Transcriptome Profiling Reveals Molecular Responses to Salt Stress in Common Vetch (Vicia sativa L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth
2.2. Measurements of Dry Weight and Leaf Relative Water Content (RWC)
2.3. Measurements of Soluble Sugars
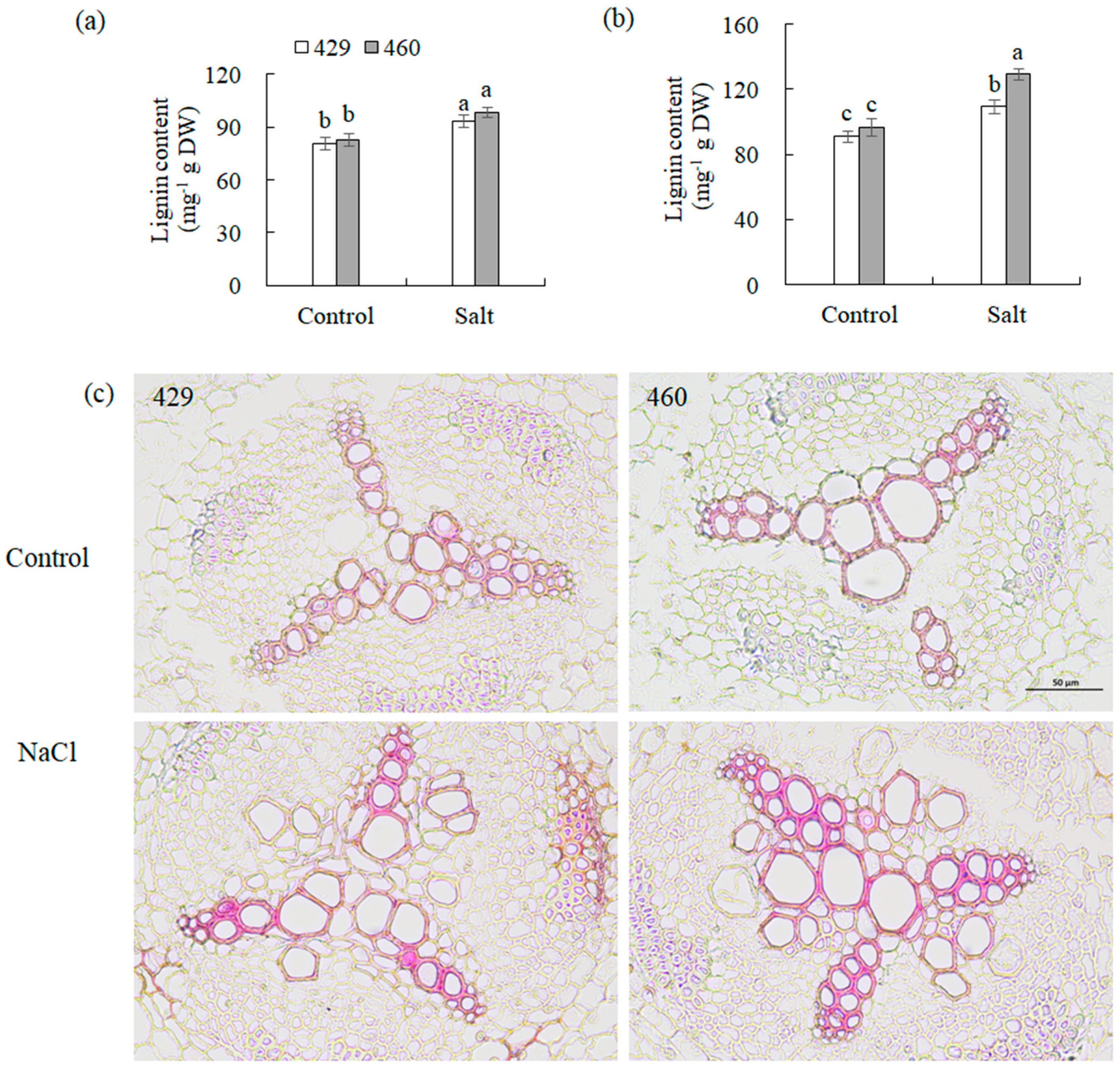
2.4. Measurement of Total Lignin Content
2.5. Lignin Staining
2.6. RNA Isolation, Library Construction, and Sequencing
2.7. Analysis of the Differentially Expressed Genes (DEGs)
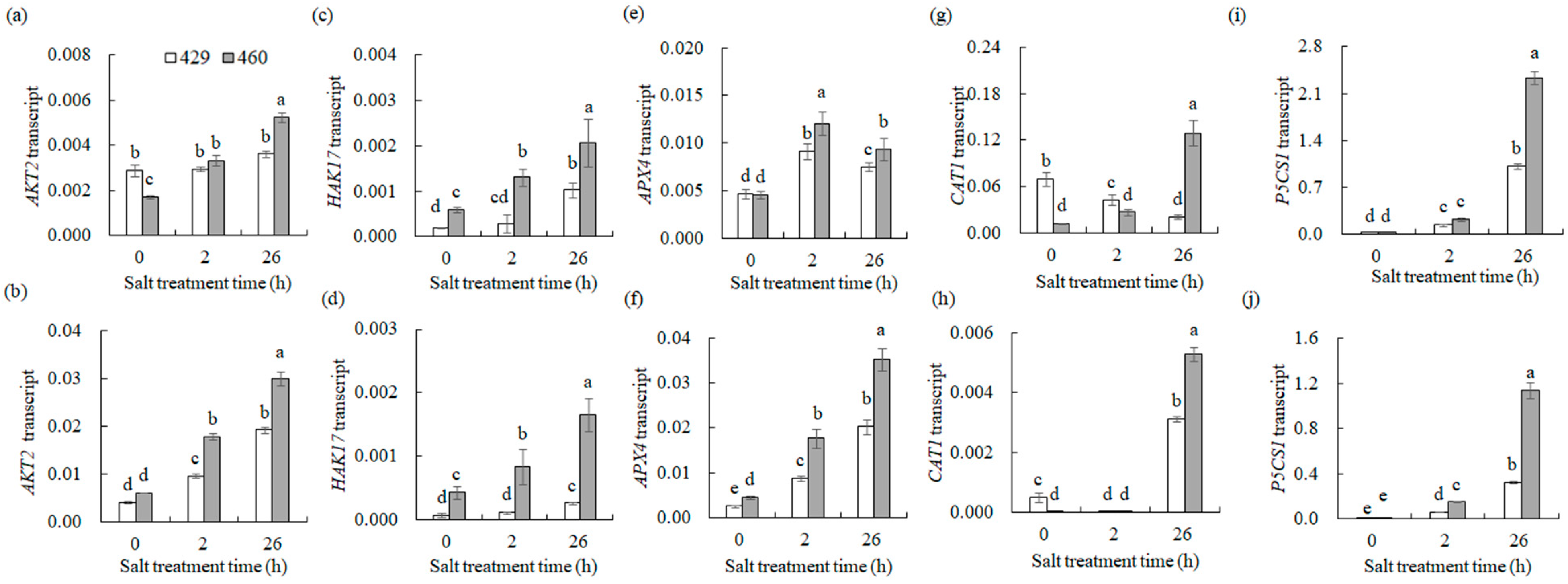
2.8. Quantitative Real-Time PCR (qPCR) Analysis
2.9. Statistical Analysis
3. Results
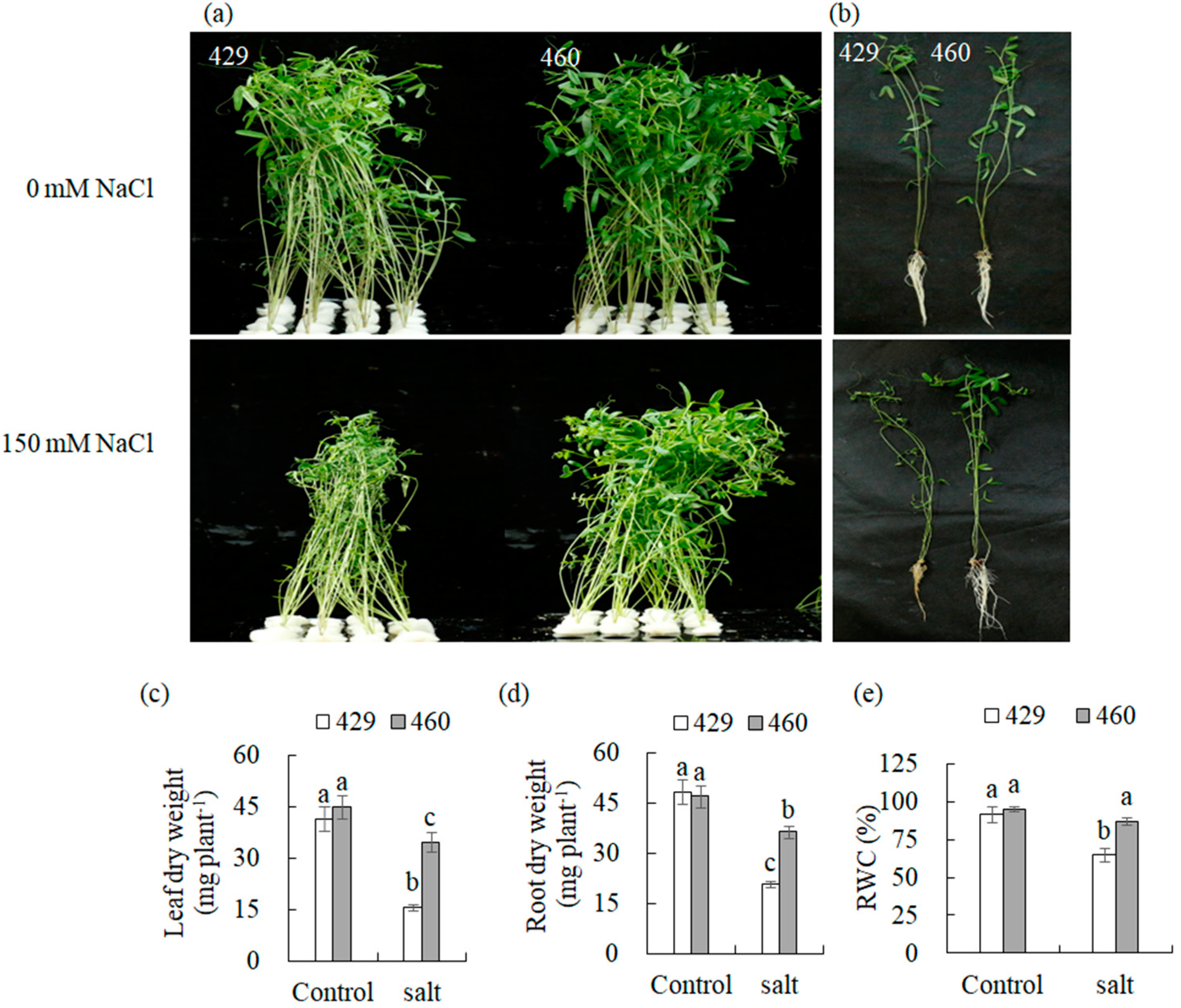
3.1. Phenotypic Differences between Salt-Tolerant (T) and Salt-Sensitive (S) Strains under Salinity Stress
3.2. RNA Sequencing, Transcriptome Assembly, and Annotation
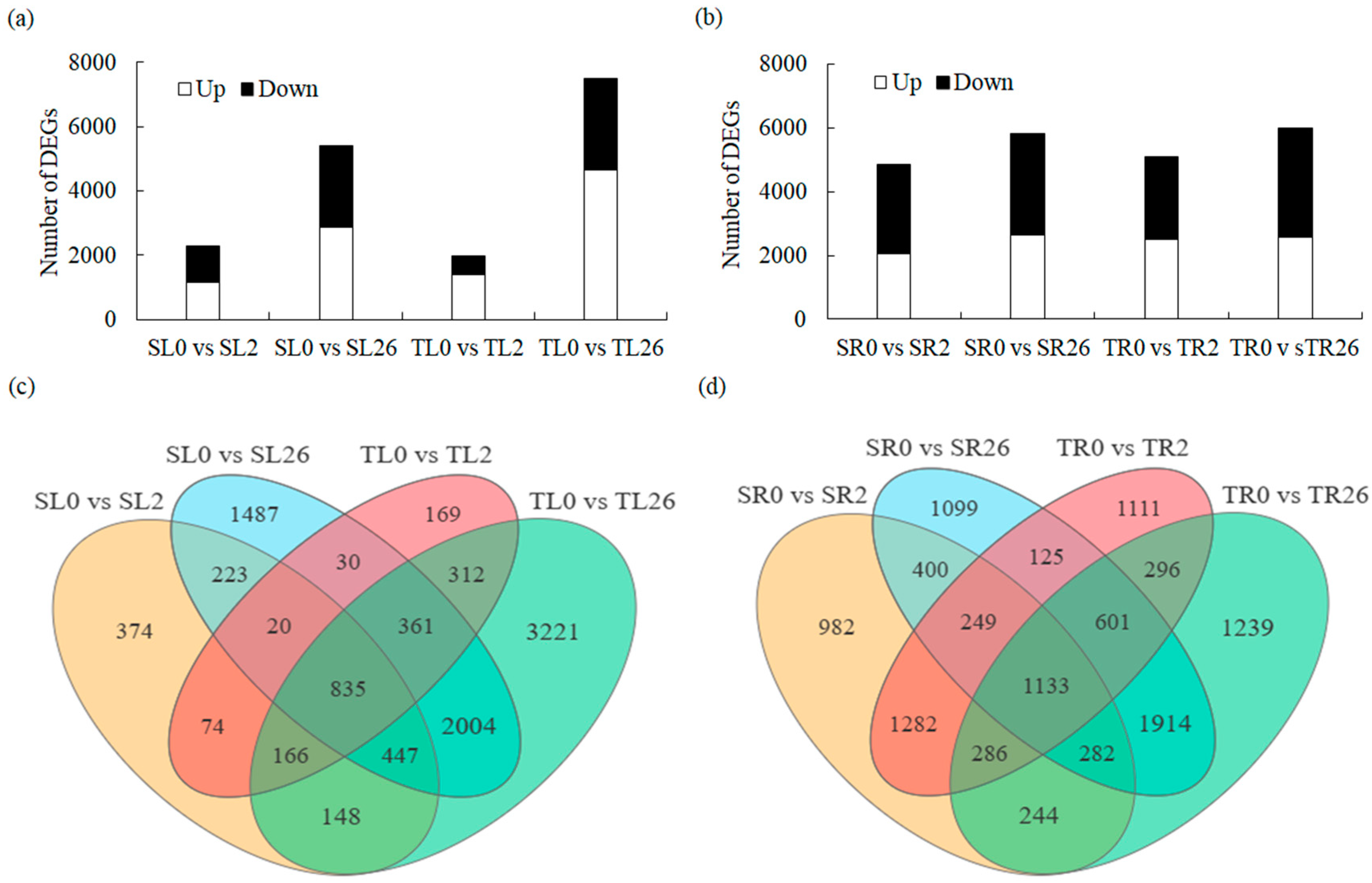
3.3. DEG Identification and Analysis in Leaves and Roots
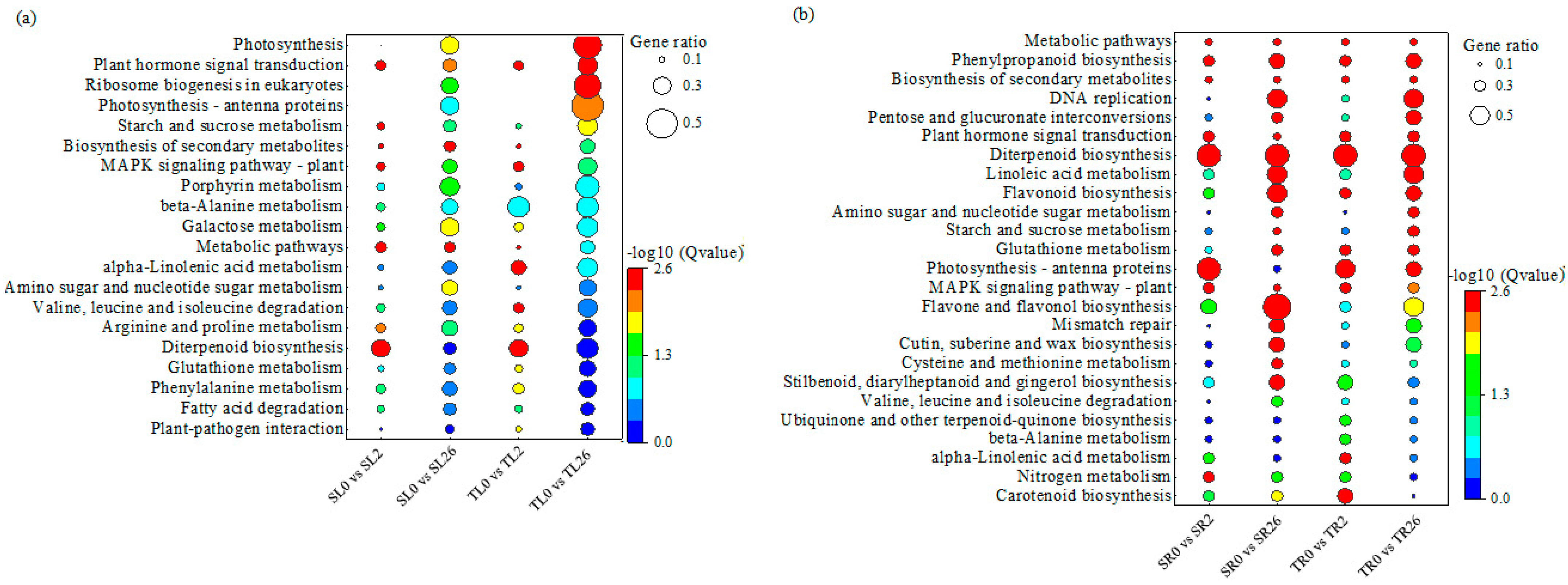
3.4. Analysis of KEGG Pathway Enrichment of DEGs in Response to Salt Stress
3.5. Plant Hormone and MAPK Signaling Pathways Are Common for Common Vetch in Response to Salt Stress
3.6. Phenylpropanoid Biosynthesis Is Involved in Response to Salt Stress in Common Vetch
3.7. Analysis of Salt-Responsive DEGs Associated with the Higher Salt Tolerance in 460 Than in 429
3.7.1. Alpha-Linolenic Acid Metabolism
3.7.2. Photosynthesis-Antenna Pathway
3.7.3. Carotenoid Biosynthesis
3.7.4. Starch and Sucrose Metabolism
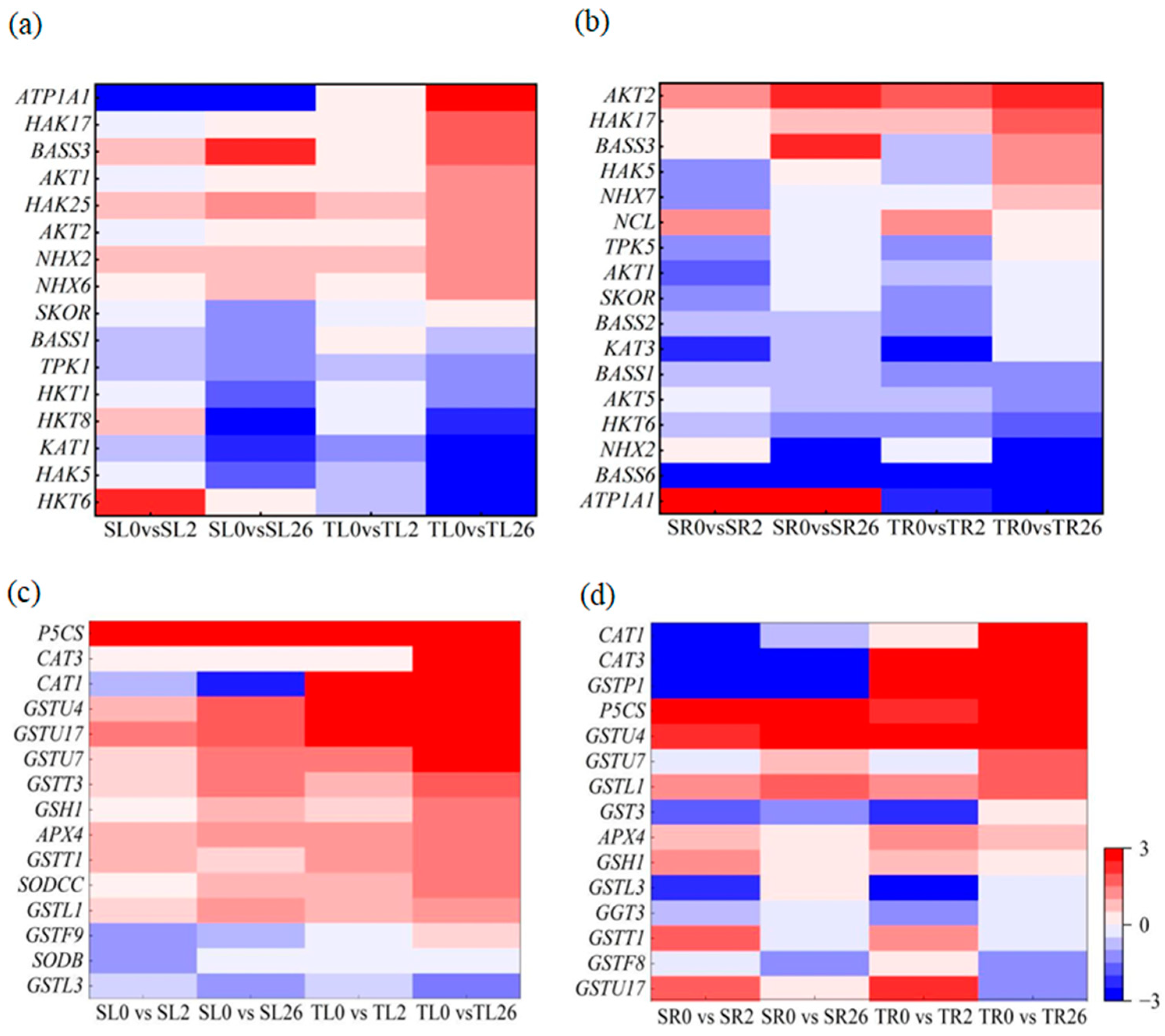
3.7.5. Na+ and K+ Transport and ROS Scavenging
4. Discussion
4.1. Plant Hormone and MAPK Signaling Were Associated with Salt Tolerance in Common Vetch
4.2. Phenylpropanoid Biosynthesis Was Associated with Salt Tolerance in Common Vetch
4.3. The Carbohydrate and Energy Metabolisms Were Associated with the Differential Salt Tolerance between Two Lines
4.4. Na+ and K+ Transport and ROS Scavenging Were Associated with the Differential Salt Tolerance between the Two Lines
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhou, H.P.; Shi, H.F.; Yang, Y.Q.; Feng, X.X.; Chen, X.; Xiao, F.; Lin, H.H.; Guo, Y. Insights into plant salt stress signaling and tolerance. J. Genet. Genomics 2023, 51, 16–34. [Google Scholar] [CrossRef]
- Almeida, D.M.; Oliveira, M.M.; Saibo, N.J.M. Regulation of Na+ and K+ homeostasis in plants: Towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 2018, 40, 326–345. [Google Scholar] [CrossRef]
- Guo, H.Y.; Wang, Y.C.; Wang, L.Q.; Hu, P.; Wang, Y.M.; Jia, Y.Y.; Zhang, C.R.; Zhang, Y.M.; Wang, C.; Yang, C.P. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol. J. 2017, 15, 107–121. [Google Scholar] [CrossRef]
- Koch, K. Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 2004, 7, 235–246. [Google Scholar] [CrossRef]
- Opassiri, R.; Pomthong, B.; Akiyama, T.; Nakphaichit, M.; Onkoksoong, T.; Ketudat, C.M.; Ketudat, C.J.R. A stress-induced rice (Oryza sativa L.) beta-glucosidase represents a new subfamily of glycosyl hydrolase family 5 containing a fascin-like domain. Biochem. J. 2007, 408, 241–249. [Google Scholar] [CrossRef]
- Monroe, J.D.; Storm, A.R. Review: The Arabidopsis β-amylase (BAM) gene family: Diversity of form and function. Plant Sci. 2018, 276, 163–170. [Google Scholar] [CrossRef]
- Jia, X.M.; Zhu, Y.F.; Hu, Y.; Zhang, R.; Cheng, L.; Zhu, Z.L.; Zhao, T.; Zhang, X.Y.; Wang, Y.X. Integrated physiologic, proteomic, and metabolomic analyses of Malus halliana adaptation to saline-alkali stress. Hortic. Res. 2019, 6, 91. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.T.; Huang, R.S.; Han, Y.Y.; Zhang, Z.Y.; Chen, Y.Y.; Shi, H.F.; Guo, Z.F. A novel salt responsive PvHAK16 negatively regulates salt tolerance in transgenic Arabidopsis thaliana. Environ. Exp. Bot. 2022, 194, 104689. [Google Scholar] [CrossRef]
- Kim, H.J.; Triplett, B. Involvement of extracellular Cu/Zn superoxide dismutase in cotton fiber primary and secondary cell wall biosynthesis. Plant Signal. Behav. 2008, 3, 1119–1121. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.F.; Gao, X.L.; Nan, Z.B.; Zhang, Z.X. Potential value of the common vetch Vicia sativa L. as an animal feed stuff, a review. J. Anim. Physiol. Anim. Nutr. 2017, 101, 807–823. [Google Scholar] [CrossRef] [PubMed]
- Dalias, P.; Neocleous, D. Comparative analysis of the nitrogen effect of common agricultural practices and rotation systems in a rain fed mediterranean environment. Plants 2017, 6, 61. [Google Scholar] [CrossRef] [PubMed]
- Bet, C.D.; do Prado Cordoba, L.; Ribeiro, L.S.; Schnitzler, E. Common vetch Vicia sativa as a new starch source, its thermal, rheological and structural properties after acid hydrolysis. Food Biophys. 2016, 11, 275–282. [Google Scholar] [CrossRef]
- Nguyen, V.; Riley, S.; Nagel, S.; Fisk, I.; Searle, I.R. Common vetch, a drought tolerant, high protein neglected leguminous crop with potential as a sustainable food source. Front. Plant Sci. 2020, 11, 818. [Google Scholar] [CrossRef]
- Mikić, A.; Mihailović, V.; Ćupina, B.; Milić, D.; Katić, S.; Karagić, D.; D’Ottavio, P.; Kraljević-Balalić, M. Forage yield components and classification of common vetch Vicia sativa L. cultivars of diverse geographic origin. Grass Forage Sci. 2014, 69, 315–322. [Google Scholar] [CrossRef]
- Dong, R.; Jahufer, M.Z.Z.; Dong, D.K.; Liu, W.X.; Wang, Y.R.; Liu, Z.P. Characterisation of the morphological variation for seed traits among 537 germplasm accessions of common vetch Vicia sativa L. using digital image analysis. N. Z. J. Agr. Res. 2016, 59, 422–435. [Google Scholar] [CrossRef]
- Córdoba, E.M.; Fernández-Aparicio, M.; González-Verdejo, C.I.; López-Grau, C.; del Valle Muñoz-Muñoz, M.; Nadal, S. Search for resistant genotypes to Cuscuta campestris infection in two legume species, Vicia sativa and Vicia ervilia. Plants 2021, 104, 738. [Google Scholar] [CrossRef]
- Shirasawa, K.; Kosugi, S.; Sasaki, K.; Ghelfi, A.; Okazaki, K.; Toyoda, A.; Hirakawa, H.; Isobe, S. Genome features of common vetch Vicia sativa in natural habitats. Plant Direct. 2021, 5, e352. [Google Scholar] [CrossRef]
- Ma, L.; Wang, X.; Yan, M.; Liu, F.; Zhang, S.X.; Wang, X.M. Genome survey sequencing of common vetch (Vicia sativa L.) and genetic diversity analysis of Chinese germplasm with genomic SSR markers. Mol. Biol. Rep. 2022, 49, 313–320. [Google Scholar] [CrossRef]
- Dong, R.; Dong, D.K.; Luo, D.; Zhou, Q.; Chai, X.T.; Zhang, J.Y.; Xie, W.G.; Dong, Y.; Wang, Y.R.; Liu, Z.P. Transcriptome analyses reveal candidate pod shattering-associated genes involved in the pod ventral sutures of common vetch Vicia sativa L. Front. Plant Sci. 2017, 8, 649. [Google Scholar] [CrossRef]
- Rui, H.; Zhang, X.; Shinwari, K.I.; Zheng, L.; Shen, Z. Comparative transcriptomic analysis of two Vicia sativa L. collections with contrasting responses to cadmium stress reveals the important role of metal transporters in cadmium tolerance. Plant Soil. 2018, 423, 241–255. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, Q.X.; Xu, W.Z.; Zhang, J.H.; Wang, X.; Nie, G.; Yao, L.; Wang, H.; Lin, C.W. De novo assembly and discovery of genes that involved in drought tolerance in the common vetch. Int. J. Mol. Sci. 2019, 20, 328. [Google Scholar] [CrossRef]
- Dong, R.; Luo, B.; Tang, L.; Wang, Q.X.; Lu, Z.J.; Chen, C.; Yang, F.; Wang, S.; He, J. A comparative transcriptomic analysis reveals a coordinated mechanism activated in response to cold acclimation in common vetch (Vicia sativa L.). BMC Genom. 2022, 23, 814. [Google Scholar] [CrossRef]
- Lin, X.S.; Wang, Q.X.; Min, X.Y.; Liu, W.X.; Liu, Z.P. Comparative transcriptomic analysis of root and leaf transcript profiles reveals the coordinated mechanisms in response to salinity stress in common vetch. Int. J. Mol. Sci. 2022, 23, 8477. [Google Scholar] [CrossRef]
- Sun, Y.M.; Lu, Y.W.; Xi, H.J.; Zhao, N.; Guo, Z.F. Transcriptomic analysis revealed the candidate metabolic pathways and genes associated with cold tolerance in a mutant without anthocyanin accumulation in common vetch (Vicia sativa L.). Plant Physiol. Biochem. 2023, 200, 107770. [Google Scholar] [CrossRef]
- Sun, Y.M.; Li, J.; Xing, J.C.; Yu, X.; Lu, Y.W.; Xu, W.K.; Zhao, N.; Guo, Z.F. Evaluation of salt tolerance in common vetch (Vicia sativa L.) germplasms and the physiological responses to salt stress. J. Plant Physiol. 2022, 278, 153811. [Google Scholar] [CrossRef] [PubMed]
- Epstein, E. Mineral Nutrition of Plants: Principles and Perspectives; Pimprenta: New York, NY, USA, 1972; 412p. [Google Scholar]
- Barrs, H.D.; Weatherly, P.E. A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust. J. Biol. Sci. 1962, 15, 413–428. [Google Scholar] [CrossRef]
- Sun, Q.G.; Huang, R.S.; Zhu, H.F.; Sun, Y.M.; Guo, Z.F. A novel Medicago truncatula calmodulin-like protein (MtCML42) regulates cold tolerance and flowering time. Plant J. 2021, 108, 1069–1082. [Google Scholar] [CrossRef]
- Song, C.; Li, X.L.; Jia, B.; Liu, L.; Wei, P.P.; Manzoor, M.A.; Wang, F.; Li, B.Y.; Wang, G.L.; Chen, C.W.; et al. Comparative transcriptomics unveil the crucial genes involved in coumarin biosynthesis in peucedanum praeruptorum Dunn. Front. Plant Sci. 2022, 13, 899819. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, K.; Hwang, H.; Bhatnagar, N.; Kim, D.Y.; Yoon, I.S.; Byun, M.O.; Kim, S.T.; Jung, K.H.; Kim, B.G. Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J. Exp. Bot. 2008, 65, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Kuang, Y.F.; Lin, Y.P.; Guo, Y.C.; Li, H.Y.; Fan, P.G.; Li, S.H.; Liang, Z.C. Overexpression of grape ABA receptor gene VaPYL4 enhances tolerance to multiple abiotic stresses in Arabidopsis. BMC Plant Biol. 2022, 22, 271. [Google Scholar] [CrossRef]
- Nie, K.; Zhao, H.; Wang, X.; Niu, Y.; Zhou, H.; Zheng, Y. The MIEL1-ABI5/MYB30 regulatory module fine tunes abscisic acid signaling during seed germination. J. Integr. Plant Biol. 2022, 64, 930–941. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Q.; Shen, J.; Nie, K.; Zheng, Y. MIW1 participates in ABA signaling through the regulation of MYB30 in Arabidopsis. Plant Sci. 2023, 332, 111717. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, R.J.; Han, T.T.; Cai, W.; Fu, Z.W.; Lu, Y.T. Salt stress reduces root meristem size by nitric oxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiol. 2015, 168, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Moe-Lange, J.; Hennet, L.; Feldman, L.J. Salt stress affects the redox status of Arabidopsis root meristems. Front. Plant Sci. 2016, 7, 81. [Google Scholar] [CrossRef]
- Li, G.; Ye, Y.X.; Ren, X.Q.; Qi, M.Y.; Zhao, H.Y.; Zhou, Q. The rice Aux/IAA transcription factor gene OsIAA18 enhances salt and osmotic tolerance in Arabidopsis. Biol. Plant. 2020, 64, 454–464. [Google Scholar] [CrossRef]
- Zhang, A.Y.; Yang, X.; Lu, J.; Song, F.Y.; Sun, J.H.; Wang, C.; Lian, J.; Zhao, L.L.; Zhao, B.C. OsIAA20, an Aux/IAA protein, mediates abiotic stress tolerance in rice through an ABA pathway. Plant Sci. 2021, 308, 110903. [Google Scholar] [CrossRef]
- Zhang, X.X.; Liu, P.; Qing, C.Y.; Yang, C.; Shen, Y.; Ma, L.L. Comparative transcriptome analyses of maize seedling root responses to salt stress. PeerJ 2021, 9, e10765. [Google Scholar] [CrossRef]
- Kumar, K.; Raina, S.K.; Sultan, S.M. Arabidopsis MAPK signaling pathways and their cross talks in abiotic stress response. J. Plant Biochem. 2020, 29, 700–714. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, S. Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 2022, 64, 301–341. [Google Scholar] [CrossRef]
- Kumar, K.; Sinha, A.K. Overexpression of constitutively active mitogen activated protein kinase kinase 6 enhances tolerance to salt stress in rice. Rice 2013, 6, 25. [Google Scholar] [CrossRef]
- Xu, J.; Li, Y.; Wang, Y.; Liu, H.X.; Lei, L.; Yang, H.L.; Liu, G.Q.; Ren, D.T. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 2008, 283, 26996–27006. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.J.; Nie, J.N.; Cao, C.Y.; Jin, Y.K.; Yan, M.; Wang, F.Z.; Liu, J.; Xiao, Y.; Liang, Y.H.; Zhang, W.H. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. N. Phytol. 2010, 188, 762–773. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.M.; Sui, C.C.; Yu, Y.Y.; Liu, X.L.; Li, B.; Sun, Q.H. Grape VvMAPK9 positively regulates salt tolerance in Arabidopsis and grape callus through regulating the antioxidative system. Plant Cell Tissue Organ Cult. 2022, 148, 609–622. [Google Scholar] [CrossRef]
- Kang, S.M.; Xiao, L.P.; Meng, L.P.; Jounob, P.M. Isolation and structural characterization of lignin from cotton stalk treated in an ammonia hydrothermal system. Mol. Sci. 2012, 7, 15209–15226. [Google Scholar] [CrossRef] [PubMed]
- Sonali, S.; Arun, L.M. Physiological and genomic basis of mechanical-functional trade-off in plant vasculature. Front. Plant Sci. 2014, 3, 224. [Google Scholar]
- Kelij, S.; Majd, A.; Nematzade, G.; Jounob, P. Activation of lignin biosynthetic enzymes during internodal development of aeluropus littoralis exposed to NaCl. J. Genet. Res. 2015, 1, 19–24. [Google Scholar]
- Han, X.j.; Zhao, Y.Q.; Chen, Y.J.; Xu, J.; Jiang, C.; Wang, X.Q.; Zhuo, R.Y.; Lu, M.Z.; Zhang, J. Lignin biosynthesis and accumulation in response to abiotic stresses in woody plants. For. Res. 2022, 2, 9. [Google Scholar] [CrossRef]
- Shafi, A.; Zahoor, I. Plant Survival and Tolerance Under High Salinity: Primary and Secondary Cell Wall-Sensing Mechanism. In Salt Stress, Microbes, and Plant Interactions: Causes and Solution; Akhtar, M., Ed.; Springer: Singapore, 2019. [Google Scholar]
- Li, Y.X.; Zhang, T.T.; Kang, Y.; Wang, P.; Yu, W.; Wang, J.; Li, W.; Jiang, X.Y.; Zhou, Y. Integrated metabolome, transcriptome analysis, and multi-flux full-length sequencing offer novel insights into the function of lignin biosynthesis as a Sesuvium portulacastrum response to salt stress. Int. J. Biol. Macromol. 2023, 237, 124222. [Google Scholar] [CrossRef]
- Cao, B.; Li, N.; Xu, K. Crosstalk of phenylpropanoid biosynthesis with hormone signaling in Chinese cabbage is key to counteracting salt stress. Environ. Exp. Bot. 2020, 179, 104209. [Google Scholar] [CrossRef]
- Rosa, M.; Prado, C.; Podazza, G.; Interdonato, R.; González, J.A.; Hilal, M.; Prado, F.E. Soluble sugars—Metabolism, sensing and abiotic stress: A complex network in the life of plants. Plant Signal Behav. 2009, 4, 388–393. [Google Scholar] [CrossRef]
- Li, C.; Qi, Y.; Zhao, C.; Wang, X.; Zhang, Q. Transcriptome profiling of the salt stress response in the leaves and roots of halophytic Eutrema salsugineum. Front. Genet. 2021, 12, 770742. [Google Scholar] [CrossRef]
- Yuan, Q.H.; Xie, F.; Huang, W.; Hu, M.; Yan, Q.; Chen, Z.M.; Zheng, Y.; Liu, L. The review of alpha-linolenic acid: Sources, metabolism, and pharmacology. Phytother. Res. 2022, 36, 164–188. [Google Scholar] [CrossRef] [PubMed]
- Sui, N.; Tian, S.; Wang, W.; Wang, M.; Fan, H. Overexpression of glycerol-3-phosphate acyltransferase from Suaeda salsa improves salt tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1337. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Jing, W.; Zhang, W. The mitogen-activated protein kinase cascade MKK1-MPK4 mediates salt signaling in rice. Plant Sci. 2014, 227, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Havaux, M. Carotenoid oxidation products as stress signals in plants. Plant J. 2014, 79, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; He, S.Z.; Liu, D.G.; Patil, G.B.; Zhai, H.; Wang, F.B.; Stephenson, T.J.; Wang, Y.N.; Wang, B.; Valliyodan, B.; et al. A sweet potato geranylgeranyl pyrophosphate synthase gene, IbGGPS, increases carotenoid content and enhances osmotic stress tolerance in Arabidopsis thaliana. PLoS ONE 2015, 10, e0137623. [Google Scholar]
- Zhuang, K.Y.; Kong, F.Y.; Zhang, S.; Meng, C.; Yang, M.M.; Liu, Z.B.; Wang, Y.; Ma, N.N.; Meng, Q.W. Whirly1 enhances tolerance to chilling stress in tomato via protection of photosystem II and regulation of starch degradation. N. Phytol. 2018, 221, 1998–2012. [Google Scholar] [CrossRef] [PubMed]
- Sasi, S.; Venkatesh, J.; Daneshi, R.F.; Gururani, M.A. Photosystem II extrinsic proteins and their putative role in abiotic stress tolerance in higher plants. Plants 2018, 7, 100. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, M.; Liu, A.; Zhou, R.; Li, D.; Dossa, K.; Wang, L.; Zhang, Y.; Gong, H.; Zhang, X.; et al. Comparative proteomic analysis of two sesame genotypes with contrasting salinity tolerance in response to salt stress. J. Proteom. 2019, 201, 73–83. [Google Scholar] [CrossRef]
- Kronzucker, H.J.; Britto, D.T. Sodium transport in plants, a critical review. N. Phytol. 2011, 189, 54–81. [Google Scholar] [CrossRef]
- Lebaudy, A.; Véry, A.A.; Sentenac, H. K+ channel activity in plants: Genes, regulations and functions. FEBS Lett. 2007, 581, 2357–2366. [Google Scholar] [CrossRef]
- Chérel, I.; Michard, E.; Platet, N.; Mouline, K.; Alcon, C.; Sentenac, H.; Thibaud, J.B. Physical and functional interaction of the Arabidopsis K+ channel AKT2 and phosphatase AtPP2CA. Plant Cell 2002, 14, 1133–1146. [Google Scholar] [CrossRef]
- Sandmann, M.; Skłodowski, K.; Gajdanowicz, P.; Michard, E.; Rocha, M.; Gomez-Porras, J.L.; González, W.; Corrêa, L.G.G.; Ramírez-Aguilar, S.J.; Cuin, T.A.; et al. The K+ battery-regulating Arabidopsis K+ channel AKT2 is under the control of multiple post-translational steps. Plant Signal. Behav. 2011, 6, 558–562. [Google Scholar] [CrossRef]
- Yang, T.Y.; Zhang, S.; Hu, Y.B.; Wu, F.C.; Hu, Q.D.; Chen, G.; Cai, J.; Wu, T.; Moran, N.; Yu, L.; et al. The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 2014, 166, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.M.; Tang, Q.; Cai, J.; Xu, B.C.; Yu, L. Rice OsHAK16 functions in potassium uptake andtranslocation in shoot, maintaining potassium homeostasis and salt tolerance. Planta 2019, 250, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Nieves-Cordones, M.; Alemán, F.; Martínez, V.; Rubio, F. The Arabidopsis thaliana HAK5 K+ transporter is required for plant growth and K+ acquisition from low K+ solutions under saline conditions. Mol. Plant. 2010, 3, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.X.; Hodson, J.N.; Williams, J.P.; Blumwald, E. Engineering salt-tolerant Brassica plants, characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc. Natl. Acad. Sci. USA 2001, 98, 12832–12836. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Breusegem, F.V. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.X.; Li, H.Y.; Pang, H.C.; Fu, J.M. Responses of antioxidant gene, protein and enzymes to salinity stress in two genotypes of perennial ryegrass Lolium perenne differing in salt tolerance. J. Plant Physiol. 2012, 169, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yang, A.; Zhang, W.H. Comparative studies on tolerance of rice genotypes differing in their tolerance to moderate salt stress. BMC Plant Biol. 2017, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Qin, Y.; Hu, X.; Li, G.; Ding, H.; Xiong, X.; Wang, W. Transcriptome analysis uncovers the gene expression profile of salt-stressed potato (Solanum tuberosum L.). Sci. Rep. 2020, 10, 5411. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Zhao, N.; Sun, H.; Xu, S.; Lu, Y.; Xi, H.; Guo, Z.; Shi, H. Transcriptome Profiling Reveals Molecular Responses to Salt Stress in Common Vetch (Vicia sativa L.). Plants 2024, 13, 714. https://doi.org/10.3390/plants13050714
Sun Y, Zhao N, Sun H, Xu S, Lu Y, Xi H, Guo Z, Shi H. Transcriptome Profiling Reveals Molecular Responses to Salt Stress in Common Vetch (Vicia sativa L.). Plants. 2024; 13(5):714. https://doi.org/10.3390/plants13050714
Chicago/Turabian StyleSun, Yanmei, Na Zhao, Hongjian Sun, Shan Xu, Yiwen Lu, Haojie Xi, Zhenfei Guo, and Haifan Shi. 2024. "Transcriptome Profiling Reveals Molecular Responses to Salt Stress in Common Vetch (Vicia sativa L.)" Plants 13, no. 5: 714. https://doi.org/10.3390/plants13050714
APA StyleSun, Y., Zhao, N., Sun, H., Xu, S., Lu, Y., Xi, H., Guo, Z., & Shi, H. (2024). Transcriptome Profiling Reveals Molecular Responses to Salt Stress in Common Vetch (Vicia sativa L.). Plants, 13(5), 714. https://doi.org/10.3390/plants13050714







