Landscape Heterogeneity Drives Genetic Diversity in the Highly Dispersive Moss Funaria hygrometrica Hedw.
Abstract
1. Introduction
2. Results
2.1. Genetic Variability, DNA Polymorphism, Haplotype Frequency, and Geographical Distribution
2.1.1. Nuclear ITS1
2.1.2. Nuclear ITS2
2.1.3. Chloroplast rps3-rpl16
2.1.4. Mitochondrial rpl5-rpl16
2.2. Genotypic Diversity
2.3. Geographical vs. Altitudinal Distribution of Haplotype Groups
2.4. Genetic Differentiation, Migration, and Linkage Disequilibrium
2.4.1. Genetic Differentiation and Population Diversity
2.4.2. Migration and Genetic Diversity
2.4.3. Linkage Disequilibrium Analysis
2.5. Landscape Heterogeneity in Sierra Nevada Mountains
3. Discussion
4. Materials and Methods
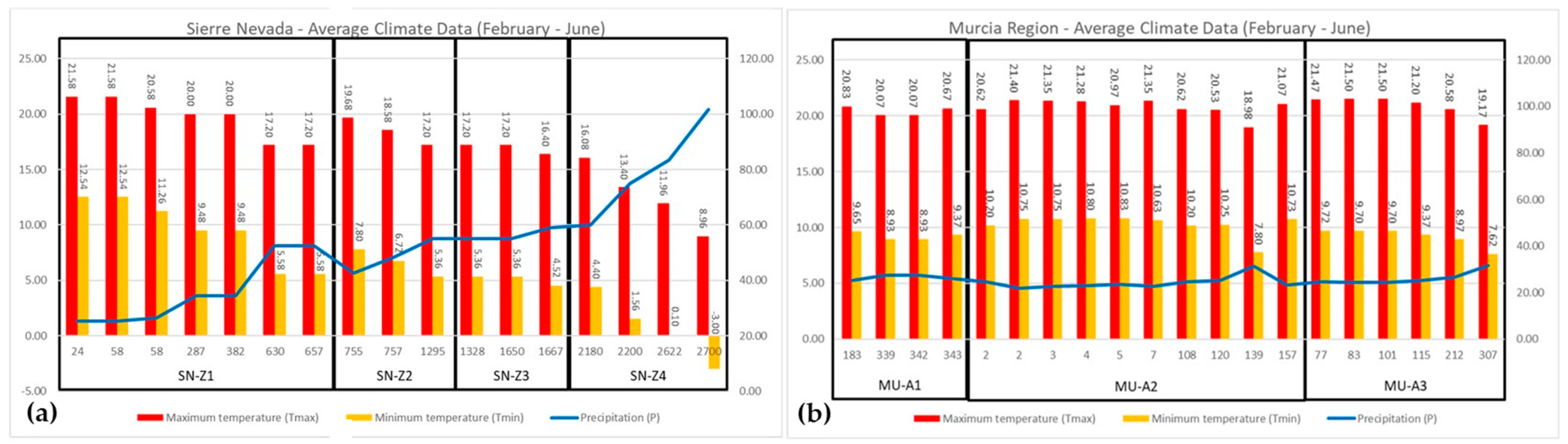
4.1. Sampling Regions, Plant Material, and Environmental Conditions
4.2. In Vitro Cultivation, DNA Extraction, and PCR Amplification
4.3. Genetic Variation, Diversity, and DNA Polymorphism Analyses
4.4. Genetic Differentiation and Migration
4.5. Landscape vs. Genetic Diversity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Medina, N.G.; Draper, I.; Lara, F. Biogeography of Mosses and Allies: Does Size Matter? In Biogeography of Microscopic Organisms; Fontaneto, D., Ed.; Cambridge University Press: Cambridge, UK, 2011; pp. 209–233. ISBN 978-0-521-76670-8. [Google Scholar]
- Patiño, J.; Bisang, I.; Goffinet, B.; Hedenäs, L.; McDaniel, S.; Pressel, S.; Stech, M.; Ah-Peng, C.; Bergamini, A.; Caners, R.T.; et al. Unveiling the Nature of a Miniature World: A Horizon Scan of Fundamental Questions in Bryology. J. Bryol. 2022, 44, 1–34. [Google Scholar] [CrossRef]
- Frahm, J.-P. Diversity, Dispersal and Biogeography of Bryophytes (Mosses). Biodivers Conserv 2008, 17, 277–284. [Google Scholar] [CrossRef]
- Parihar, N.S. An Introduction to Embryophyta Volume I: Bryophyta; Central Book Depot: Allahabad, India, 1962. [Google Scholar]
- Muñoz, J.; Felicísimo, Á.M.; Cabezas, F.; Burgaz, A.R.; Martínez, I. Wind as a Long-Distance Dispersal Vehicle in the Southern Hemisphere. Science 2004, 304, 1144–1147. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.J.; Szövényi, P.; Shaw, B. Bryophyte Diversity and Evolution: Windows into the Early Evolution of Land Plants. Am. J Bot. 2011, 98, 352–369. [Google Scholar] [CrossRef]
- Gibby, M. Bryophytes and Pteridophytes: Spore-Bearing Land Plants. In The Living Planet; Maclean, N., Ed.; Cambridge University Press: Cambridge, UK, 2023; pp. 37–64. ISBN 978-1-108-75882-6. [Google Scholar]
- Zanatta, F.; Engler, R.; Collart, F.; Broennimann, O.; Mateo, R.G.; Papp, B.; Muñoz, J.; Baurain, D.; Guisan, A.; Vanderpoorten, A. Bryophytes Are Predicted to Lag behind Future Climate Change despite Their High Dispersal Capacities. Nat. Commun. 2020, 11, 5601. [Google Scholar] [CrossRef] [PubMed]
- Biersma, E.M.; Convey, P.; Wyber, R.; Robinson, S.A.; Dowton, M.; Van de Vijver, B.; Linse, K.; Jackson, J.A. Latitudinal Biogeographic Structuring in the Globally Distributed Moss Ceratodon purpureus. Front. Plant Sci. 2020, 11, 502359. [Google Scholar] [CrossRef]
- Shaw, A.J.; Werner, O.; Ros, R.M. Intercontinental Mediterranean Disjunct Mosses: Morphological and Molecular Patterns. Am. J. Bot. 2003, 90, 540–550. [Google Scholar] [CrossRef]
- McDaniel, S.F.; Shaw, A.J. Selective Sweeps and Intercontinental Migration in the Cosmopolitan Moss Ceratodon purpureus (Hedw.) Brid. Mol. Ecol. 2005, 14, 1121–1132. [Google Scholar] [CrossRef]
- Pisa, S.; Biersma, E.M.; Convey, P.; Patiño, J.; Vanderpoorten, A.; Werner, O.; Ros, R.M. The Cosmopolitan Moss Bryum argenteum in Antarctica: Recent Colonisation or in Situ Survival? Polar Biol 2014, 37, 1469–1477. [Google Scholar] [CrossRef]
- Shaw, A.J.; Albright, D.L. Potential for the Evolution of Heavy Metal Tolerance in Bryum argenteum, a Moss. II. Generalized Tolerances among Diverse Populations. Bryologist 1990, 93, 187. [Google Scholar] [CrossRef]
- Reynolds, L.A.; Nicholas McLetchie, D. Short Distances between Extreme Microhabitats Do Not Result in Ecotypes in Syntrichia caninervis. J. Bryol. 2011, 33, 148–153. [Google Scholar] [CrossRef]
- Pisa, S.; Werner, O.; Vanderpoorten, A.; Magdy, M.; Ros, R.M. Elevational Patterns of Genetic Variation in the Cosmopolitan Moss Bryum argenteum (Bryaceae). Am. J. Bot. 2013, 100, 2000–2008. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Jiang, Y.; Song, X.; Tang, J.; Kou, J.; Fan, Y.; Shao, X. Temperature, Not Precipitation, Drives the Morphological Traits of Didymodon Rigidulus in Tibet. Ecol. Indic. 2021, 133, 108401. [Google Scholar] [CrossRef]
- Crum, H.A. The Geographic Origins of the Mosses of North America’s Eastern Deciduous Forest. J. Hattori Bot. Lab. 1972, 35, 269–298. [Google Scholar]
- Ochyra, R.; Bednarek-Ochyra, H.; Smith, R.I.L. Illustrated Moss Flora of Antarctica; Cambridge University Press: Cambridge, UK, 2008; ISBN 0-521-81402-2. [Google Scholar]
- Taylor, P.J.; Eppley, S.M.; Jesson, L.K. Sporophytic Inbreeding Depression in Mosses Occurs in a Species with Separate Sexes but Not in a Species with Combined Sexes. Am. J. Bot. 2007, 94, 1853–1859. [Google Scholar] [CrossRef]
- Bassi, P.; Basile, A.; Sorbo, S. Behaviour of Repetitive Non-Coding DNA in Response to Heavy Metal Stress in the Protonemata of Funaria hygrometrica. Plant Biosyst. Int. J. Deal. All Asp. Plant Biol. 2015, 149, 315–321. [Google Scholar] [CrossRef]
- Magdy, M.; Werner, O.; McDaniel, S.F.; Goffinet, B.; Ros, R.M. Genomic Scanning Using AFLP to Detect Loci under Selection in the Moss Funaria hygrometrica along a Climate Gradient in the Sierra Nevada Mountains, Spain. Plant Biol. J. 2016, 18, 280–288. [Google Scholar] [CrossRef]
- Itouga, M.; Hayatsu, M.; Sato, M.; Tsuboi, Y.; Kato, Y.; Toyooka, K.; Suzuki, S.; Nakatsuka, S.; Kawakami, S.; Kikuchi, J.; et al. Protonema of the Moss Funaria Hygrometrica Can Function as a Lead (Pb) Adsorbent. PLoS ONE 2017, 12, e0189726. [Google Scholar] [CrossRef]
- Kirbis, A.; Waller, M.; Ricca, M.; Bont, Z.; Neubauer, A.; Goffinet, B.; Szövényi, P. Transcriptional Landscapes of Divergent Sporophyte Development in Two Mosses, Physcomitrium (physcomitrella) Patens and Funaria hygrometrica. Front. Plant Sci. 2020, 11, 747. [Google Scholar] [CrossRef]
- Johri, M.-M. Caulonema Differentiation in Funaria protonema. Int. J. Dev. Biol. 2020, 64, 21–28. [Google Scholar] [CrossRef]
- Nieto-Lugilde, M.; Werner, O.; McDaniel, S.F.; Ros, R.M. Environmental Variation Obscures Species Diversity in Southern European Populations of the Moss Genus Ceratodon. Taxon 2018, 67, 673–692. [Google Scholar] [CrossRef]
- Nieto-Lugilde, M.; Werner, O.; McDaniel, S.F.; Koutecký, P.; Kučera, J.; Rizk, S.M.; Ros, R.M. Peripatric Speciation Associated with Genome Expansion and Female-biased Sex Ratios in the Moss Genus Ceratodon. Am. J. Bot. 2018, 105, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Magdy, M.; Ou, L.; Yu, H.; Chen, R.; Zhou, Y.; Hassan, H.; Feng, B.; Taitano, N.; van der Knaap, E.; Zou, X.; et al. Pan-Plastome Approach Empowers the Assessment of Genetic Variation in Cultivated Capsicum Species. Hortic. Res. 2019, 6, 108. [Google Scholar] [CrossRef] [PubMed]
- Magdy, M.; Ouyang, B. The Complete Mitochondrial Genome of the Chiltepin Pepper (Capsicum annuum var. glabriusculum), the Wild Progenitor of Capsicum annuum L. Mitochondrial DNA Part B 2020, 5, 683–684. [Google Scholar] [CrossRef] [PubMed]
- Peterman, W.E. ResistanceGA: An R Package for the Optimization of Resistance Surfaces Using Genetic Algorithms. Methods Ecol. Evol. 2018, 9, 1638–1647. [Google Scholar] [CrossRef]
- Molero-Mesa, J.; Pérez-Raya, F.; Tendero, F.V. Parque Natural de Sierra Nevada: Paisaje Fauna Flora Itinerarios; Rueda Editorial: Madrid, Spain, 1992; ISBN 84-7207-067-0. [Google Scholar]
- Rams, S.; Werner, O.; Ros, R.M. Updated Checklist of the Bryophytes from the Sierra Nevada Mountains (S of Spain). Cryptogam. Bryol. 2014, 35, 261–311. [Google Scholar] [CrossRef]
- Korpelainen, H.; Jägerbrand, A.; Von Cräutlein, M. Genetic Structure of Mosses Pleurozium schreberi (Willd. Ex Brid.) Mitt. and Racomitrium lanuginosum (Hedw.) Brid. along Altitude Gradients in Hokkaido, Japan. J. Bryol. 2012, 34, 309–312. [Google Scholar] [CrossRef]
- Shaw, J. Genetic Variation for Tolerance to Copper and Zinc within and among Populations of the Moss, Funaria hygrometrica Hedw. New Phytol. 1988, 109, 211–222. [Google Scholar] [CrossRef]
- Shaw, A.J. The Genetic Structure of Sporophytic and Gamtophytic Populations of the Moss, Funaria hygrometrica Hedw. Evolution 1991, 45, 1260–1274. [Google Scholar] [CrossRef]
- Natcheva, R.; Cronberg, N. Maternal Transmission of Cytoplasmic DNA in Interspecific Hybrids of Peat Mosses, Sphagnum (Bryophyta). J. Evol. Biol. 2007, 20, 1613–1616. [Google Scholar] [CrossRef]
- Terao, H. Maternal Inheritance in the Soy Bean. Am. Nat. 1918, 52, 51–56. [Google Scholar] [CrossRef]
- Yang, T.W.; Yang, Y.A.; Xiong, Z. Paternal Inheritance of Chloroplast DNA in Interspecific Hybrids in the Genus Larrea (Zygophyllaceae). Am. J. Bot. 2000, 87, 1452–1458. [Google Scholar] [CrossRef]
- McCauley, D.E.; Sundby, A.K.; Bailey, M.F.; Welch, M.E. Inheritance of Chloroplast DNA Is Not Strictly Maternal in Silene vulgaris (Caryophyllaceae): Evidence from Experimental Crosses and Natural Populations. Am. J. Bot. 2007, 94, 1333–1337. [Google Scholar] [CrossRef]
- Svab, Z.; Maliga, P. Exceptional Transmission of Plastids and Mitochondria from the Transplastomic Pollen Parent and Its Impact on Transgene Containment. Proc. Natl. Acad. Sci. USA 2007, 104, 7003–7008. [Google Scholar] [CrossRef]
- Hartl, D.L.; Clark, A.G. Principles of Population Genetics; Sinauer Associates: Sunderland, MA, USA, 1989. [Google Scholar]
- Haig, D. Living Together and Living Apart: The Sexual Lives of Bryophytes. Phil. Trans. R. Soc. B 2016, 371, 20150535. [Google Scholar] [CrossRef]
- Longton, R.E. Reproductive Biology. New Man. Bryol. 1983, 1, 386–462. [Google Scholar]
- Van Der Velde, M.; During, H.J.; Van De Zande, L.; Bijlsma, R. The Reproductive Biology of Polytrichum formosum: Clonal Structure and Paternity Revealed by Microsatellites. Mol. Ecol. 2001, 10, 2423–2434. [Google Scholar] [CrossRef]
- Eppley, S.M.; Taylor, P.J.; Jesson, L.K. Self-Fertilization in Mosses: A Comparison of Heterozygote Deficiency between Species with Combined versus Separate Sexes. Heredity 2007, 98, 38–44. [Google Scholar] [CrossRef]
- Wen, C.S.; Hsiao, J.Y. Altitudinal Genetic Differentiation and Diversity of Taiwan Lily (Lilium longiflorum var. formosanum; Liliaceae) Using RAPD Markers and Morphological Characters. Int. J. Plant Sci. 2001, 162, 287–295. [Google Scholar] [CrossRef]
- Ohsawa, T.; Tsuda, Y.; Saito, Y.; Sawada, H.; Ide, Y. Altitudinal Genetic Diversity and Differentiation of Quercus Crispula in the Chichibu Mountains, Central Japan. Int. J. Plant Sci. 2007, 168, 333–340. [Google Scholar] [CrossRef]
- Gómez Ortiz, A.; Palacios Estremera, D.; Palade, B.; Vázquez Selem, L.; Salvador Franch, F.; Tanarro García, L.M.; Oliva Franganillo, M. La Evolución Glaciar de Sierra Nevada y La Formación de Glaciares Rocosos. BAGE 2013, 61, 139–162. [Google Scholar] [CrossRef][Green Version]
- Cacho, I.; Grimalt, J.O.; Canals, M.; Sbaffi, L.; Shackleton, N.J.; Schönfeld, J.; Zahn, R. Variability of the Western Mediterranean Sea Surface Temperature during the Last 25,000 Years and Its Connection with the Northern Hemisphere Climatic Changes. Paleoceanography 2001, 16, 40–52. [Google Scholar] [CrossRef]
- Wetterdienst, D. Klimadaten Deutschland. Messstation Düsseldorf. 2009. Available online: https://www.dwd.de/DE/leistungen/klimadatendeutschland/klimadatendeutschland.html (accessed on 1 October 2023).
- McDaniel, S.F.; Willis, J.H.; Shaw, A.J. A Linkage Map Reveals a Complex Basis for Segregation Distortion in an Interpopulation Cross in the Moss Ceratodon purpureus. Genetics 2007, 176, 2489–2500. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, S.F.; Willis, J.H.; Shaw, A.J. The Genetic Basis of Developmental Abnormalities in Interpopulation Hybrids of the Moss Ceratodon purpureus. Genetics 2008, 179, 1425–1435. [Google Scholar] [CrossRef] [PubMed]
- Soltis, P.S.; Soltis, D.E. The Role of Hybridization in Plant Speciation. Annu. Rev. Plant Biol. 2009, 60, 561–588. [Google Scholar] [CrossRef]
- Rivas-Martínez, S.; Rivas-Sáenz, S. Worldwide Bioclimatology Manual and Guide; Universidad Complutense de Madrid: Madrid, Spain, 2016. [Google Scholar]
- Gil, J.A. Flora y Vegetación Briofítica de Sierra Nevada. Monogr. Flora Y Veg. Bética 1988, 3, 63–72. [Google Scholar]
- Susana, R.; Rosa, M.R.; María, J.C.; Juan, G. Checklist de los briófitos de Sierra Nevada (Andalucía, España). Boletín Soc. Española Briología 2001, 18, 137–164. [Google Scholar] [CrossRef]
- Rams Sánchez, S. Estudios Briológicos Sobre Flora, Vegetación, Taxonomía y Conservación En Sierra Nevada (Andalucía, S de España). Ph.D. Thesis, Murcia University, Murcia, Spain, 2008. Available online: https://dialnet.unirioja.es/servlet/tesis?codigo=311327 (accessed on 10 August 2024).
- WorldClim: Global Climate Free Climate Data for Ecological Modeling and GIS. Available online: https://www.worldclim.org/ (accessed on 1 October 2023).
- Smith, A.J.E.; Smith, R. The Moss Flora of Britain and Ireland, 2nd ed.; Cambridge University Press: Cambridge, UK, 2004; ISBN 978-0-521-54672-0. [Google Scholar]
- Sabovljevic, M.; Bijelovic, A.; Dragicevic, I. In Vitro Culture of Mosses: Aloina aloides (KF Schultz) Kindb., Brachythecium velutinum (Hedw.) BS & G., Ceratodon purpureus (Hedw.) Brid., Eurhynchium praelongum (Hedw.) BS & G. and Grimmia pulvinata (Hedw.) Sm. Turk. J. Bot. 2003, 27, 441–446. [Google Scholar]
- Douzery, E.J.P.; Pridgeon, A.M.; Kores, P.; Linder, H.P.; Kurzweil, H.; Chase, M.W. Molecular Phylogenetics of Diseae (Orchidaceae): A Contribution from Nuclear Ribosomal ITS Sequences. Am. J. Bot. 1999, 86, 887–899. [Google Scholar] [CrossRef]
- Liu, Y.; Moskwa, N.L.; Goffinet, B. Development of Eight Mitochondrial Markers for Funariaceae (Musci) and Their Amplification Success in Other Mosses. Am. J. Bot. 2012, 99, e62–e65. [Google Scholar] [CrossRef][Green Version]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. In Proceedings of the Nucleic Acids Symposium Series, Oxford, UK, 1 November 1999; Volume 41, pp. 95–98. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Paradis, E. Pegas: An R Package for Population Genetics with an Integrated–Modular Approach. Bioinformatics 2010, 26, 419–420. [Google Scholar] [CrossRef] [PubMed]
- R Core Team R: A Language and Environment for Statistical Computing; 2013. Available online: https://www.scirp.org/reference/ReferencesPapers?ReferenceID=1787696 (accessed on 10 August 2024).
- Beerli, P.; Palczewski, M. Unified Framework to Evaluate Panmixia and Migration Direction Among Multiple Sampling Locations. Genetics 2010, 185, 313–326. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, İ.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Pearson, K.X. On the Criterion That a given System of Deviations from the Probable in the Case of a Correlated System of Variables Is Such That It Can Be Reasonably Supposed to Have Arisen from Random Sampling. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1900, 50, 157–175. [Google Scholar] [CrossRef]
- Fisher, R.A. On the Interpretation of χ2 from Contingency Tables, and the Calculation of P. J. R. Stat. Soc. 1922, 85, 87. [Google Scholar] [CrossRef]
- Warnes, G.R.; Bolker, B.M.; Lumley, T.; Johnson, R.T.; Jain, N.; Schwartz, M.A.; Rogers, J. Gmodels: Various R Programming Tools for Model Fitting. Available online: https://cran.r-project.org/package=gmodels (accessed on 1 October 2023).
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very High Resolution Interpolated Climate Surfaces for Global Land Areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Anderson, M.J. PCO: A FORTRAN Computer Program for Principal Coordinate Analysis; Department of Statistics, University of Auckland: Auckland, New Zealand, 2003. [Google Scholar]
- Peakall, R.; Smouse, P.E. genalex 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]

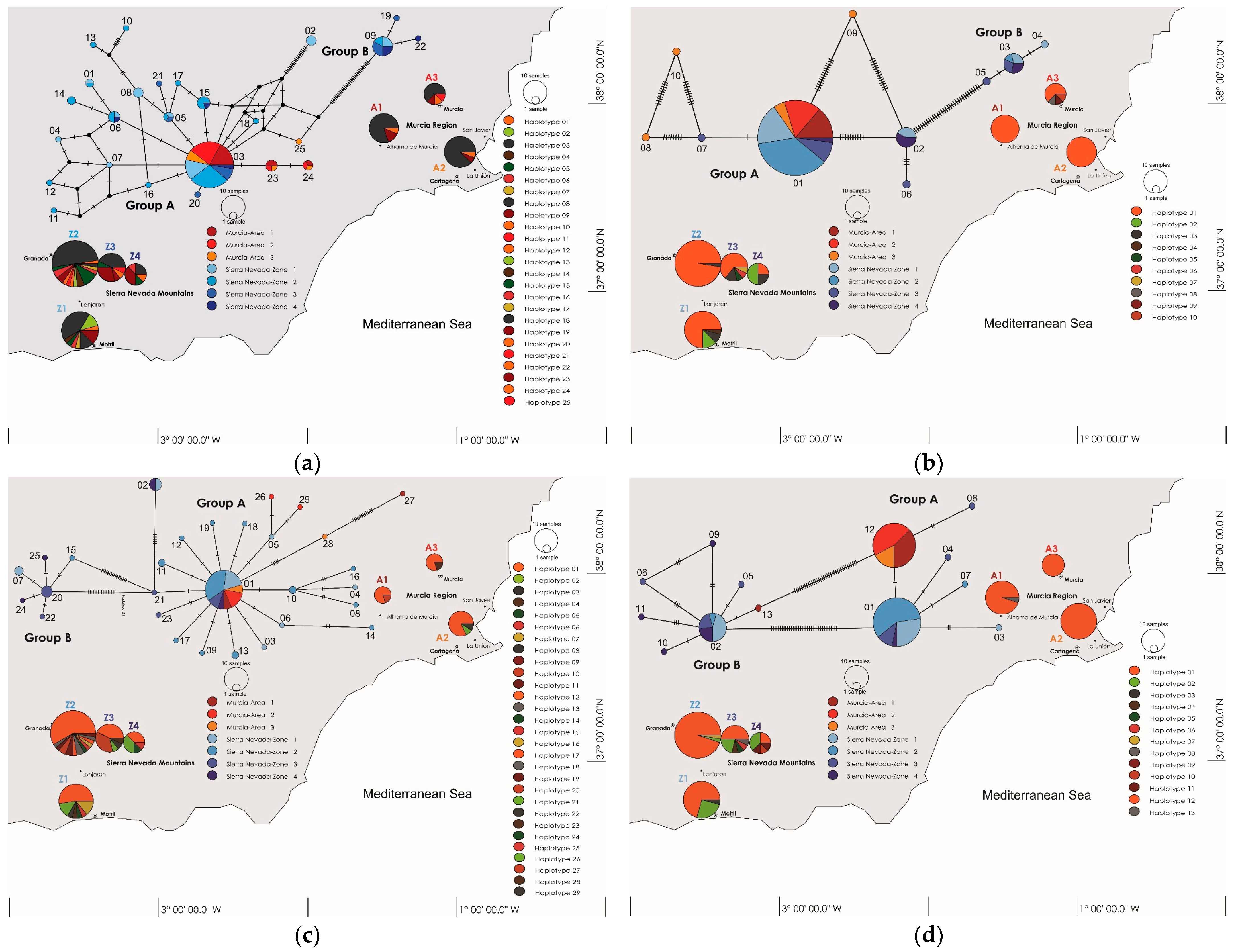
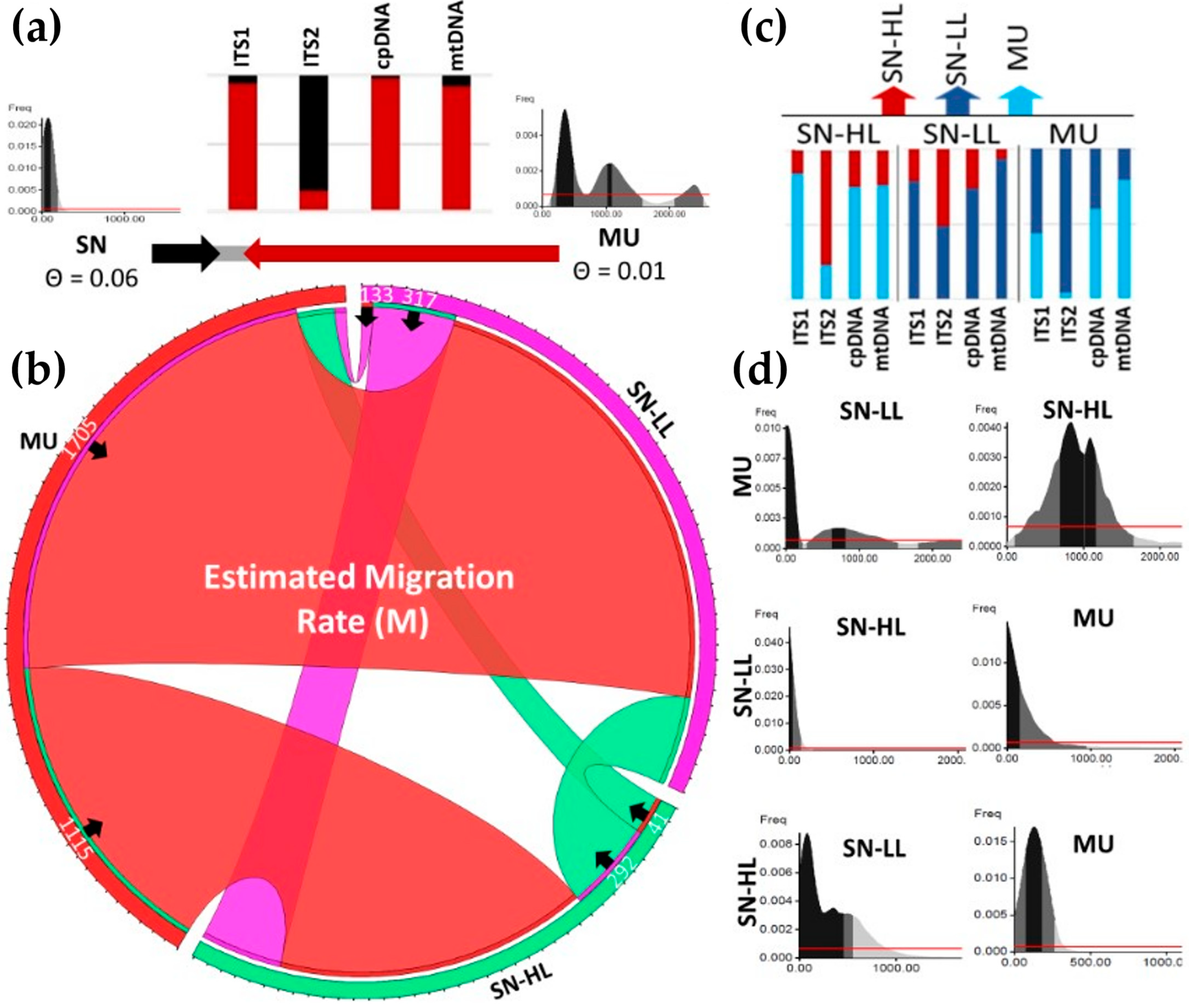
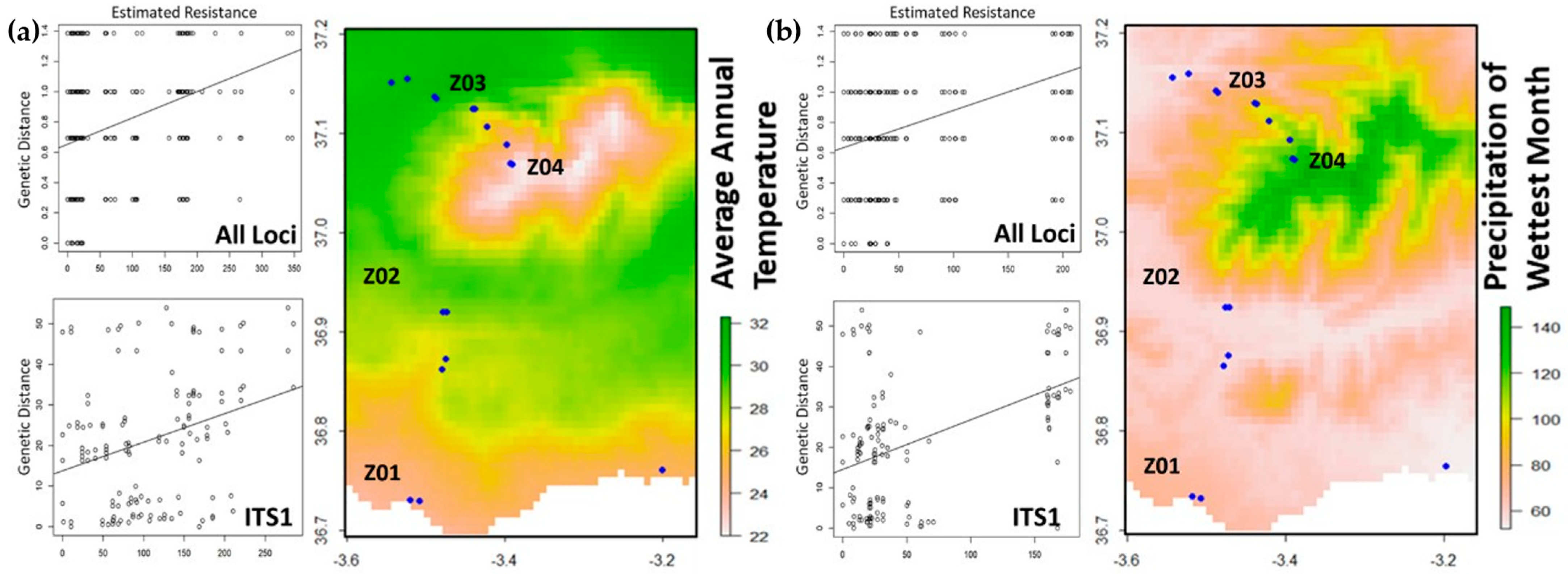
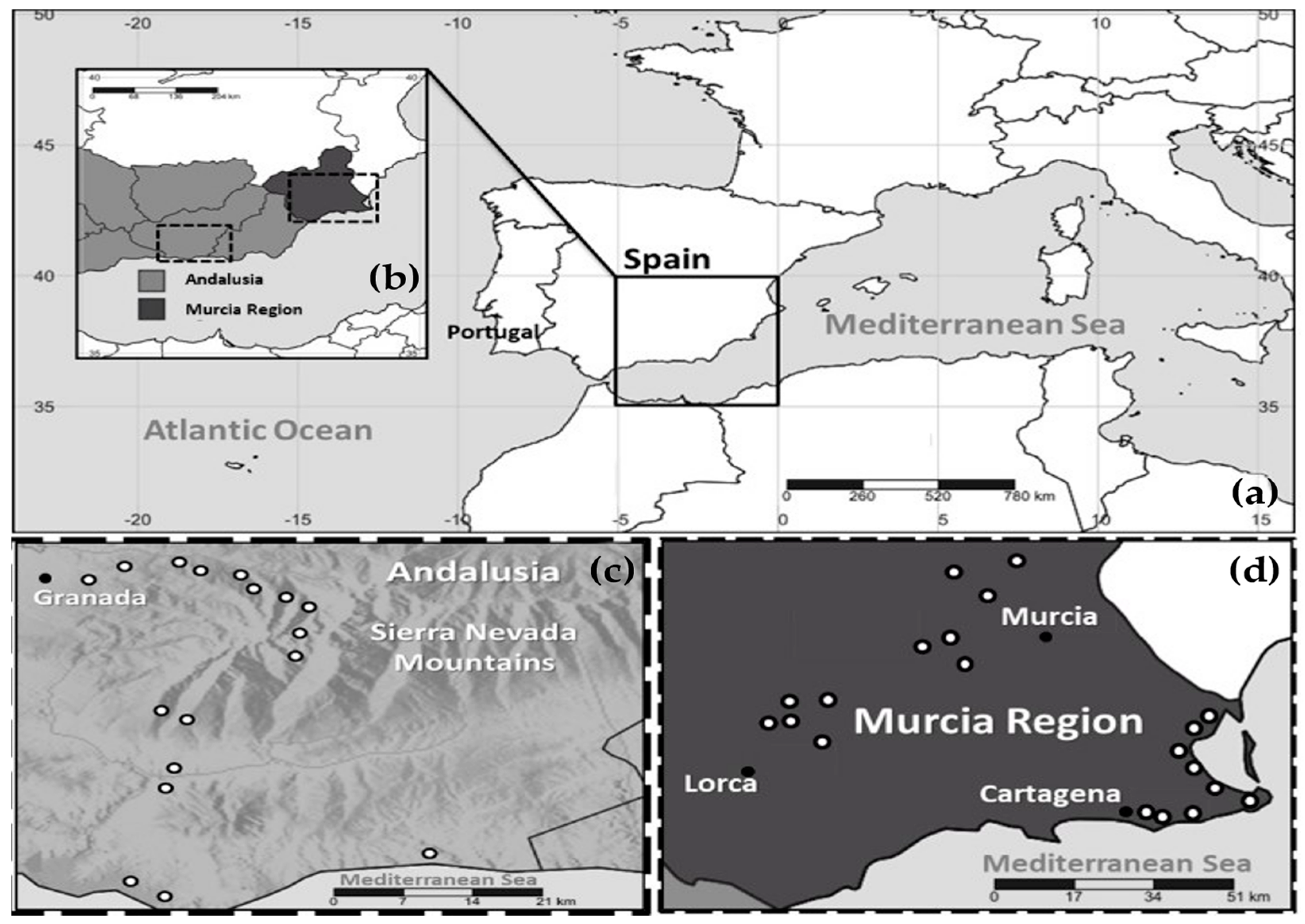
| Locus | Region | Sequences Count a | Amplification rate % | Group A Length (bp)/Count/Pairwise Identity % | Group B Length (bp)/Count/Pairwise Identity % | Total Pairwise Identity % | h | Hd | π% | Nm | Tajima’s D |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ITS1 | SN | 22/62 | 100 | 244–249/70/98.70 | 254–255/14/99.70 | 93.70 | 22 | 0.78 | 0.51 | - | - |
| MU | 40 | 75.47 | 248/40/99.70 | − | 99.70 | 4 | 0.35 | 0.14 | - | - | |
| Total/ Alignment | 124 | 90.51 | 281 | 95.40 | 25 | 0.67 | 0.40 | 3.02 | −1.83 * | ||
| ITS2 | SN | 22/62 | 100 | 311–312/17/93.00 | 316/67/99.99 | 96.60 | 7 | 0.37 | 0.88 | - | - |
| MU | 38 | 71.70 | 309/01/- | 316/37/99.70 | 99.50 | 4 | 0.15 | 0.42 | - | - | |
| Total/ Alignment | 122 | 89.05 | 331 | 97.60 | 10 | 0.31 | 0.75 | 4.97 | −1.74 NS | ||
| cpDNA | SN | 22/62 | 94.05 | 778–779/12/99.80 | 794–798/67/99.50 | 99.10 | 25 | 0.72 | 0.20 | - | - |
| MU | 21 | 39.62 | 795–797/20/99.90 | 802/01/- | 99.80 | 5 | 0.35 | 0.02 | - | - | |
| Total/ Alignment | 100 | 72.99 | 805 | 95.40 | 29 | 0.66 | 0.16 | 4.76 | −2.18 ** | ||
| mtDNA | SN | 22/57 | 100 | 790–791/66/99.98 | 836/18/99.90 | 98.10 | 11 | 0.43 | 0.03 | - | - |
| MU | 52 | 98.11 | 792/51/100 | 836/01/- | 100.0 | 2 | 0.04 | 0.00 | - | - | |
| Total/ Alignment | 136 | 99.27 | 837 | 98.6 | 13 | 0.65 | 0.02 | 2.71 | −2.32 ** | ||
| Haplotype Combinations * | Count | |||||
|---|---|---|---|---|---|---|
| Total for SN | SN-LL | SN-HL | Total for MU | |||
| SN-Z1 | SN-Z2 | SN-Z3 | SN-Z4 | |||
| AAAA | 56 | 15 | 34 | 6 | 1 | 18 |
| AA - A | 5 | 3 | 2 | 0 | 0 | 20 |
| A - - A | 0 | 0 | 0 | 0 | 0 | 2 |
| - - AA | 0 | 0 | 0 | 0 | 0 | 2 |
| - - A - | 0 | 0 | 0 | 0 | 0 | 1 |
| - - - A | 0 | 0 | 0 | 0 | 0 | 9 |
| BBBB | 7 | 3 | 1 | 2 | 1 | 0 |
| BAAA | 2 | 0 | 1 | 1 | 0 | 0 |
| ABAA | 1 | 0 | 0 | 1 | 0 | 0 |
| AABA | 1 | 0 | 0 | 0 | 1 | 0 |
| AAAB | 1 | 0 | 0 | 0 | 1 | 0 |
| - - - B | 0 | 0 | 0 | 0 | 0 | 1 |
| AABB | 6 | 3 | 0 | 2 | 1 | 0 |
| BABB | 3 | 0 | 0 | 1 | 2 | 0 |
| BBAB | 1 | 0 | 0 | 0 | 1 | 0 |
| BABA | 1 | 0 | 0 | 1 | 0 | 0 |
| Total | 84 | 24 | 38 | 14 | 8 | 53 |
| Comparison | ITS1 | ITS2 | cpDNA | mtDNA |
|---|---|---|---|---|
| SN-HL vs. SN-LL | 0.00038 * 0.00113 * | 0.03397 0.04850 | 0.00057 * 0.00176 * | 0.00014 * 0.00041 * |
| SN-HL vs. MU | 1.2132 × 10−05 * 2.4519 × 10−05 * | 0.01111 * 0.01984 * | 0.00922 * 0.00378 * | 2.9226 × 10−07 * 1.7011 × 10−06 * |
| SN-LL vs. MU | 0.06551 0.17003 | 0.38055 0.64619 | 0.24415 0.56754 | 0.05116 0.06905 |
| Contrast | Loci | ΦCT | ΦSC | ΦST |
|---|---|---|---|---|
| Geographical | ITS1 | 0.02 NS | 0.18 * | 0.20 * |
| ITS2 | 0.07 NS | 0.09 NS | 0.15 * | |
| cpDNA | 0.04 NS | 0.19 * | 0.22 ** | |
| mtDNA | 0.07 NS | 0.32 * | 0.22 ** | |
| Altitudinal | ITS1 | 0.41 ** | 0.00 NS | 0.40 ** |
| ITS2 | 0.22 ** | 0.03 NS | 0.24 ** | |
| cpDNA | 0.32 ** | 0.04 NS | 0.35 ** | |
| mtDNA | 0.47 ** | 0.16 * | 0.55 ** | |
| Geographical and altitudinal | ITS1 | 0.03 NS | 0.04 NS | 0.07 * |
| ITS2 | 0.14 * | 0.01 NS | 0.15 * | |
| cpDNA | 0.21 * | 0.04 NS | 0.25 ** | |
| mtDNA | 0.27 * | 0.16 * | 0.39 ** |
| Loci | All Samples | Samples below 1300 m a.s.l. | Samples above 1300 m a.s.l. | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ITS1 | ITS2 | cpDNA | mtDNA | ITS1 | ITS2 | cpDNA | mtDNA | ITS1 | ITS2 | cpDNA | mtDNA | |
| ITS1 | ||||||||||||
| ITS2 | 0.00 | 0.00 | 0.04 | |||||||||
| cpDNA | 0.06 | 0.00 | 0.00 | 0.00 | 0.49 | 0.50 | ||||||
| mtDNA | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.31 | 0.21 | 0.22 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magdy, M.; Werner, O.; Patiño, J.; Ros, R.M. Landscape Heterogeneity Drives Genetic Diversity in the Highly Dispersive Moss Funaria hygrometrica Hedw. Plants 2024, 13, 2785. https://doi.org/10.3390/plants13192785
Magdy M, Werner O, Patiño J, Ros RM. Landscape Heterogeneity Drives Genetic Diversity in the Highly Dispersive Moss Funaria hygrometrica Hedw. Plants. 2024; 13(19):2785. https://doi.org/10.3390/plants13192785
Chicago/Turabian StyleMagdy, Mahmoud, Olaf Werner, Jairo Patiño, and Rosa María Ros. 2024. "Landscape Heterogeneity Drives Genetic Diversity in the Highly Dispersive Moss Funaria hygrometrica Hedw." Plants 13, no. 19: 2785. https://doi.org/10.3390/plants13192785
APA StyleMagdy, M., Werner, O., Patiño, J., & Ros, R. M. (2024). Landscape Heterogeneity Drives Genetic Diversity in the Highly Dispersive Moss Funaria hygrometrica Hedw. Plants, 13(19), 2785. https://doi.org/10.3390/plants13192785







