Expression of FAD and SAD Genes in Developing Seeds of Flax Varieties under Different Growth Conditions
Abstract
1. Introduction
2. Results
2.1. Results of Transcriptome Sequencing of Flax Seeds
2.2. Expression of FAD and SAD Genes in Flax Seeds
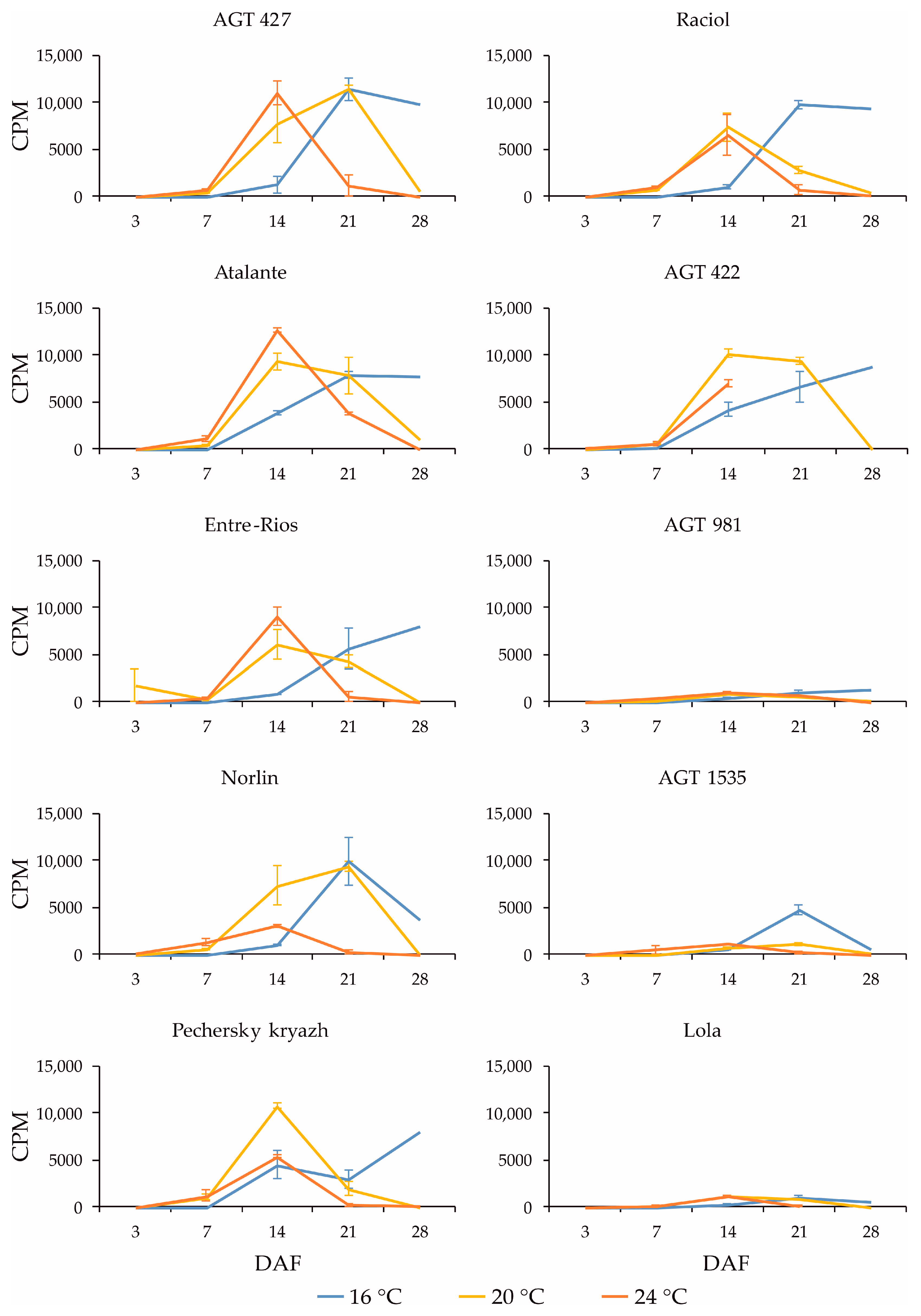
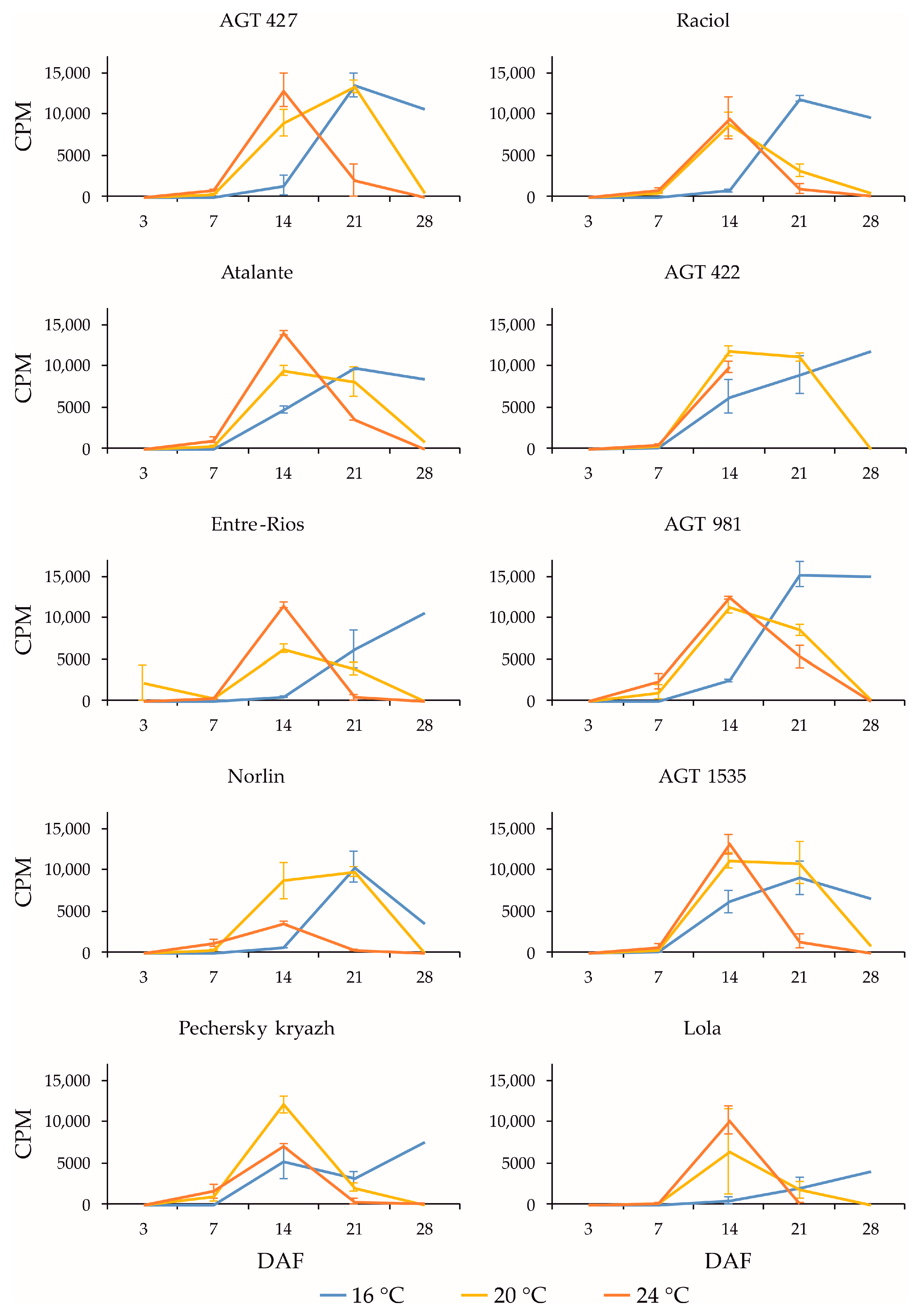
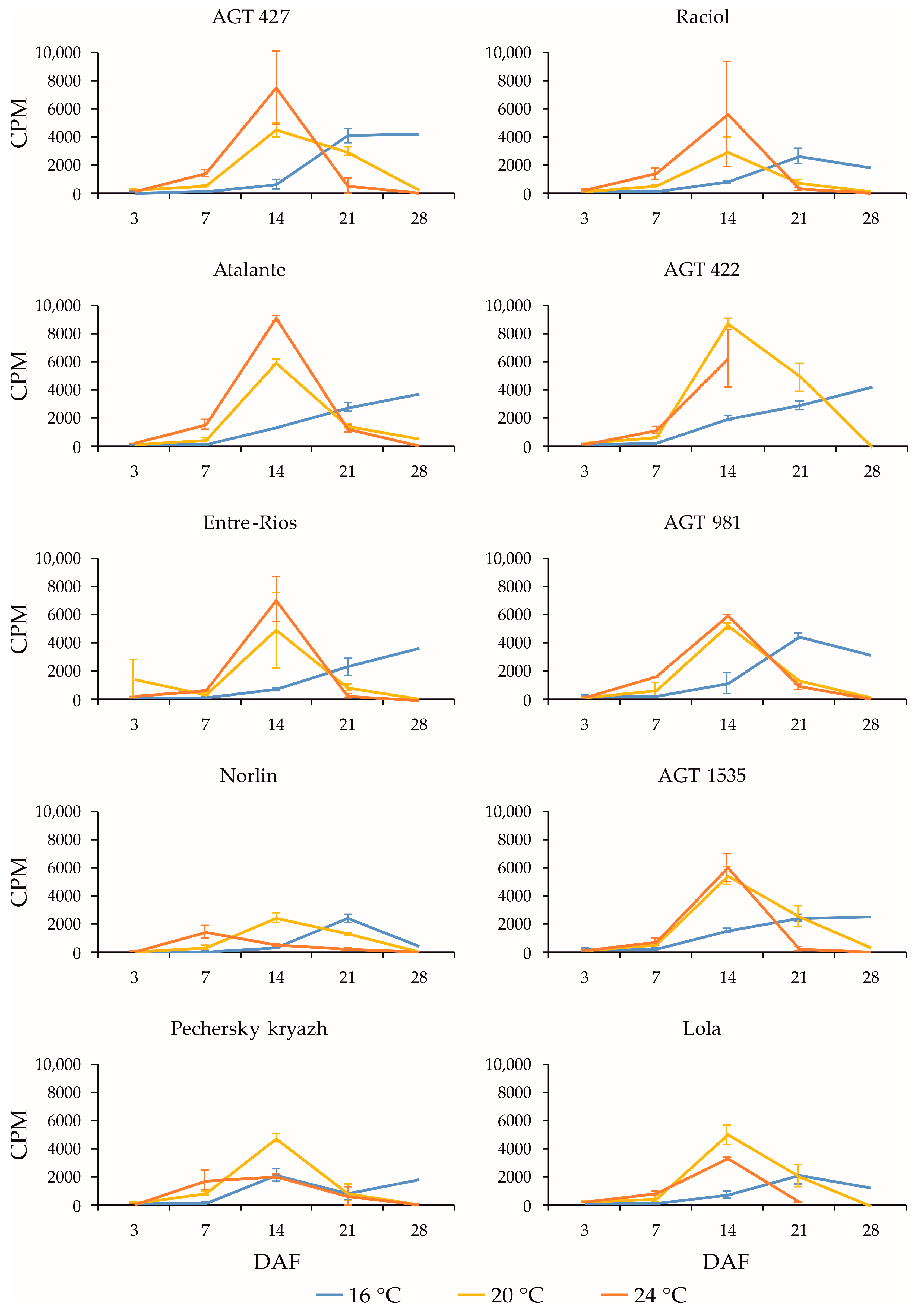
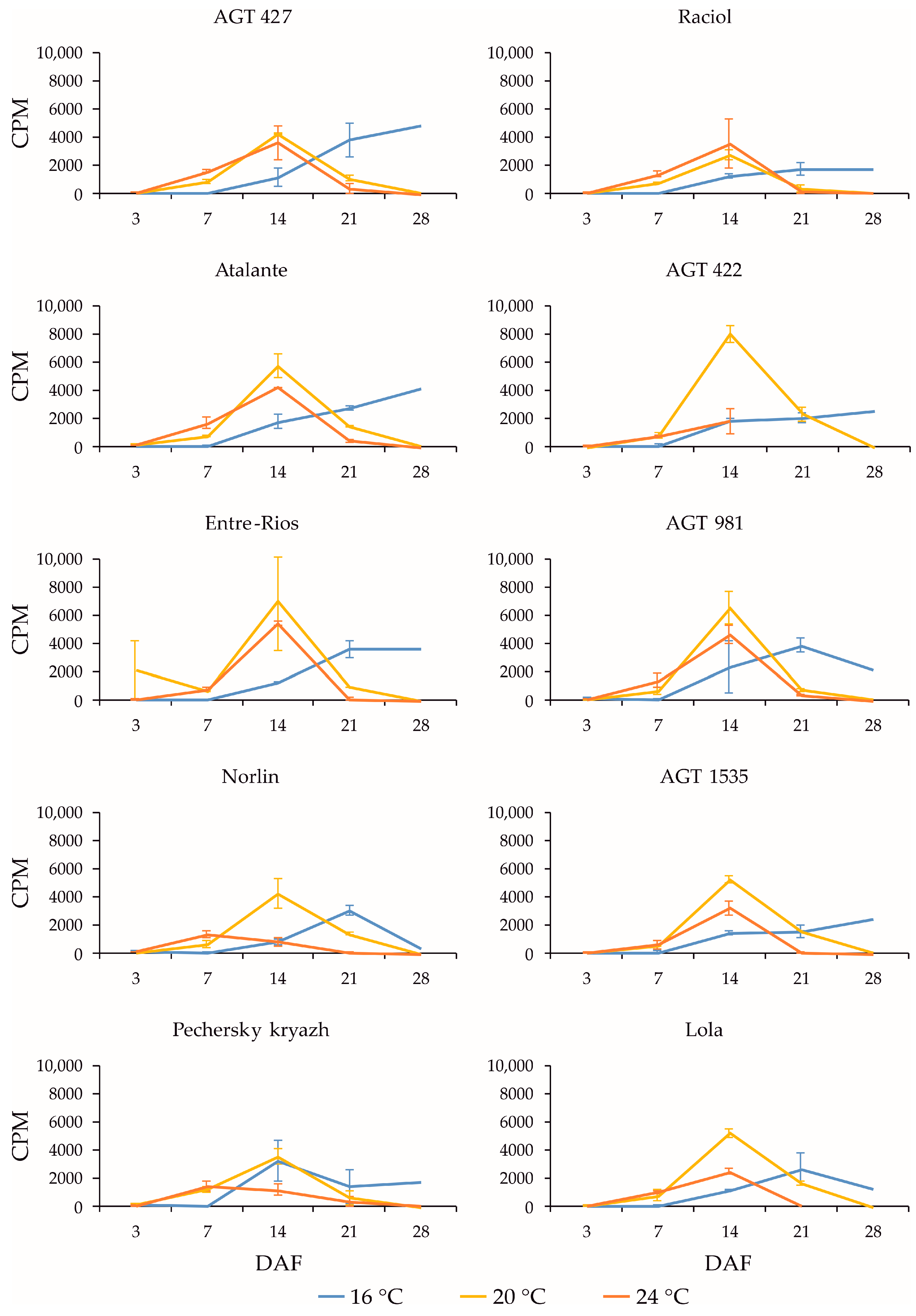
2.3. Expression Profiles of FAD3a and FAD3b Genes during Flax Seed Development under Different Temperature and Watering Conditions
2.4. Expression Profiles of FAD2b-2, FAD2a-1, and FAD2a-2 Genes during Flax Seed Development under Different Temperature and Watering Conditions
2.5. Expression Levels of SAD Genes during Flax Seed Development under Different Temperature and Watering Conditions
3. Discussion
4. Materials and Methods
4.1. Flax Varieties
4.2. Growth Conditions
4.3. Collection of Plant Material and RNA Isolation
4.4. Preparation and Sequencing of cDNA Libraries
4.5. Expression Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parikh, M.; Maddaford, T.G.; Austria, J.A.; Aliani, M.; Netticadan, T.; Pierce, G.N. Dietary Flaxseed as a Strategy for Improving Human Health. Nutrients 2019, 11, 1171. [Google Scholar] [CrossRef]
- Saini, R.K.; Prasad, P.; Sreedhar, R.V.; Akhilender Naidu, K.; Shang, X.; Keum, Y.S. Omega-3 Polyunsaturated Fatty Acids (PUFAs): Emerging Plant and Microbial Sources, Oxidative Stability, Bioavailability, and Health Benefits-A Review. Antioxidants 2021, 10, 1627. [Google Scholar] [CrossRef]
- Al-Madhagy, S.; Ashmawy, N.S.; Mamdouh, A.; Eldahshan, O.A.; Farag, M.A. A comprehensive review of the health benefits of flaxseed oil in relation to its chemical composition and comparison with other omega-3-rich oils. Eur. J. Med. Res. 2023, 28, 240. [Google Scholar] [CrossRef]
- Campos, J.R.; Severino, P.; Ferreira, C.S.; Zielinska, A.; Santini, A.; Souto, S.B.; Souto, E.B. Linseed Essential Oil-Source of Lipids as Active Ingredients for Pharmaceuticals and Nutraceuticals. Curr. Med. Chem. 2019, 26, 4537–4558. [Google Scholar] [CrossRef] [PubMed]
- Cloutier, S.; Ragupathy, R.; Niu, Z.; Duguid, S. SSR-based linkage map of flax (Linum usitatissimum L.) and mapping of QTLs underlying fatty acid composition traits. Mol. Breed. 2011, 28, 437–451. [Google Scholar] [CrossRef]
- Muir, A.D.; Westcott, N.D. Flax: The Genus Linum; CRC Press: Boca Raton, FL, USA, 2003. [Google Scholar]
- Dmitriev, A.A.; Kezimana, P.; Rozhmina, T.A.; Zhuchenko, A.A.; Povkhova, L.V.; Pushkova, E.N.; Novakovskiy, R.O.; Pavelek, M.; Vladimirov, G.N.; Nikolaev, E.N.; et al. Genetic diversity of SAD and FAD genes responsible for the fatty acid composition in flax cultivars and lines. BMC Plant Biol. 2020, 20, 301. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, X.R.; Zhang, Z.J.; Dribnenki, P.; Singh, S.; Green, A. Development of high oleic oil crop platform in flax through RNAi-mediated multiple FAD2 gene silencing. Plant Cell Rep. 2015, 34, 643–653. [Google Scholar] [CrossRef]
- Brutch, N.; Porokhoviniva, E.; Shelenga, T. Perspectives of the creation of oil flax varieties for the specialized purpose. Agrar. Report. South-East 2016, 1, 50–52. [Google Scholar]
- Porokhovinova, E.; Shelenga, T.; Kosykh, L.; Sanin, A.; Kazarina, A.; Kutuzova, S.; Pavlov, A.; Brutch, N. Biochemical diversity of fatty acid composition in flax from VIR genetic collection and effect of environment on its development. Ecol. Genet. 2016, 14, 13. [Google Scholar] [CrossRef][Green Version]
- Jain, R.K.; Thompson, R.G.; Taylor, D.C.; MacKenzie, S.L.; McHughen, A.; Rowland, G.G.; Tenaschuk, D.; Coffey, M. Isolation and characterization of two promoters from linseed for genetic engineering. Crop Sci. 1999, 39, 1696–1701. [Google Scholar] [CrossRef]
- Singh, S.; McKinney, S.; Green, A. Sequence of a cDNA from Linum usitatissimum encoding the stearoyl-acyl carrier protein desaturase. Plant Physiol. 1994, 104, 1075. [Google Scholar] [CrossRef][Green Version]
- Fofana, B.; Duguid, S.; Cloutier, S. Cloning of fatty acid biosynthetic genes β-ketoacyl CoA synthase, fatty acid elongase, stearoyl-ACP desaturase, and fatty acid desaturase and analysis of expression in the early developmental stages of flax (Linum usitatissimum L.) seeds. Plant Sci. 2004, 166, 1487–1496. [Google Scholar] [CrossRef]
- Khadake, R.M.; Ranjekar, P.K.; Harsulkar, A.M. Cloning of a novel omega-6 desaturase from flax (Linum usitatissimum L.) and its functional analysis in Saccharomyces cerevisiae. Mol. Biotechnol. 2009, 42, 168–174. [Google Scholar] [CrossRef]
- Krasowska, A.; Dziadkowiec, D.; Polinceusz, A.; Plonka, A.; Łukaszewicz, M. Cloning of flax oleic fatty acid desaturase and its expression in yeast. J. Am. Oil Chem. Soc. 2007, 84, 809–816. [Google Scholar] [CrossRef]
- Vrinten, P.; Hu, Z.; Munchinsky, M.A.; Rowland, G.; Qiu, X. Two FAD3 desaturase genes control the level of linolenic acid in flax seed. Plant Physiol. 2005, 139, 79–87. [Google Scholar] [CrossRef]
- Banik, M.; Duguid, S.; Cloutier, S. Transcript profiling and gene characterization of three fatty acid desaturase genes in high, moderate, and low linolenic acid genotypes of flax (Linum usitatissimum L.) and their role in linolenic acid accumulation. Genome 2011, 54, 471–483. [Google Scholar] [CrossRef]
- You, F.M.; Li, P.; Kumar, S.; Ragupathy, R.; Li, Z.; Fu, Y.-B.; Cloutier, S. Genome-wide identification and characterization of the gene families controlling fatty acid biosynthesis in flax (Linum usitatissimum L). J. Proteom. Bioinform. 2014, 7, 310–326. [Google Scholar]
- Dvorianinova, E.M.; Zinovieva, O.L.; Pushkova, E.N.; Zhernova, D.A.; Rozhmina, T.A.; Povkhova, L.V.; Novakovskiy, R.O.; Sigova, E.A.; Turba, A.A.; Borkhert, E.V.; et al. Key FAD2, FAD3, and SAD Genes Involved in the Fatty Acid Synthesis in Flax Identified Based on Genomic and Transcriptomic Data. Int. J. Mol. Sci. 2023, 24, 14885. [Google Scholar] [CrossRef]
- Rajwade, A.V.; Kadoo, N.Y.; Borikar, S.P.; Harsulkar, A.M.; Ghorpade, P.B.; Gupta, V.S. Differential transcriptional activity of SAD, FAD2 and FAD3 desaturase genes in developing seeds of linseed contributes to varietal variation in alpha-linolenic acid content. Phytochemistry 2014, 98, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Thambugala, D.; Cloutier, S. Fatty acid composition and desaturase gene expression in flax (Linum usitatissimum L.). J. Appl. Genet. 2014, 55, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Rajwade, A.V.; Joshi, R.S.; Kadoo, N.Y.; Gupta, V.S. Sequence characterization and in silico structure prediction of fatty acid desaturases in linseed varieties with differential fatty acid composition. J. Sci. Food Agric. 2016, 96, 4896–4906. [Google Scholar] [CrossRef] [PubMed]
- Thambugala, D.; Duguid, S.; Loewen, E.; Rowland, G.; Booker, H.; You, F.M.; Cloutier, S. Genetic variation of six desaturase genes in flax and their impact on fatty acid composition. TAG. Theor. Appl. Genet. Theor. Und Angew. Genet. 2013, 126, 2627–2641. [Google Scholar] [CrossRef] [PubMed]
- Porokhovinova, E.A.; Shelenga, T.V.; Matveeva, T.V.; Pavlov, A.V.; Grigorieva, E.A.; Brutch, N.B. Polymorphism of genes controlling low level of linolenic acid in lines from VIR flax genetic collection. Ecol. Genet. 2019, 17, 5–19. [Google Scholar] [CrossRef]
- Povkhova, L.V.; Pushkova, E.N.; Rozhmina, T.A.; Zhuchenko, A.A.; Frykin, R.I.; Novakovskiy, R.O.; Dvorianinova, E.M.; Gryzunov, A.A.; Borkhert, E.V.; Sigova, E.A.; et al. Development and Complex Application of Methods for the Identification of Mutations in the FAD3A and FAD3B Genes Resulting in the Reduced Content of Linolenic Acid in Flax Oil. Plants 2022, 12, 95. [Google Scholar] [CrossRef] [PubMed]
- Radovanovic, N.; Thambugala, D.; Duguid, S.; Loewen, E.; Cloutier, S. Functional characterization of flax fatty acid desaturase FAD2 and FAD3 isoforms expressed in yeast reveals a broad diversity in activity. Mol. Biotechnol. 2014, 56, 609–620. [Google Scholar] [CrossRef]
- Xie, D.; Dai, Z.; Yang, Z.; Tang, Q.; Deng, C.; Xu, Y.; Wang, J.; Chen, J.; Zhao, D.; Zhang, S.; et al. Combined genome-wide association analysis and transcriptome sequencing to identify candidate genes for flax seed fatty acid metabolism. Plant Sci. Int. J. Exp. Plant Biol. 2019, 286, 98–107. [Google Scholar] [CrossRef] [PubMed]
- You, F.M.; Xiao, J.; Li, P.; Yao, Z.; Jia, G.; He, L.; Kumar, S.; Soto-Cerda, B.; Duguid, S.D.; Booker, H.M.; et al. Genome-Wide Association Study and Selection Signatures Detect Genomic Regions Associated with Seed Yield and Oil Quality in Flax. Int. J. Mol. Sci. 2018, 19, 2303. [Google Scholar] [CrossRef] [PubMed]
- Marketta, S.; Juha-Matti, P.; Merja, E.; Ari, K.; Lauri, J.; Veli, H. Yield, SDG lignan, cadmium, lead, oil and protein contents of linseed (Linum usitatissimum L.) cultivated in trials and at different farm conditions in the south-western part of Finland. Agric. Food Sci. 2013, 22, 296–306. [Google Scholar] [CrossRef]
- Dybing, C.D.; Zimmerman, D.C. Fatty Acid accumulation in maturing flaxseeds as influenced by environment. Plant Physiol. 1966, 41, 1465–1470. [Google Scholar] [CrossRef]
- Fofana, B.; Cloutier, S.; Duguid, S.; Ching, J.; Rampitsch, C. Gene expression of stearoyl-ACP desaturase and delta12 fatty acid desaturase 2 is modulated during seed development of flax (Linum usitatissimum). Lipids 2006, 41, 705–712. [Google Scholar] [CrossRef]
- Khadake, R.; Khonde, V.; Mhaske, V.; Ranjekar, P.; Harsulkar, A. Functional and bioinformatic characterisation of sequence variants of Fad3 gene from flax. J. Sci. Food Agric. 2011, 91, 2689–2696. [Google Scholar] [CrossRef]
- Dash, P.K.; Rai, R.; Mahato, A.K.; Gaikwad, K.; Singh, N.K. Transcriptome Landscape at Different Developmental Stages of a Drought Tolerant Cultivar of Flax (Linum usitatissimum). Front. Chem. 2017, 5, 82. [Google Scholar] [CrossRef]
- Dmitriev, A.A.; Kudryavtseva, A.V.; Krasnov, G.S.; Koroban, N.V.; Speranskaya, A.S.; Krinitsina, A.A.; Belenikin, M.S.; Snezhkina, A.V.; Sadritdinova, A.F.; Kishlyan, N.V.; et al. Gene expression profiling of flax (Linum usitatissimum L.) under edaphic stress. BMC Plant Biol. 2016, 16, 237. [Google Scholar] [CrossRef]
- Galindo-Gonzalez, L.; Deyholos, M.K. RNA-seq Transcriptome Response of Flax (Linum usitatissimum L.) to the Pathogenic Fungus Fusarium oxysporum f. sp. lini. Front. Plant Sci. 2016, 7, 1766. [Google Scholar] [CrossRef]
- Dmitriev, A.A.; Krasnov, G.S.; Rozhmina, T.A.; Novakovskiy, R.O.; Snezhkina, A.V.; Fedorova, M.S.; Yurkevich, O.Y.; Muravenko, O.V.; Bolsheva, N.L.; Kudryavtseva, A.V.; et al. Differential gene expression in response to Fusarium oxysporum infection in resistant and susceptible genotypes of flax (Linum usitatissimum L.). BMC Plant Biol. 2017, 17, 253. [Google Scholar] [CrossRef] [PubMed]
- Dmitriev, A.A.; Novakovskiy, R.O.; Pushkova, E.N.; Rozhmina, T.A.; Zhuchenko, A.A.; Bolsheva, N.L.; Beniaminov, A.D.; Mitkevich, V.A.; Povkhova, L.V.; Dvorianinova, E.M.; et al. Transcriptomes of Different Tissues of Flax (Linum usitatissimum L.) Cultivars With Diverse Characteristics. Front. Genet. 2020, 11, 565146. [Google Scholar] [CrossRef]
- Dmitriev, A.A.; Krasnov, G.S.; Rozhmina, T.A.; Zyablitsin, A.V.; Snezhkina, A.V.; Fedorova, M.S.; Pushkova, E.N.; Kezimana, P.; Novakovskiy, R.O.; Povkhova, L.V.; et al. Flax (Linum usitatissimum L.) response to non-optimal soil acidity and zinc deficiency. BMC Plant Biol. 2019, 19, 54. [Google Scholar] [CrossRef]
- Petrova, A.; Kozlova, L.; Gorshkov, O.; Nazipova, A.; Ageeva, M.; Gorshkova, T. Cell Wall Layer Induced in Xylem Fibers of Flax Upon Gravistimulation Is Similar to Constitutively Formed Cell Walls of Bast Fibers. Front. Plant Sci. 2021, 12, 660375. [Google Scholar] [CrossRef]
- Gorshkov, O.; Chernova, T.; Mokshina, N.; Gogoleva, N.; Suslov, D.; Tkachenko, A.; Gorshkova, T. Intrusive Growth of Phloem Fibers in Flax Stem: Integrated Analysis of miRNA and mRNA Expression Profiles. Plants 2019, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Gorshkova, T.; Chernova, T.; Mokshina, N.; Gorshkov, V.; Kozlova, L.; Gorshkov, O. Transcriptome Analysis of Intrusively Growing Flax Fibers Isolated by Laser Microdissection. Sci. Rep. 2018, 8, 14570. [Google Scholar] [CrossRef] [PubMed]
- Gorshkov, O.; Mokshina, N.; Ibragimova, N.; Ageeva, M.; Gogoleva, N.; Gorshkova, T. Phloem fibres as motors of gravitropic behaviour of flax plants: Level of transcriptome. Funct. Plant Biol. 2018, 45, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Gorshkov, O.; Mokshina, N.; Gorshkov, V.; Chemikosova, S.; Gogolev, Y.; Gorshkova, T. Transcriptome portrait of cellulose-enriched flax fibres at advanced stage of specialization. Plant Mol. Biol. 2017, 93, 431–449. [Google Scholar] [CrossRef]
- Gao, P.; Qiu, S.; Ma, X.; Parkin, I.A.P.; Xiang, D.; Datla, R. Spatiotemporal Transcriptomic Atlas of Developing Embryos and Vegetative Tissues in Flax. Plants 2022, 11, 2031. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Zou, Y.; Guo, X.; Li, H.; Lu, H. Fatty acid desaturases (FADs) modulate multiple lipid metabolism pathways to improve plant resistance. Mol. Biol. Rep. 2022, 49, 9997–10011. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Liu, F.; Sun, X.; Wang, B.; Liu, J.; Ni, X.; Hu, C.; Deng, G.; Tong, Z.; Zhang, Y.; et al. Genome-wide identification of FAD gene family and their contributions to the temperature stresses and mutualistic and parasitic fungi colonization responses in banana. Int. J. Biol. Macromol. 2022, 204, 661–676. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wei, J.; He, L.; Zhang, Y.; Zhao, Y.; Xu, X.; Wei, Y.; Ge, S.; Ding, D.; Liu, M.; et al. Identification of Fatty Acid Desaturases in Maize and Their Differential Responses to Low and High Temperature. Genes 2019, 10, 445. [Google Scholar] [CrossRef] [PubMed]
- Yurchenko, O.P.; Park, S.; Ilut, D.C.; Inmon, J.J.; Millhollon, J.C.; Liechty, Z.; Page, J.T.; Jenks, M.A.; Chapman, K.D.; Udall, J.A.; et al. Genome-wide analysis of the omega-3 fatty acid desaturase gene family in Gossypium. BMC Plant Biol. 2014, 14, 312. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.J.; Cao, N.; Zhang, Z.G.; Shang, Q.M. Characterization of the Fatty Acid Desaturase Genes in Cucumber: Structure, Phylogeny, and Expression Patterns. PLoS ONE 2016, 11, e0149917. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, S.; Zoong-Lwe, Z.S.; Gandhi, N.; Welti, R.; Fallen, B.; Smith, J.R.; Rustgi, S. Comparative Lipidomic Analysis Reveals Heat Stress Responses of Two Soybean Genotypes Differing in Temperature Sensitivity. Plants 2020, 9, 457. [Google Scholar] [CrossRef]
- D’Angeli, S.; Altamura, M.M. Unsaturated Lipids Change in Olive Tree Drupe and Seed during Fruit Development and in Response to Cold-Stress and Acclimation. Int. J. Mol. Sci. 2016, 17, 1889. [Google Scholar] [CrossRef]
- Wang, L.; Stegemann, J.P. Extraction of high quality RNA from polysaccharide matrices using cetyltrimethylammonium bromide. Biomaterials 2010, 31, 1612–1618. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Krasnov, G.S.; Dmitriev, A.A.; Kudryavtseva, A.V.; Shargunov, A.V.; Karpov, D.S.; Uroshlev, L.A.; Melnikova, N.V.; Blinov, V.M.; Poverennaya, E.V.; Archakov, A.I.; et al. PPLine: An Automated Pipeline for SNP, SAP, and Splice Variant Detection in the Context of Proteogenomics. J. Proteome Res. 2015, 14, 3729–3737. [Google Scholar] [CrossRef]
- Dmitriev, A.A.; Pushkova, E.N.; Novakovskiy, R.O.; Beniaminov, A.D.; Rozhmina, T.A.; Zhuchenko, A.A.; Bolsheva, N.L.; Muravenko, O.V.; Povkhova, L.V.; Dvorianinova, E.M.; et al. Genome Sequencing of Fiber Flax Cultivar Atlant Using Oxford Nanopore and Illumina Platforms. Front. Genet. 2020, 11, 590282. [Google Scholar] [CrossRef]

| Variety | OLE, % | LIO, % | LIN, % | FAD3a Mutation G to A (Lu7:16092348) | FAD3b Mutation C to T (Lu12:1035655) | FAD3a Mutation C to T (Lu7:16090340) |
|---|---|---|---|---|---|---|
| AGT 427 | 13.1 | 13.4 | 64.3 | − | − | − |
| Atalante | 16.4 | 14.7 | 58.2 | − | − | − |
| Entre-Rios | 20.1 | 16.5 | 52.5 | − | − | − |
| Norlin | 22.2 | 14.5 | 54.3 | − | − | − |
| Pechersky kryazh | 31.8 | 10.2 | 52.0 | − | − | − |
| Raciol | 15.5 | 39.2 | 35.0 | − | + | − |
| AGT 422 | 18.3 | 35.5 | 32.6 | − | + | − |
| AGT 981 | 18.5 | 67.2 | 3.2 | + | + | − |
| AGT 1535 | 18.8 | 64.0 | 4.9 | + | + | − |
| Lola | 12.9 | 68.0 | 9.4 | − | − | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pushkova, E.N.; Povkhova, L.V.; Dvorianinova, E.M.; Novakovskiy, R.O.; Rozhmina, T.A.; Gryzunov, A.A.; Sigova, E.A.; Zhernova, D.A.; Borkhert, E.V.; Turba, A.A.; et al. Expression of FAD and SAD Genes in Developing Seeds of Flax Varieties under Different Growth Conditions. Plants 2024, 13, 956. https://doi.org/10.3390/plants13070956
Pushkova EN, Povkhova LV, Dvorianinova EM, Novakovskiy RO, Rozhmina TA, Gryzunov AA, Sigova EA, Zhernova DA, Borkhert EV, Turba AA, et al. Expression of FAD and SAD Genes in Developing Seeds of Flax Varieties under Different Growth Conditions. Plants. 2024; 13(7):956. https://doi.org/10.3390/plants13070956
Chicago/Turabian StylePushkova, Elena N., Liubov V. Povkhova, Ekaterina M. Dvorianinova, Roman O. Novakovskiy, Tatiana A. Rozhmina, Aleksey A. Gryzunov, Elizaveta A. Sigova, Daiana A. Zhernova, Elena V. Borkhert, Anastasia A. Turba, and et al. 2024. "Expression of FAD and SAD Genes in Developing Seeds of Flax Varieties under Different Growth Conditions" Plants 13, no. 7: 956. https://doi.org/10.3390/plants13070956
APA StylePushkova, E. N., Povkhova, L. V., Dvorianinova, E. M., Novakovskiy, R. O., Rozhmina, T. A., Gryzunov, A. A., Sigova, E. A., Zhernova, D. A., Borkhert, E. V., Turba, A. A., Yablokov, A. G., Bolsheva, N. L., Dmitriev, A. A., & Melnikova, N. V. (2024). Expression of FAD and SAD Genes in Developing Seeds of Flax Varieties under Different Growth Conditions. Plants, 13(7), 956. https://doi.org/10.3390/plants13070956








