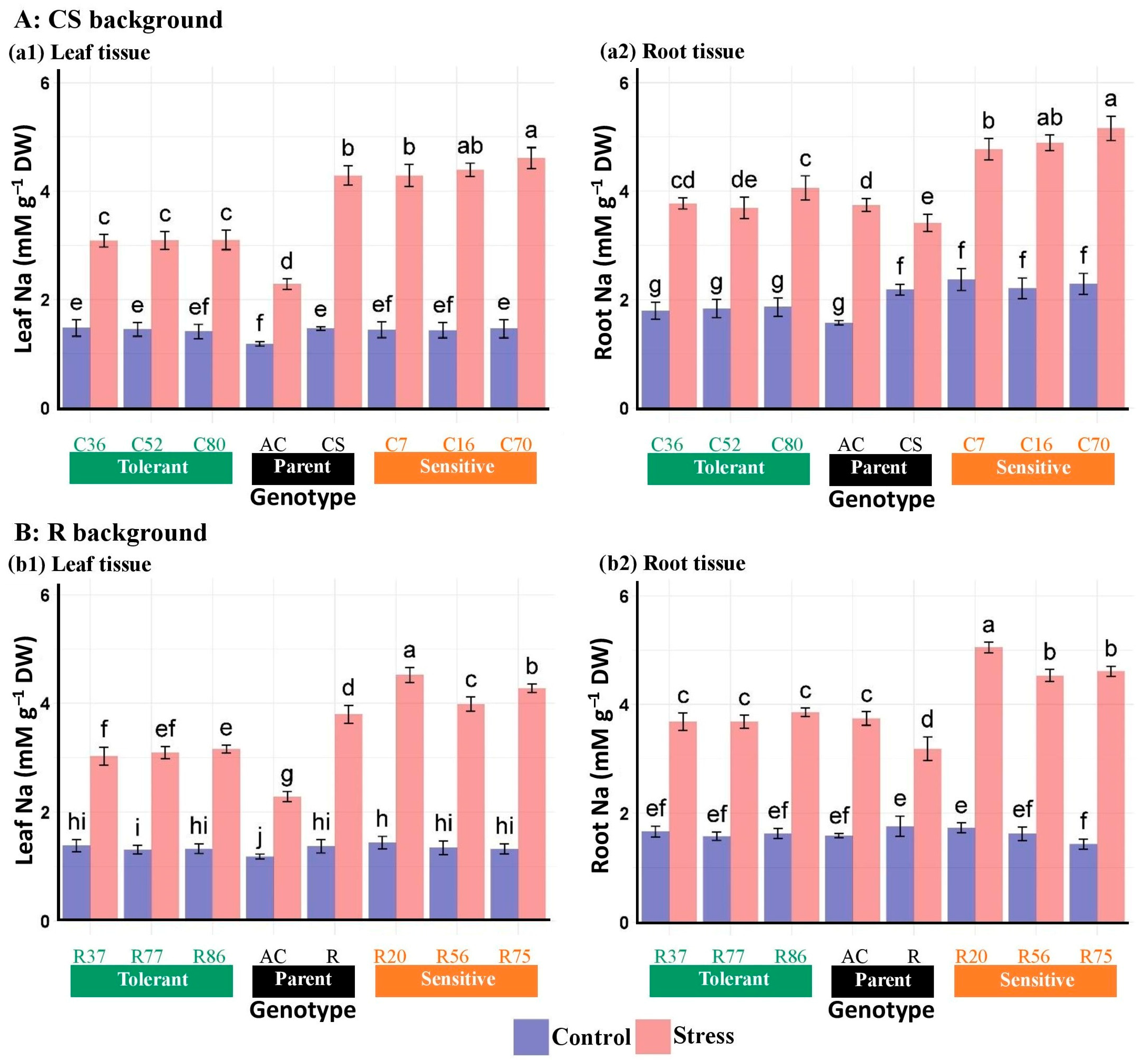
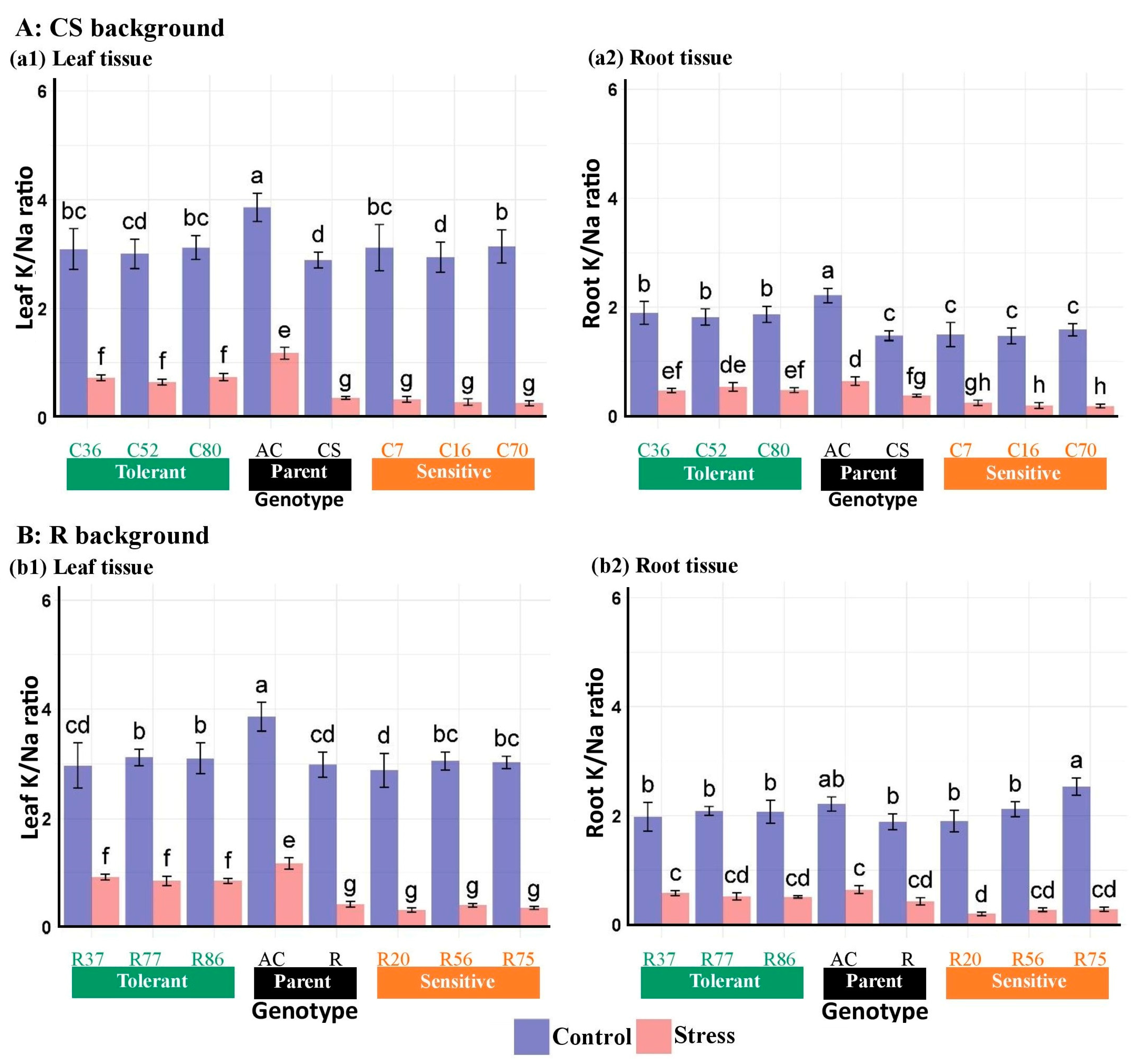
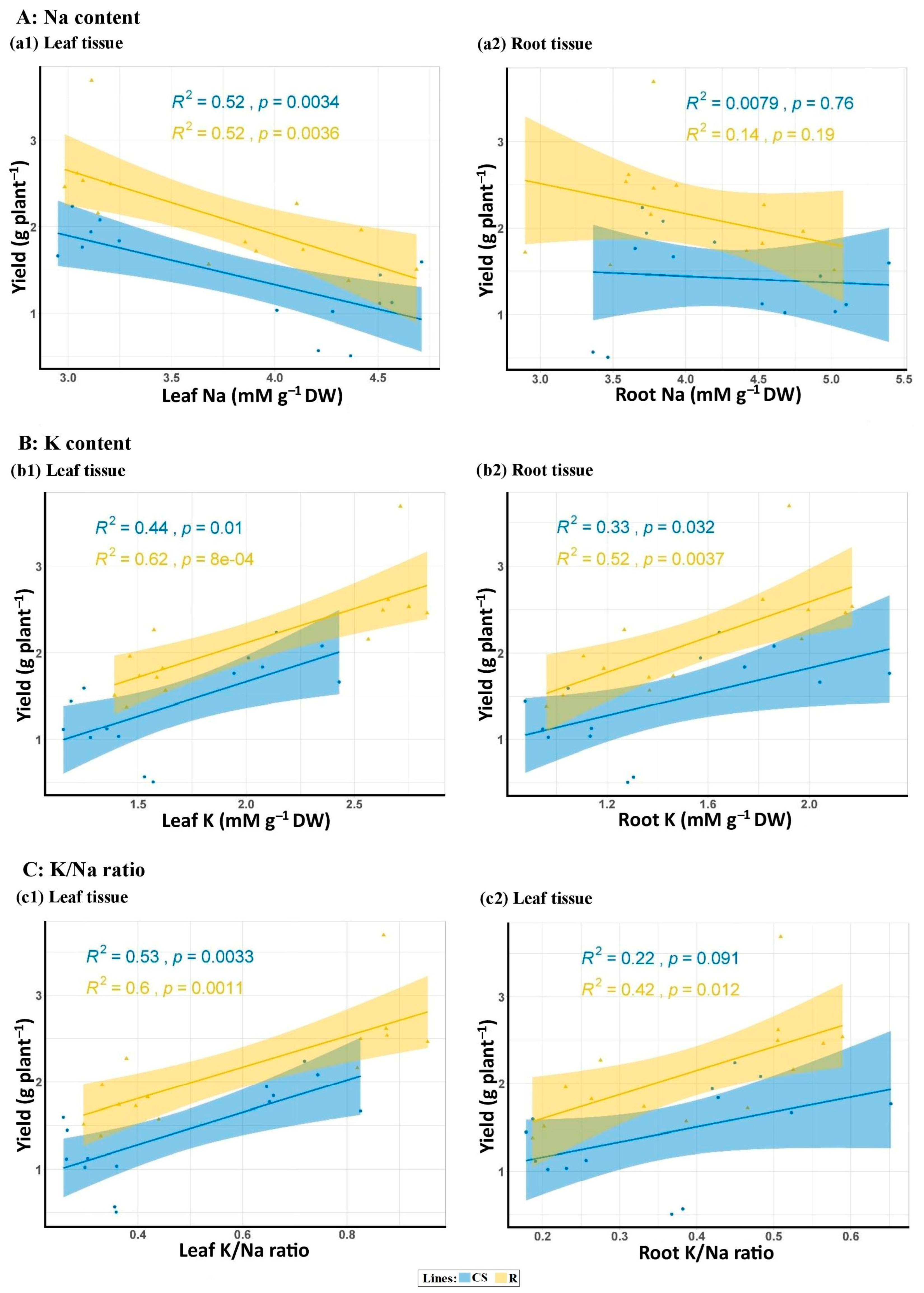
3.2.7. Na and K Concentrations
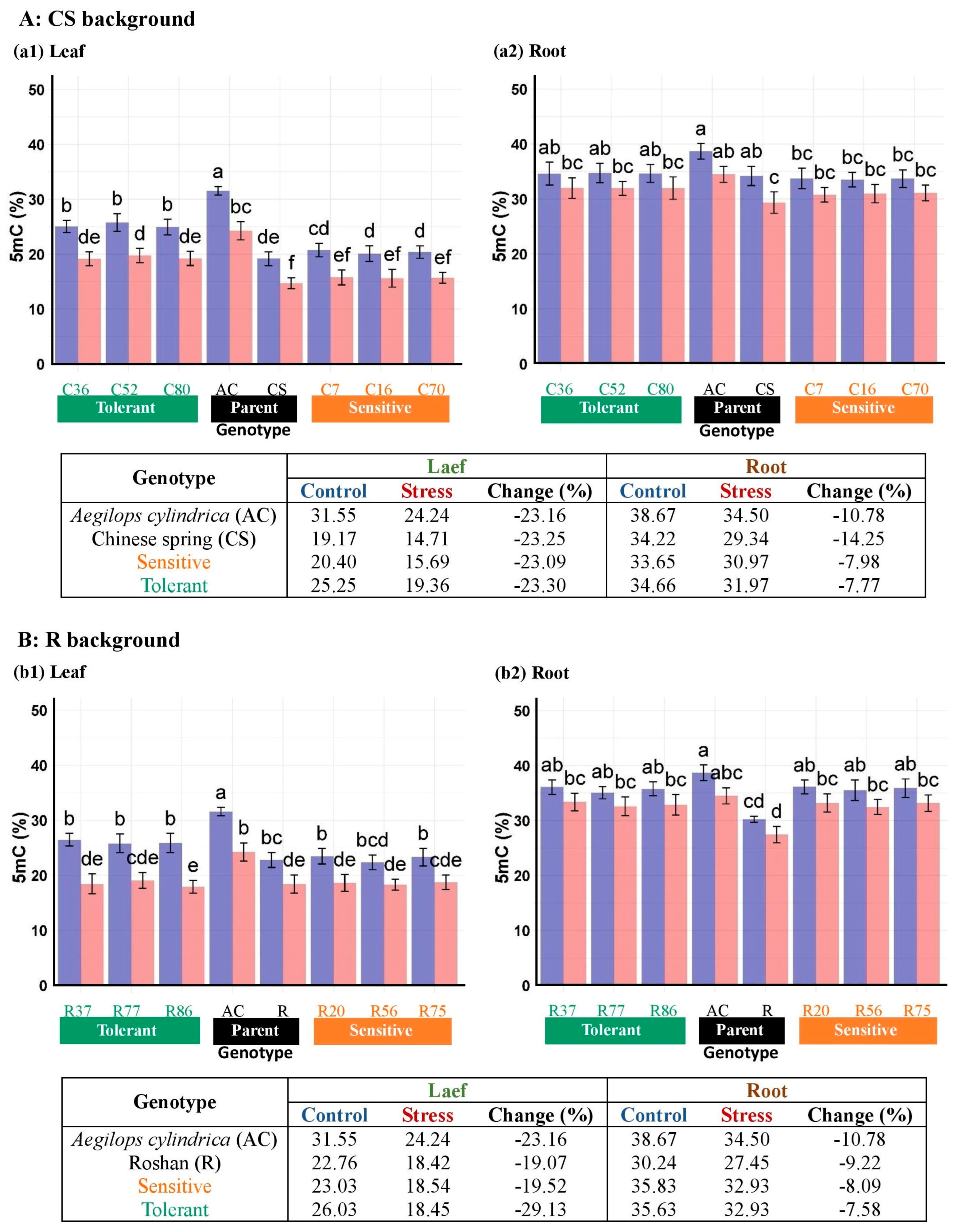
3.3. DNA Methylation Responses of Introgressed Lines to Salinity
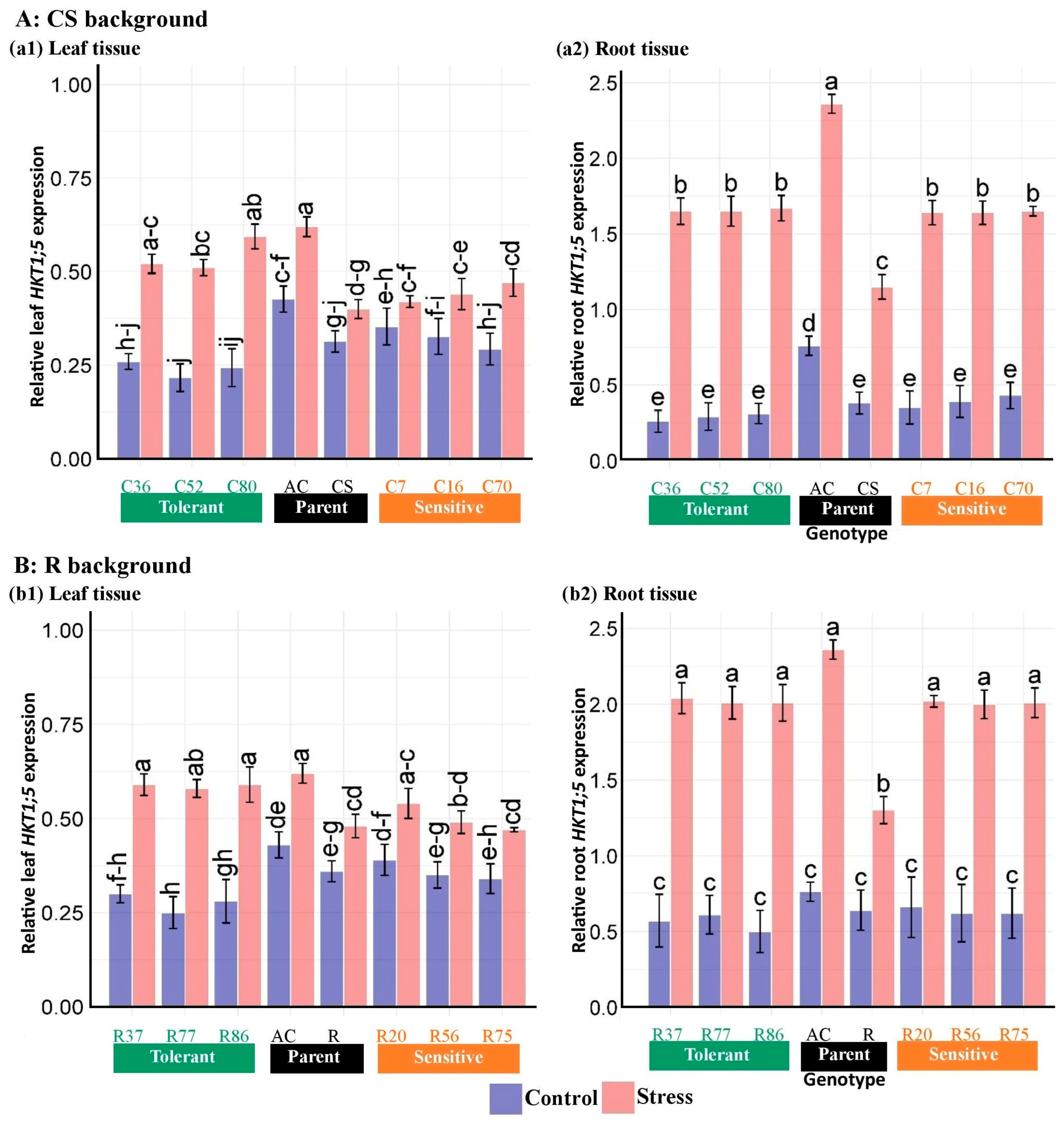
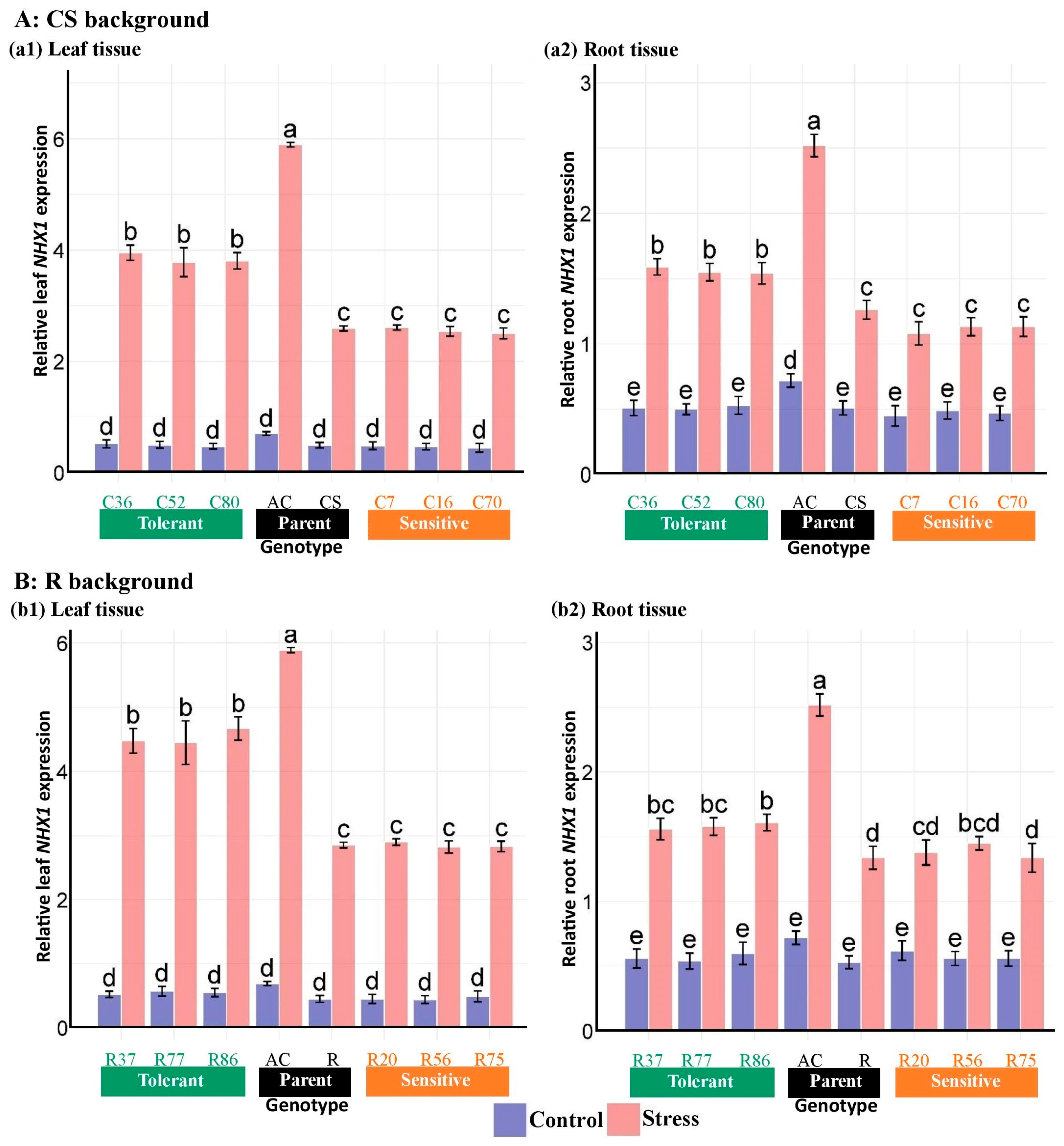
3.4. Expression of Salinity Tolerance Genes in the Introgressed Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Arzani, A.; Ashraf, M. Smart engineering of genetic resources for enhanced salinity tolerance in crop plants. Crit. Rev. Plant Sci. 2016, 35, 146–189. [Google Scholar] [CrossRef]
- Homaee, M.; Feddes, R.; Dirksen, C. A macroscopic water extraction model for nonuniform transient salinity and water stress. Soil Sci. Soc. Am. J. 2002, 66, 1764–1772. [Google Scholar] [CrossRef]
- Munns, R.; Gilliham, M. Salinity tolerance of crops–what is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhou, M.; Shabala, L.; Shabala, S. Linking osmotic adjustment and stomatal characteristics with salinity stress tolerance in contrasting barley accessions. Funct. Plant Biol. 2014, 42, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Ulhassan, Z.; Brestic, M.; Zivcak, M.; Zhou, W.; Allakhverdiev, S.I.; Yang, X.; Safdar, M.E.; Yang, W.; Liu, W. Photosynthesis research under climate change. Photosynth. Res. 2021, 150, 5–19. [Google Scholar] [CrossRef]
- Francois, L.; Maas, E.; Donovan, T.; Youngs, V. Effect of salinity on grain yield and quality, vegetative growth, and germination of semi-dwarf and durum wheat. Agron. J. 1986, 78, 1053–1058. [Google Scholar] [CrossRef]
- Arabbeigi, M.; Arzani, A.; Majidi, M.M. Expression profiles of P5CS and DREB2 genes under salt stress in Aegilops cylindrica. Russ. J. Plant Physiol. 2019, 66, 583–590. [Google Scholar] [CrossRef]
- Huang, S.; Spielmeyer, W.; Lagudah, E.S.; Munns, R. Comparative mapping of HKT genes in wheat, barley, and rice, key determinants of Na+ transport, and salt tolerance. J. Exp. Bot. 2008, 59, 927–937. [Google Scholar] [CrossRef]
- Kiani, R.; Arzani, A.; Mirmohammady Maibody, S. Polyphenols, flavonoids, and antioxidant activity involved in salt tolerance in wheat, Aegilops cylindrica and their amphidiploids. Front. Plant Sci. 2021, 12, 646221. [Google Scholar] [CrossRef] [PubMed]
- Arzani, A.; Ashraf, M. Cultivated ancient wheats (Triticum spp.): A potential source of health-beneficial food products. Compr. Rev. Food Sci. Food Saf. 2017, 16, 477–488. [Google Scholar] [CrossRef]
- Kiani, R.; Arzani, A.; Mirmohammady Maibody, S.; Rahimmalek, M.; Razavi, K. Morpho-physiological and gene expression responses of wheat by Aegilops cylindrica amphidiploids to salt stress. Plant Cell Tissue Organ Cult. 2021, 144, 619–639. [Google Scholar] [CrossRef]
- Singh, P.K.; Gautam, S. Role of salicylic acid on physiological and biochemical mechanism of salinity stress tolerance in plants. Acta Physiol. Plant. 2013, 35, 2345–2353. [Google Scholar] [CrossRef]
- Soundararajan, P.; Manivannan, A.; Jeong, B.R. Different antioxidant defense systems in halophytes and glycophytes to overcome salinity stress. In Sabkha Ecosystems: Volume VI: Asia/Pacific; Springer: Cham, Switzerland, 2019; pp. 335–347. [Google Scholar]
- Liang, W.; Ma, X.; Wan, P.; Liu, L. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Darko, E.; Khalil, R.; Dobi, Z.; Kovács, V.; Szalai, G.; Janda, T.; Molnár, I. Addition of Aegilops biuncialis chromosomes 2M or 3M improves the salt tolerance of wheat in different way. Sci. Rep. 2020, 10, 22327. [Google Scholar] [CrossRef] [PubMed]
- Kiani, R.; Arzani, A.; Habibi, F. Physiology of salinity tolerance in Aegilops cylindrica. Acta Physiol. Plant. 2015, 37, 135. [Google Scholar] [CrossRef]
- Pan, T.; Liu, M.; Kreslavski, V.D.; Zharmukhamedov, S.K.; Nie, C.; Yu, M.; Kuznetsov, V.V.; Allakhverdiev, S.I.; Shabala, S. Non-stomatal limitation of photosynthesis by soil salinity. Crit. Rev. Environ. Sci. Technol. 2021, 51, 791–825. [Google Scholar] [CrossRef]
- Shabala, S. Signalling by potassium: Another second messenger to add to the list? J. Exp. Bot. 2017, 68, 4003–4007. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, X.; Giraldo, J.P.; Shabala, S. It is not all about sodium: Revealing tissue specificity and signalling roles of potassium in plant responses to salt stress. Plant Soil 2018, 431, 1–17. [Google Scholar] [CrossRef]
- Duan, H.-R.; Ma, Q.; Zhang, J.-L.; Hu, J.; Bao, A.-K.; Wei, L.; Wang, Q.; Luan, S.; Wang, S.-M. The inward-rectifying K+ channel SsAKT1 is a candidate involved in K+ uptake in the halophyte Suaeda salsa under saline condition. Plant Soil 2015, 395, 173–187. [Google Scholar] [CrossRef]
- Yan, K.; Shao, H.; Shao, C.; Chen, P.; Zhao, S.; Brestic, M.; Chen, X. Physiological adaptive mechanisms of plants grown in saline soil and implications for sustainable saline agriculture in coastal zone. Acta Physiol. Plant. 2013, 35, 2867–2878. [Google Scholar] [CrossRef]
- Ebrahim, F.; Arzani, A.; Rahimmalek, M.; Sun, D.; Peng, J. Salinity tolerance of wild barley Hordeum vulgare ssp. spontaneum. Plant Breed. 2020, 139, 304–316. [Google Scholar] [CrossRef]
- Rubio, F.; Nieves-Cordones, M.; Horie, T.; Shabala, S. Doing ‘business as usual’comes with a cost: Evaluating energy cost of maintaining plant intracellular K+ homeostasis under saline conditions. New Phytol. 2020, 225, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- Arabbeigi, M.; Arzani, A.; Majidi, M.M.; Kiani, R.; Sayed Tabatabaei, B.E.; Habibi, F. Salinity tolerance of Aegilops cylindrica genotypes collected from hyper-saline shores of Uremia Salt Lake using physiological traits and SSR markers. Acta Physiol. Plant. 2014, 36, 2243–2251. [Google Scholar] [CrossRef]
- Ariño-Estrada, G.; Mitchell, G.S.; Saha, P.; Arzani, A.; Cherry, S.R.; Blumwald, E.; Kyme, A.Z. Imaging salt uptake dynamiCS in plants using PET. Sci. Rep. 2019, 9, 18626. [Google Scholar] [CrossRef]
- Arabbeigi, M.; Arzani, A.; Majidi, M.M.; Sayed-Tabatabaei, B.E.; Saha, P. Expression pattern of salt tolerance-related genes in Aegilops cylindrica. Physiol. Mol. Biol. Plants 2018, 24, 61–73. [Google Scholar] [CrossRef]
- Abdulraheem, M.I.; Xiong, Y.; Moshood, A.Y.; Cadenas-Pliego, G.; Zhang, H.; Hu, J. Mechanisms of plant epigenetic regulation in response to plant stress: Recent discoveries and implications. Plants 2024, 13, 163. [Google Scholar] [CrossRef]
- Hoseini, M.; Arzani, A. Epigenetic adaptation to drought and salinity in crop plants. J. Plant Mol. Breed. 2023, 11, 1–16. [Google Scholar]
- Kumar, S.; Seem, K.; Kumar, S.; Singh, A.; Krishnan, S.G.; Mohapatra, T. DNA methylome analysis provides insights into gene regulatory mechanism for better performance of rice under fluctuating environmental conditions: Epigenomics of adaptive plasticity. Planta 2024, 259, 4. [Google Scholar] [CrossRef]
- Li, B.; Cai, H.; Liu, K.; An, B.; Wang, R.; Yang, F.; Zeng, Z.; Jiao, C.; Xu, Y. DNA methylation alterations and their association with high temperature tolerance in rice anthesis. J. Plant Growth Regul. 2023, 42, 780–794. [Google Scholar] [CrossRef]
- Şahin, Z.; Ağar, G.; Yiğider, E.; Aydın, M. Effects of Selenium on DNA Methylation and Genomic Instability Induced by Drought Stress in Wheat (Triticum aestivum L.). Türkiye Tarımsal Araştırmalar Derg. 2024, 11, 26–37. [Google Scholar] [CrossRef]
- Stadnik, B.; Tobiasz-Salach, R.; Mazurek, M. Effect of silicon on oat salinity tolerance: Analysis of the epigenetic and physiological response of plants. Agriculture 2022, 13, 81. [Google Scholar] [CrossRef]
- Tobiasz-Salach, R.; Mazurek, M.; Jacek, B. Physiological, biochemical, and epigenetic reaction of maize (Zea mays L.) to cultivation in conditions of varying soil salinity and foliar application of silicon. Int. J. Mol. Sci. 2023, 24, 1141. [Google Scholar] [CrossRef] [PubMed]
- Jiabu, D.; Yu, M.; Xu, Q.; Yang, H.; Mu, W.; Basang, Y. Genome-wide DNA methylation dynamics during drought responsiveness in Tibetan hulless barley. J. Plant Growth Regul. 2023, 42, 4391–4401. [Google Scholar] [CrossRef]
- Naderi, S.; Maali-Amiri, R.; Sadeghi, L.; Hamidi, A. Physio-biochemical and DNA methylation analysis of the defense response network of wheat to drought stress. Plant Physiol. Biochem. 2024, 209, 108516. [Google Scholar] [CrossRef]
- Kiani, R.; Arzani, A.; Maibody, S.; Rahimmalek, M.; Ayers, T. Hybridization of wheat and Aegilops cylindrica: Development, karyomorphology, DNA barcoding and salt tolerance of the amphidiploids. J. Plant Biochem. Biotechnol. 2021, 30, 943–959. [Google Scholar] [CrossRef]
- Fernandez, G.C.J. Effective selection criteria for assessing plant stress tolerance. In Adaptation of Food Crops to Temperature and Water Stress; Kuo, C.G., Ed.; AVRDC: Shanhua, Taiwan, 1993; pp. 257–270. [Google Scholar]
- Rosielle, A.; Hamblin, J. Theoretical aspects of selection for yield in stress and non-stress environment 1. Crop Sci. 1981, 21, 943–946. [Google Scholar] [CrossRef]
- Fischer, R.A.; Maurer, R. Drought resistance in spring wheat cultivars. I. Grain yield responses. Aust. J. Agric. Res. 1978, 29, 897–912. [Google Scholar] [CrossRef]
- Bouslama, M.; Schapaugh, W.T., Jr. Stress tolerance in soybeans. I. Evaluation of three screening techniques for heat and drought tolerance 1. Crop Sci. 1984, 24, 933–937. [Google Scholar] [CrossRef]
- Gavuzzi, P.; Rizza, F.; Palumbo, M.; Campanile, R.G.; Ricciardi, G.L.; Borghi, B. Evaluation of field and laboratory predictors of drought and heat tolerance in winter cereals. Can. J. Plant Sci. 1997, 77, 523–531. [Google Scholar] [CrossRef]
- Schneider, K.A.; Rosales-Serna, R.; Ibarra-Perez, F.; Cazares-Enriquez, B.; Acosta-Gallegos, J.A.; Ramirez-Vallejo, P.; Wassimi, N.; Kelly, J.D. Improving common bean performance under drought stress. Crop Sci. 1997, 37, 43–50. [Google Scholar] [CrossRef]
- Hossain, A.B.S.; Sears, A.G.; Cox, T.S.; Paulsen, G.M. Desiccation tolerance and its relationship to assimilate partitioning in winter wheat. Crop Sci. 1990, 30, 622–627. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.; Teare, I. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Irigoyen, J.; Einerich, D.; Sánchez-Díaz, M. Water stress induced changes in concentrations of proline and total soluble sugars in nodulated alfalfa (Medicago sativa) plants. Physiol. Plant. 1992, 84, 55–60. [Google Scholar] [CrossRef]
- Taulavuori, E.; Hellström, E.K.; Taulavuori, K.; Laine, K. Comparison of two methods used to analyse lipid peroxidation from Vaccinium myrtillus (L.) during snow removal, reacclimation and cold acclimation. J. Exp. Bot. 2001, 52, 2375–2380. [Google Scholar] [CrossRef]
- Sharma, O.P.; Bhat, T.K. DPPH antioxidant assay revisited. Food Chem. 2009, 113, 1202–1205. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Buschmann, C. Chlorophylls and carotenoids: Measurement and characterization by UV-VIS spectroscopy. Curr. Protoc. Food Anal. Chem. 2001, 1, F4. 3.1–F4. 3.8. [Google Scholar] [CrossRef]
- Houshmand, S.; Arzani, A.; Maibody SA, M.; Feizi, M. Evaluation of salt-tolerant genotypes of durum wheat derived from in vitro and field experiments. Field Crops Res. 2005, 91, 345–354. [Google Scholar] [CrossRef]
- Kump, B.; Javornik, B. Evaluation of genetic variability among common buckwheat (Fagopyrum esculentum Moench) populations by RAPD markers. Plant Sci. 1996, 114, 149–158. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Arzani, A. Improving salinity tolerance in crop plants: A biotechnological view. In Vitro Cell. Dev. Biol.-Plant 2008, 44, 373–383. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Henry, R.J.; Nevo, E. Exploring natural selection to guide breeding for agriculture. Plant Biotechnol. J. 2014, 12, 655–662. [Google Scholar] [CrossRef]
- Zeibig, F.; Kilian, B.; Frei, M. The grain quality of wheat wild relatives in the evolutionary context. Theor. Appl. Genet. 2022, 135, 4029–4048. [Google Scholar] [CrossRef]
- Gharaghanipor, N.; Arzani, A.; Rahimmalek, M.; Ravash, R. Physiological and transcriptome indicators of salt tolerance in wild and cultivated barley. Front. Plant Sci. 2022, 13, 819282. [Google Scholar] [CrossRef]
- Kumar, S.; Beena, A.; Awana, M.; Singh, A. Physiological, biochemical, epigenetic and molecular analyses of wheat (Triticum aestivum) genotypes with contrasting salt tolerance. Front. Plant Sci. 2017, 8, 280121. [Google Scholar] [CrossRef]
- Romero-Aranda, M.R.; Jurado, O.; Cuartero, J. Silicon alleviates the deleterious salt effect on tomato plant growth by improving plant water status. J. Plant Physiol. 2006, 163, 847–855. [Google Scholar] [CrossRef]
- Radi, A.A.; Farghaly, F.A.; Hamada, A.M. Physiological and biochemical responses of salt-tolerant and salt-sensitive wheat and bean cultivars to salinity. J. Biol. Earth Sci. 2013, 3, 72–88. [Google Scholar]
- Zeeshan, M.; Lu, M.; Sehar, S.; Holford, P.; Wu, F. Comparison of biochemical, anatomical, morphological, and physiological responses to salinity stress in wheat and barley genotypes deferring in salinity tolerance. Agronomy 2020, 10, 127. [Google Scholar] [CrossRef]
- Ehtaiwesh, A.; Sunoj, V.S.J.; Djanaguiraman, M.; Prasad, P.V.V. Response of winter wheat genotypes to salinity stress under controlled environments. Front. Plant Sci. 2024, 15, 1396498. [Google Scholar] [CrossRef]
- Arzani, A. Manipulating programmed cell death pathways for enhancing salinity tolerance in crops. In Salinity Responses and Tolerance in Plants, Volume 2: Exploring RNAi, Genome Editing and Systems Biology; Springer: Cham, Switzerland, 2018; pp. 93–118. [Google Scholar]
- Kaur, G.; Asthir, B.; Bains, N. Modulation of proline metabolism under drought and salt stress conditions in wheat seedlings. Indian J. Biochem. Biophys 2018, 55, 114–124. [Google Scholar]
- Matković Stojšin, M.; Petrović, S.; Banjac, B.; Zečević, V.; Roljević Nikolić, S.; Majstorović, H.; Đorđević, R.; Knežević, D. Assessment of genotype stress tolerance as an effective way to sustain wheat production under salinity stress conditions. Sustainability 2022, 14, 6973. [Google Scholar] [CrossRef]
- Rodrigues, M.J.; Monteiro, I.; Castañeda-Loaiza, V.; Placines, C.; Oliveira, M.C.; Reis, C.; Caperta, A.D.; Soares, F.; Pousão-Ferreira, P.; Pereira, C. Growth performance, in vitro antioxidant properties and chemical composition of the halophyte Limonium algarvense Erben are strongly influenced by the irrigation salinity. Ind. Crops Prod. 2020, 143, 111930. [Google Scholar] [CrossRef]
- Sadak, M.S.; Talaat, I.M. Attenuation of negative effects of saline stress in wheat plant by chitosan and calcium carbonate. Bull. Natl. Res. Cent. 2021, 45, 136. [Google Scholar] [CrossRef]
- Stojšin, M.M.; Petrović, S.; Dimitrijević, M.; Malenčić, D.; Zečević, V.; Banjac, B.; Knežević, D. Effect of salinity stress on antioxidant activity and grain yield of different wheat genotypes. Turk. J. Field Crops 2022, 27, 33–40. [Google Scholar] [CrossRef]
- Saddiq, M.S.; Iqbal, S.; Hafeez, M.B.; Ibrahim, A.M.; Raza, A.; Fatima, E.M.; Baloch, H.; Jahanzaib Woodrow, P.; Ciarmiello, L.F. Effect of salinity stress on physiological changes in winter and spring wheat. Agronomy 2021, 11, 1193. [Google Scholar] [CrossRef]
- Wu, H.; Shabala, L.; Zhou, M.; Su, N.; Wu, Q.; Ul-Haq, T.; Zhu, J.; Mancuso, S.; Azzarello, E.; Shabala, S. Root vacuolar Na+ sequestration but not exclusion from uptake correlates with barley salt tolerance. Plant J. 2019, 100, 55–67. [Google Scholar] [CrossRef]
- van Bezouw, R.F.H.M.; Janssen, E.M.; Ashrafuzzaman, M.; Ghahramanzadeh, R.; Kilian, B.; Graner, A.; Visser, R.G.F.; van der Linden, C.G. Shoot sodium exclusion in salt stressed barley (Hordeum vulgare L.) is determined by allele specific increased expression of HKT1;5. J. Plant Physiol. 2019, 241, 153029. [Google Scholar] [CrossRef]
- Byrt, C.S.; Xu, B.; Krishnan, M.; Lightfoot, D.J.; Athman, A.; Jacobs, A.K.; Watson-Haigh, N.S.; Plett, D.; Munns, R.; Tester, M.; et al. The Na+ transporter, Ta hkt 1; 5-d, limits shoot Na+ accumulation in bread wheat. Plant J. 2014, 80, 516–526. [Google Scholar] [CrossRef]
- Oyiga, B.C.; Sharma, R.C.; Baum, M.; Ogbonnaya, F.C.; Léon, J.; Ballvora, A. Allelic variations and differential expressions detected at quantitative trait loci for salt stress tolerance in wheat. Plant Cell Environ. 2018, 41, 919–935. [Google Scholar] [CrossRef]
- Rus, A.; Lee, B.-H.; Munoz-Mayor, A.; Sharkhuu, A.; Miura, K.; Zhu, J.-K.; Bressan, R.A.; Hasegawa, P.M. AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol. 2004, 136, 2500–2511. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, G.; Shabala, S.; Véry, A.A.; Hariharan, G.N.; Somasundaram, S.; Pulipati, S.; Sellamuthu, G.; Harikrishnan, M.; Kumkum, K.; Shabala, L.; et al. To exclude or to accumulate? Revealing the role of the sodium HKT1;5 transporter in plant adaptive responses to varying soil salinity. Plant Physiol. Biochem. 2021, 169, 333–342. [Google Scholar] [CrossRef]
- Nawaz, I.; Iqbal, M.; Hakvoort, H.W.; Bliek, M.; de Boer, B.; Schat, H. Expression levels and promoter activities of candidate salt tolerance genes in halophytic and glycophytic Brassicaceae. Environ. Exp. Bot. 2014, 99, 59–66. [Google Scholar] [CrossRef]
- Xu, B.; Hrmova, M.; Gilliham, M. High affinity Na+ transport by wheat HKT1;5 is blocked by K+. Plant Direct 2020, 4, e00275. [Google Scholar] [CrossRef] [PubMed]
- Arzani, A.; Kumar, S.; Mansour, M.M.F. Salt tolerance in plants: Molecular and functional adaptations. Front. Plant Sci. 2023, 14, 1280788. [Google Scholar] [CrossRef]
- Ashapkin, V.V.; Kutueva, L.I.; Aleksandrushkina, N.I.; Vanyushin, B.F. Epigenetic mechanisms of plant adaptation to biotic and abiotic stresses. Int. J. Mol. Sci. 2020, 21, 7457. [Google Scholar] [CrossRef]
- Steward, N.; Ito, M.; Yamaguchi, Y.; Koizumi, N.; Sano, H. Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J. Biol. Chem. 2002, 277, 37741–37746. [Google Scholar] [CrossRef]



| Source of Variation | df | Mean Square | |||||
|---|---|---|---|---|---|---|---|
| Grain Yield | Grain Weight | Grain per Spike | Spikes Per Plant | Spike Length | Plant Height | ||
| “Chinese Spring” background | |||||||
| Year (Y) | 1 | 2.77 ** | 1.53 ** | 408.76 ** | 228.32 ** | 1.99 ns | 74,324.90 ** |
| Stress (S) | 1 | 187.41 ** | 78.41 ** | 13,386.65 ** | 31.72 ** | 73.58 ** | 1846.71 ** |
| Genotype (G) | 72 | 2.31 ** | 1.23 ** | 157.43 ** | 22.43 ** | 2.75 ** | 160.18 ** |
| Y × S | 1 | 0.04 ns | 0.43 ns | 456.56 ** | 4.69 ns | 0.09 ns | 62.15 ns |
| Y × G | 72 | 0.03 ns | 0.04 ns | 1.52 ns | 7.7 ** | 0.07 ns | 117.27 ** |
| S × G | 72 | 1.42 ** | 0.63 ** | 78.65 ** | 14.38 ** | 0.77 ns | 12.48 ns |
| Y × S × G | 72 | 0.02 ns | 0.01 ns | 2.99 ns | 9.20 ** | 0.02 ns | 5.48 ns |
| Residual | 576 | 0.08 | 0.14 | 15.01 | 1.59 | 1.57 | 37.75 |
| CV (%) | 20.87 | 17.66 | 17.20 | 37.09 | 16.1 | 8.50 | |
| “Roshan” background | |||||||
| Year (Y) | 1 | 0.06 ns | 5.94 ** | 18.46 ns | 0.83 ns | 1.53 ns | 2211.56 ** |
| Stress (S) | 1 | 448.59 ** | 26.12 ** | 18,897.31 ** | 48.48 ** | 237.16 ** | 99,573.21 ** |
| Genotype (G) | 84 | 2.95 ** | 1.84 ** | 125.26 ** | 3.11 ** | 1.73 ns | 137.00 ** |
| Y × S | 1 | 5.77 ** | 3.00 * | 324.32 ** | 7.07 ** | 17.14 ** | 1.96 ns |
| Y × G | 84 | 0.16 ns | 0.25 ns | 15.05 ns | 0.35 ns | 0.43 ns | 13.99 ns |
| S × G | 84 | 1.76 ** | 1.20 ** | 114.50 ** | 2.18 ** | 1.86 ns | 108.68 ** |
| Y × S × G | 84 | 0.05 ns | 0.15 ns | 9.48 ns | 0.10 ns | 0.86 ns | 28.68 ns |
| Residual | 672 | 0.28 | 0.60 | 44.94 | 0.51 | 1.70 | 46.12 |
| CV (%) | 23.62 | 22.79 | 22.21 | 31.07 | 12.72 | 8.99 | |
| “Chinese Spring” Background | |||||
| Yield | 0.29 *** | 0.39 *** | 0.37 *** | 0.18 *** | 0.32 *** |
| 0.37 *** | GW | 0.20 *** | −0.13 *** | 0.40 *** | 0.38 *** |
| 0.35 *** | 0.50 *** | GpS | −0.15 *** | 0.31 *** | 0.21 *** |
| 0.44 *** | −0.40 *** | −0.49 *** | SpP | −0.13 ** | 0.04 |
| 0.20 *** | 0.57 *** | 0.48 *** | −0.34 *** | SL | 0.57 *** |
| 0.33 *** | 0.60 *** | 0.50 *** | −0.26 *** | 0.72 *** | PH |
| “Roshan” Background | |||||
| Yield | 0.14 ** | 0.08 | 0.72 *** | 0.27 *** | 0.30 *** |
| −0.23 *** | GW | −0.38 *** | −0.15 *** | 0.22 *** | 0.22 *** |
| 0.52 *** | −0.10 * | GpS | −0.34 *** | 0.44 *** | 0.22 *** |
| 0.61 *** | −0.65 *** | −0.09 * | SpP | −0.18 *** | −0.02 |
| 0.48 *** | −0.07 | 0.34 *** | 0.20 *** | SL | 0.64 *** |
| 0.64 *** | −0.15 *** | 0.40 *** | 0.34 *** | 0.64 *** | PH |
| Background | Group | Genotypes | Years | Yn | Ys | YSI | YI | HM | STI | GMP | MRP | MP | TOL | YLI | SSI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chinese Spring (CS) | Sensitive | C7 | 1 | 3.19 | 1.04 | 0.32 | 1.14 | 1.56 | 1.00 | 1.82 | 2.90 | 2.11 | 2.16 | 0.68 | 1.35 |
| 2 | 3.46 | 1.12 | 0.32 | 1.12 | 1.70 | 1.03 | 1.97 | 2.90 | 2.29 | 2.34 | 0.68 | 1.40 | |||
| C16 | 1 | 3.65 | 1.02 | 0.28 | 1.13 | 1.60 | 1.13 | 1.93 | 3.14 | 2.34 | 2.63 | 0.72 | 1.44 | ||
| 2 | 4.06 | 1.12 | 0.28 | 1.11 | 1.75 | 1.20 | 2.13 | 3.20 | 2.59 | 2.94 | 0.72 | 1.50 | |||
| C70 | 1 | 3.91 | 1.44 | 0.37 | 1.60 | 2.11 | 1.71 | 2.38 | 3.75 | 2.68 | 2.47 | 0.63 | 1.26 | ||
| 2 | 4.13 | 1.59 | 0.39 | 1.59 | 2.30 | 1.74 | 2.57 | 3.72 | 2.86 | 2.54 | 0.61 | 1.27 | |||
| Recurrent parent | Chinese spring | 1 | 2.15 | 0.51 | 0.24 | 0.56 | 0.82 | 0.33 | 1.05 | 1.75 | 1.33 | 1.64 | 0.76 | 1.53 | |
| 2 | 2.43 | 0.57 | 0.23 | 0.57 | 0.92 | 0.37 | 1.17 | 1.82 | 1.50 | 1.86 | 0.77 | 1.59 | |||
| Tolerant | C36 | 1 | 2.39 | 2.24 | 0.94 | 2.47 | 2.31 | 1.62 | 2.31 | 3.79 | 2.31 | 0.15 | 0.06 | 0.12 | |
| 2 | 2.58 | 2.08 | 0.81 | 2.07 | 2.30 | 1.42 | 2.32 | 3.40 | 2.33 | 0.50 | 0.19 | 0.40 | |||
| C52 | 1 | 2.12 | 1.94 | 0.92 | 2.15 | 2.03 | 1.25 | 2.03 | 3.31 | 2.03 | 0.17 | 0.08 | 0.16 | ||
| 2 | 2.31 | 1.77 | 0.76 | 1.76 | 2.00 | 1.08 | 2.02 | 2.95 | 2.04 | 0.54 | 0.24 | 0.49 | |||
| C80 | 1 | 1.94 | 1.66 | 0.86 | 1.84 | 1.79 | 0.98 | 1.79 | 2.90 | 1.80 | 0.27 | 0.14 | 0.28 | ||
| 2 | 2.23 | 1.84 | 0.82 | 1.83 | 2.02 | 1.09 | 2.03 | 2.98 | 2.04 | 0.39 | 0.18 | 0.37 | |||
| Roshan (R) | Sensitive | R20 | 1 | 5.50 | 1.38 | 0.25 | 0.91 | 2.20 | 0.85 | 2.75 | 2.76 | 3.44 | 4.13 | 0.75 | 1.51 |
| 2 | 5.58 | 1.51 | 0.27 | 0.92 | 2.38 | 1.06 | 2.90 | 2.90 | 3.55 | 4.07 | 0.73 | 1.75 | |||
| R56 | 1 | 4.59 | 1.82 | 0.40 | 1.21 | 2.61 | 0.94 | 2.89 | 2.75 | 3.21 | 2.77 | 0.60 | 1.22 | ||
| 2 | 4.43 | 2.27 | 0.51 | 1.38 | 3.00 | 1.27 | 3.17 | 2.95 | 3.35 | 2.16 | 0.49 | 1.17 | |||
| R75 | 1 | 4.87 | 1.73 | 0.36 | 1.15 | 2.56 | 0.95 | 2.91 | 2.78 | 3.30 | 3.13 | 0.64 | 1.30 | ||
| 2 | 4.94 | 1.96 | 0.40 | 1.20 | 2.81 | 1.22 | 3.11 | 2.95 | 3.45 | 2.97 | 0.60 | 1.44 | |||
| Recurrent parent | Roshan | 1 | 3.49 | 1.57 | 0.45 | 1.04 | 2.16 | 0.62 | 2.34 | 2.21 | 2.53 | 1.93 | 0.55 | 1.11 | |
| 2 | 3.25 | 1.72 | 0.53 | 1.05 | 2.25 | 0.70 | 2.36 | 2.20 | 2.48 | 1.53 | 0.47 | 1.13 | |||
| Tolerant | R37 | 1 | 2.94 | 2.46 | 0.84 | 1.64 | 2.68 | 0.81 | 2.69 | 2.62 | 2.70 | 0.48 | 0.16 | 0.33 | |
| 2 | 2.59 | 2.54 | 0.98 | 1.55 | 2.56 | 0.83 | 2.56 | 2.47 | 2.56 | 0.05 | 0.02 | 0.05 | |||
| R77 | 1 | 3.16 | 2.62 | 0.83 | 1.74 | 2.86 | 0.93 | 2.87 | 2.80 | 2.89 | 0.54 | 0.17 | 0.35 | ||
| 2 | 2.93 | 2.16 | 0.74 | 1.32 | 2.49 | 0.80 | 2.52 | 2.36 | 2.55 | 0.78 | 0.26 | 0.63 | |||
| R86 | 1 | 4.36 | 3.69 | 0.85 | 2.45 | 4.00 | 1.81 | 4.01 | 3.92 | 4.03 | 0.67 | 0.15 | 0.31 | ||
| 2 | 3.20 | 2.50 | 0.78 | 1.52 | 2.80 | 1.01 | 2.83 | 2.66 | 2.85 | 0.71 | 0.22 | 0.53 |
| Source of Variation | df | Mean Square | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proline (Leaf) | Proline (Root) | TSS (Leaf) | TSS (Root) | RWC (Leaf) | MDA (Leaf) | DPPH (Leaf) | Chla | Chlb | Total chl | Car | Na (Leaf) | Na (Root) | K (Leaf) | K (Root) | K/Na (Leaf) | K/Na (Root) | ||
| “Chinese Spring” background | ||||||||||||||||||
| Year (Y) | 1 | 1.36 ns | 0.53 ns | 0.02 ns | 0.10 ns | 0.15 ns | 0.14 ns | 5310.82 ns | 0.03 ns | 0.003 ns | 0.01 ns | 1.33 ns | 0.30 ns | 0.53 ns | 0.44 ns | 0.02 ns | 0.99 ns | 0.19 ns |
| Stress (S) | 1 | 910.93 * | 100.99 * | 130.26 * | 84.97 * | 14,095.71 * | 541.9 * | 37,755,458.59 * | 1.01 * | 1.38 ** | 0.03 ns | 5.33 * | 119.91 * | 113.02 * | 143.47 * | 77.69 * | 160.16 * | 42.91 * |
| Genotype (G) | 7 | 76.1 ** | 5.15 ** | 23.97 ** | 19.84 ** | 383.18 ** | 11.21 ** | 4,195,117.78 ** | 0.027 ** | 0.036 ** | 0.12 ** | 1.25 ** | 2.52 ** | 2.27 ** | 1.22 ** | 0.96 ** | 1.03 ** | 0.54 ** |
| Y × S | 1 | 0.74 ns | 0.19 ns | 0.19 ns | 0.17 ns | 82.80 ns | 0.61 ns | 33,227.79 ns | 0.005 ns | 0.00 ns | 0.00 ns | 0.02 ns | 0.04 ns | 0.07 ns | 0.14 ns | 0.45 ns | 0.67 ns | 0.29 ns |
| Y × G | 7 | 0.04 ns | 0.01 ns | 0.18 ns | 0.25 ns | 26.02 ns | 0.09 ns | 66.03 ns | 0.00 ns | 0.00 ns | 0.00 ns | 0.00 ns | 0.04 ns | 0.06 ns | 0.02 ns | 0.11 ns | 0.01 ns | 0.02 ns |
| S × G | 7 | 50.18 ** | 1.22 ** | 3.58 ** | 1.76 ** | 225.18 ** | 13.89 ** | 1,163,351.31 ** | 0.01 ** | 0.001 * | 0.02 ** | 0.58 ** | 1.89 ** | 0.79 ** | 0.67 ** | 0.80 ** | 0.13 ** | 0.06 * |
| Y × S × G | 7 | 0.05 ns | 0.01 ns | 0.18 ns | 0.23 ns | 25.95 ns | 0.17 ns | 814.53 ns | 0.00 ns | 0.00 ns | 0.00 ns | 0.01 ns | 0.036 ns | 0.04 ns | 0.02 ns | 0.06 ns | 0.01 ns | 0.01 ns |
| Residual | 56 | 0.73 | 0.65 | 0.31 | 0.36 | 13.09 | 0.78 | 9786.05 | 0.03 | 0.01 | 0.04 | 0.42 | 0.16 | 0.21 | 0.34 | 0.34 | 0.34 | 0.10 |
| CV (%) | 13 | 18.48 | 11.06 | 15.33 | 5.21 | 14.81 | 5.62 | 14.62 | 20.49 | 11.52 | 6.4 | 15.85 | 14.85 | 19.33 | 24.03 | 31.60 | 29.86 | |
| “Roshan” background | ||||||||||||||||||
| Year (Y) | 1 | 0.44 ns | 0.09 ns | 0.06 ns | 0.49 ns | 206.08 ns | 1.49 ns | 19,993.05 ns | 0.07 ns | 0.00 ns | 0.04 ns | 1.18 ns | 0.40 ns | 0.09 ns | 0.32 ns | 0.00 ns | 0.83 ns | 0.19 ns |
| Stress (S) | 1 | 491.53 * | 33.81 * | 140.72 * | 75.4 ** | 16,432.48 * | 699.58 * | 8,533,011.28 * | 1.81 * | 1.27 * | 0.04 ns | 7.53 * | 114.75 * | 140.53 * | 90.44 * | 68.84 ** | 145.99 * | 67.48 * |
| Genotype (G) | 7 | 85.77 ** | 5.53 ** | 17.94 ** | 14.67 ** | 339.34 ** | 12.71 ** | 2,809,371 ** | 0.01 ** | 0.03 ** | 0.03 ** | 0.63 ** | 1.95 ** | 1.09 ** | 1.50 ** | 0.79 * | 1.03 ** | 0.19 ns |
| Y × S | 1 | 0.31 ns | 0.06 ns | 0.17 ns | 0.007 ns | 24.22 ns | 0.80 ns | 24,740.43 ns | 0.00 ns | 0.00 ns | 0.01 ns | 0.03 ns | 0.05 ns | 0.21 ns | 0.06 ns | 0.01 ns | 0.47 ns | 0.22 ns |
| Y × G | 7 | 0.02 ns | 0.07 ns | 1.04 ns | 0.88 ns | 25.82 ns | 0.05 ns | 443.90 ns | 0.00 ns | 0.00 ns | 0.00 ns | 0.01 ns | 0.01 ns | 0.12 ns | 0.00 ns | 0.12 ns | 0.00 ns | 0.11 ns |
| S × G | 7 | 63.36 ** | 1.23 ** | 1.12 ns | 0.71 ns | 261.04 ** | 12.78 ** | 215,121.07 ** | 0.00 ns | 0.01 ** | 0.02 ** | 0.54 ** | 1.46 ** | 1.27 ** | 1.06 ** | 0.86 ** | 0.17 ** | 0.23 * |
| Y × S × G | 7 | 0.01 ns | 0.02 ns | 1.02 ** | 0.75 ns | 28.40 ns | 0.05 ns | 2515.14 ns | 0.00 ns | 0.00 ns | 0.00 ns | 0.02 ns | 0.01 ns | 0.03 ns | 0.00 ns | 0.04 ns | 0.00 ns | 0.07 ns |
| Residual | 56 | 0.34 | 0.43 | 0.26 | 0.36 | 20.33 | 0.20 | 4540.07 | 0.02 | 0.006 | 0.02 | 0.61 | 0.09 | 0.09 | 0.30 | 0.37 | 0.25 | 0.10 |
| CV (%) | 9.58 | 15.51 | 9.60 | 14.25 | 6.41 | 6.43 | 4.33 | 10.30 | 15.07 | 8.61 | 7.51 | 12.73 | 10.59 | 17.78 | 24.28 | 26.50 | 25.35 | |
| Trait | Ae. cylindrica | CS Derived Lines | R Derived Lines | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Control (0 mM NaCl) | Stress (250 mM NaCl) | Change (%) | Control (0 mM NaCl) | Stress (250 mM NaCl) | Change (%) | Control (0 mM NaCl) | Stress (250 mM NaCl) | Change (%) | |
| Leaf Proline | 4.81 b | 20.63 a | 328.85 | 3.33 b | 8.12 a | 143.38 | 3.71 b | 6.63 a | 78.39 |
| Root Proline | 4.51 b | 7.24 a | 60.53 | 3.18 b | 5.13 a | 61.51 | 3.54 b | 4.51 a | 27.29 |
| Leaf TSS | 7.35 b | 9.17 a | 24.74 | 3.40 b | 5.80 a | 70.74 | 3.68 b | 6.19 a | 68.07 |
| Root TSS | 5.62 a | 8.22 a | 46.33 | 2.58 b | 4.36 a | 68.98 | 3.01 b | 4.66 a | 54.94 |
| RWC | 86.96 a | 75.41 a | −13.29 | 80.70 a | 54.65 b | −32.28 | 82.86 a | 54.60 b | −34.10 |
| MDA | 4.01 b | 5.62 a | 40.18 | 3.54 b | 8.74 a | 146.93 | 4.27 b | 10.21 a | 138.97 |
| DPPH | 389.80 a | 350.10 b | −10.18 | 2670.70 a | 1242.94 b | −53.46 | 2062.30 a | 1386.51 b | −32.77 |
| Chla | 1.13 b | 1.45 a | 27.92 | 1.10 b | 1.29 a | 17.33 | 1.21 b | 1.48 a | 22.15 |
| Chlb | 0.73 a | 0.53 b | −28.38 | 0.63 a | 0.38 b | −39.06 | 0.63 a | 0.40 b | −36.84 |
| Total chl | 1.87 b | 1.97 a | 5.76 | 1.72 a | 1.67 a | −3.16 | 1.85 a | 1.88 a | 1.89 |
| Car | 10.23 a | 11.61 a | 13.55 | 9.92 b | 10.26 a | 3.44 | 10.14 b | 10.58 a | 4.36 |
| Leaf Na | 1.18 b | 2.29 a | 93.82 | 1.44 b | 3.84 a | 166.44 | 1.35 b | 3.69 a | 173.06 |
| Root Na | 1.58 b | 3.74 a | 136.28 | 2.08 b | 4.25 a | 104.42 | 1.63 b | 4.09 a | 150.81 |
| Leaf K | 4.50 a | 2.66 b | −40.88 | 4.22 a | 1.69 b | −59.94 | 3.98 a | 2.03 b | −49.13 |
| Root K | 3.49 a | 2.38 b | −31.85 | 3.32 a | 1.42 b | −57.20 | 3.33 a | 1.55 b | −53.33 |
| Leaf K/Na | 3.86 a | 1.17 b | −69.66 | 3.05 a | 0.48 b | −84.28 | 3.02 a | 0.58 b | −80.66 |
| Root K/Na | 2.22 a | 0.64 b | −71.07 | 1.66 a | 0.35 b | −78.64 | 2.09 a | 0.40 b | −81.04 |
| Source of Variation | df | Mean Square | |||
|---|---|---|---|---|---|
| 5-mC | HKT1;5 | NHX1 | SOS1 | ||
| CS background | |||||
| Salt stress (S) | 1 | 442.75 ** | 13.00 ** | 91.66 ** | 6.11 ** |
| Genotype (G) | 7 | 79.60 ** | 0.23 ** | 2.41 ** | 0.27 ** |
| Tissue (T) | 1 | 3703.16 ** | 9.72 ** | 23.21 ** | 0.65 ** |
| S × G | 7 | 1.46 ns | 0.06 ** | 1.67 ** | 0.08 ** |
| S × T | 1 | 31.61 * | 7.09 ** | 24.01 ** | 0.15 ** |
| G × T | 7 | 15.58 * | 0.09 ** | 0.37 ** | 0.01 ns |
| S × G × T | 7 | 1.15 ns | 0.03 * | 0.38 ** | 0.005 ns |
| Residual | 64 | 6.73 | 0.01 | 0.02 | 0.02 |
| CV (%) | 9.63 | 15.08 | 9.85 | 17.81 | |
| R background | |||||
| Salt stress (S) | 1 | 481.19 ** | 14.55 ** | 113.58 ** | 8.41 ** |
| Genotype (G) | 7 | 51.17 ** | 0.10 ** | 2.03 ** | 0.31 ** |
| Tissue (T) | 1 | 3303.29 ** | 17.47 ** | 28.99 ** | 0.49 ** |
| S × G | 7 | 2.60 ns | 0.09 ** | 1.45 ** | 0.16 ** |
| S × T | 1 | 54.58 ** | 7.77 ** | 32.50 ** | 0.20 ** |
| G × T | 7 | 14.02 * | 0.06 * | 0.61 ** | 0.01 ns |
| S × G × T | 7 | 2.12 ns | 0.05 ns | 0.49 ** | 0.005 ns |
| Residual | 64 | 6.39 | 0.02 | 0.03 | 0.02 |
| CV (%) | 9.01 | 18.66 | 10.67 | 20.11 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoseini, M.; Arzani, A.; Saeidi, G.; Araniti, F. Agro-Physiological and DNA Methylation Responses to Salinity Stress in Wheat (Triticum aestivum L.), Aegilops cylindrica Host, and Their Introgressed Lines. Plants 2024, 13, 2673. https://doi.org/10.3390/plants13192673
Hoseini M, Arzani A, Saeidi G, Araniti F. Agro-Physiological and DNA Methylation Responses to Salinity Stress in Wheat (Triticum aestivum L.), Aegilops cylindrica Host, and Their Introgressed Lines. Plants. 2024; 13(19):2673. https://doi.org/10.3390/plants13192673
Chicago/Turabian StyleHoseini, Mohsen, Ahmad Arzani, Ghodratollah Saeidi, and Fabrizio Araniti. 2024. "Agro-Physiological and DNA Methylation Responses to Salinity Stress in Wheat (Triticum aestivum L.), Aegilops cylindrica Host, and Their Introgressed Lines" Plants 13, no. 19: 2673. https://doi.org/10.3390/plants13192673
APA StyleHoseini, M., Arzani, A., Saeidi, G., & Araniti, F. (2024). Agro-Physiological and DNA Methylation Responses to Salinity Stress in Wheat (Triticum aestivum L.), Aegilops cylindrica Host, and Their Introgressed Lines. Plants, 13(19), 2673. https://doi.org/10.3390/plants13192673







