Genome-Wide Analysis of Trehalose-6-Phosphate Phosphatase Gene Family and Their Expression Profiles in Response to Abiotic Stress in Groundnut
Abstract
:1. Introduction
2. Results
2.1. Identification of AhTPP Gene Family Members in Peanut
2.2. Phylogenetic Analysis and Chromosomal Locations of the AhTPP Gene Family
2.3. Gene Structure and Conserved Motif Analysis of the AhTPP Family
2.4. Collinear Analysis of AhTPP Genes
2.5. Prediction of Cis-Acting Elements in the Promoter Region of AhTPPs
2.6. Expression Profiles of AhTPP Genes in Different Tissues and under Abiotic Stress Treatments
2.7. Subcellular Localization and Functional Characterization of AhTPPs
3. Discussion
4. Materials and Methods
4.1. Plant Material and Stress Conditions
4.2. Identification and Characterization of TPP Genes in Peanuts
4.3. Phylogenetic Analysis
4.4. Gene Structure and Chromosomal Distribution
4.5. Gene Duplication and Collinearity Analysis
4.6. Analysis of Cis-Acting Elements and Expression Profiles of AhTPPs
4.7. RNA Extraction and RT-qPCR Based Gene Expression Analysis
4.8. Subcellular Localization
4.9. Heterologous Expression of AhTPP in Yeast
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raza, A.; Bhardwaj, S.; Atikur Rahman, M.; García-Caparrós, P.; Habib, M.; Saeed, F.; Charagh, S.; Foyer, C.H.; Siddique, K.H.M.; Varshney, R.K. Trehalose: A sugar molecule involved in temperature stress management in plants. Crop J. 2023, 12, 1–16. [Google Scholar] [CrossRef]
- Lunn, J.E. Gene families and evolution of trehalose metabolism in plants. Funct. Plant Biol. 2007, 34, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose metabolism in plants. Plant J. 2014, 79, 544–567. [Google Scholar] [CrossRef] [PubMed]
- Kerbler, S.M.L.; Armijos-Jaramillo, V.; Lunn, J.E.; Vicente, R. The trehalose 6-phosphate phosphatase family in plants. Physiol. Plant. 2023, 175, e14096. [Google Scholar] [CrossRef] [PubMed]
- Ponnu, J.; Wahl, V.; Schmid, M. Trehalose-6-phosphate: Connecting plant metabolism and development. Front. Plant Sci. 2011, 2, 70. [Google Scholar] [CrossRef] [PubMed]
- Wahl, V.; Ponnu, J.; Schlereth, A.; Arrivault, S.; Langenecker, T.; Franke, A.; Feil, R.; Lunn, J.E.; Stitt, M.; Schmid, M. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 2013, 339, 704–707. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, C.M.; Lunn, J.E. A tale of two sugars: Trehalose 6-phosphate and sucrose. Plant Physiol. 2016, 172, 7–27. [Google Scholar] [CrossRef] [PubMed]
- Carillo, P.; Feil, R.; Gibon, Y.; Satoh-Nagasawa, N.; Jackson, D.; Bläsing, O.E.; Stitt, M.; Lunn, J.E. A fluorometric assay for trehalose in the picomole range. Plant Methods 2013, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.U.; Nawaz, M.; Shah, A.N.; Raza, A.; Barbanti, L.; Skalicky, M.; Hashem, M.; Brestic, M.; Pandey, S.; Alamri, S.; et al. Trehalose: A key player in plant growth regulation and tolerance to abiotic stresses. J. Plant Growth Regul. 2023, 42, 4935–4957. [Google Scholar] [CrossRef]
- Liu, T.; Han, Y.Q.; Shi, J.L.; Liang, A.D.; Xu, D.D.; Ye, X.L.; Qi, H.Y. Abscisic acid involved in trehalose improved melon photosynthesis via regulating oxidative stress tolerance and cell morphology structure under cold stress. Environ. Exp. Bot. 2022, 202, 105042. [Google Scholar] [CrossRef]
- Zhang, Z.W.; Sun, M.; Gao, Y.M.; Luo, Y. Exogenous trehalose differently improves photosynthetic carbon assimilation capacities in maize and wheat under heat stress. J. Plant Interact. 2022, 17, 361–370. [Google Scholar] [CrossRef]
- Li, Z.G.; Luo, L.J.; Zhu, L.P. Involvement of trehalose in hydrogen sulfide donor sodium hydrosulfide-induced the acquisition of heat tolerance in maize (Zea mays L.) seedlings. Bot. Stud. 2014, 55, 20. [Google Scholar] [CrossRef] [PubMed]
- Avonce, N.; Mendoza-Vargas, A.; Morett, E.; Iturriaga, G. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 2006, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.T.; Wang, M.Y.; Huang, Y.S.; Zhu, P.; Qian, G.T.; Zhang, Y.M.; Liu, Y.Q.; Zhou, J.W.; Li, L.X. Genome-wide identification and analysis of stress response of trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase Genes in Quinoa. Int. J. Mol. Sci. 2023, 24, 6950. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ouyang, K.; Wang, K. Genome-wide identification, evolution, and expression analysis of TPS and TPP gene families in Brachypodium distachyon. Plants 2019, 8, 362. [Google Scholar] [CrossRef] [PubMed]
- Avonce, N.; Wuyts, J.; Verschooten, K.; Vandesteene, L.; Van Dijck, P. The Cytophaga hutchinsonii ChTPSP: First characterized bifunctional TPS–TPP protein as putative ancestor of all eukaryotic trehalose biosynthesis proteins. Mol. Biol. Evol. 2010, 27, 359–369. [Google Scholar] [CrossRef]
- Yang, H.L.; Liu, Y.J.; Wang, C.L.; Zeng, Q.Y. Molecular evolution of trehalose-6-phosphate synthase (TPS) gene family in Populus, Arabidopsis and Rice. PLoS ONE 2012, 7, e42438. [Google Scholar] [CrossRef] [PubMed]
- Vandesteene, L.; Lopez-Galvis, L.; Vanneste, K.; Feil, R.; Maere, S.; Lammens, W.; Rolland, F.; Lunn, J.E.; Avonce, N.; Beeckman, T.; et al. Expansive evolution of the trehalose-6-phosphate phosphatase gene family in Arabidopsis. Plant Physiol. 2012, 160, 884–896. [Google Scholar] [CrossRef] [PubMed]
- Du, L.Y.; Li, S.M.; Ding, L.; Cheng, X.X.; Kang, Z.S.; Mao, H.D. Genome-wide analysis of trehalose-6-phosphate phosphatases (TPP) gene family in wheat indicates their roles in plant development and stress response. BMC Plant Biol. 2022, 22, 120. [Google Scholar] [CrossRef]
- Lin, Q.F.; Wang, S.; Dao, Y.H.; Wang, J.Y.; Wang, K. Arabidopsis thaliana trehalose-6-phosphate phosphatase gene TPPI enhances drought tolerance by regulating stomatal apertures. J. Exp. Bot. 2020, 71, 4285–4297. [Google Scholar] [CrossRef]
- Ge, L.F.; Chao, D.Y.; Shi, M.; Zhu, M.Z.; Gao, J.P.; Lin, H.X. Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 2008, 228, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.G.; Chen, W.T.; Gao, J.; Yang, F.; Zhuang, C.X. Overexpression of the trehalose-6-phosphate phosphatase OsTPP3 increases drought tolerance in rice. Plant Biotechnol. Rep. 2019, 13, 285–292. [Google Scholar] [CrossRef]
- Nuccio, M.L.; Wu, J.; Mowers, R.; Zhou, H.P.; Meghji, M.; Primavesi, L.F.; Paul, M.J.; Chen, X.; Gao, Y.; Haque, E.; et al. Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat. Biotechnol. 2015, 33, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Arya, S.S.; Salve, A.R.; Chauhan, S. Peanuts as functional food: A review. J. Food Sci. Technol. 2016, 53, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.H.; Li, H.Q.; Gao, C.J.; Yu, W.C.; Zhang, S.C. Advances in omics research on peanut response to biotic stresses. Front. Plant Sci. 2023, 14, 1101994. [Google Scholar] [CrossRef] [PubMed]
- Puppala, N.; Nayak, S.N.; Sanz-Saez, A.; Chen, C.; Devi, M.J.; Nivedita, N.; Bao, Y.; He, G.H.; Traore, S.M.; Wright, D.A.; et al. Sustaining yield and nutritional quality of peanuts in harsh environments: Physiological and molecular basis of drought and heat stress tolerance. Front. Genet. 2023, 14, 1121462. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, W.J.; Chen, H.; Yang, M.; Wang, J.P.; Pandey, M.K.; Zhang, C.; Chang, W.C.; Zhang, L.S.; Zhang, X.T.; Tang, R.H.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; Han, Z.K.; Chen, Y.N.; Huai, D.X.; Kang, Y.P.; Wang, Z.H.; Yan, L.Y.; Jiang, H.F.; Lei, Y.; et al. Integrated transcriptomics and metabolomics analysis reveal key metabolism pathways contributing to cold tolerance in peanut. Front. Plant Sci. 2021, 12, 752474. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; Yao, R.N.; Ouyang, L.; Yu, T.T.; Yan, L.Y.; Chen, Y.N.; Huai, D.X.; Wang, Z.H.; Kang, Y.P.; et al. A systematic identification of cold tolerance genes in peanut using yeast functional screening system. Oil Crop Sci. 2023, 8, 184–190. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.J.; Zhang, X.L.; Zhou, Z.H.; Ma, Y.; Chu, D.; Wang, L.; Yang, Y.M.; Du, L.; Du, Y.L.; Du, J.D.; et al. Genome-and transcriptome-wide identification of trehalose-6-phosphate phosphatases (TPP) gene family and their expression patterns under abiotic stress and exogenous trehalose in soybean. BMC Plant Biol. 2023, 23, 641. [Google Scholar] [CrossRef]
- Wang, W.P.; Cui, H.; Xiao, X.F.; Wu, B.J.; Sun, J.L.; Zhang, Y.X.; Yang, Q.Y.; Zhao, Y.P.; Liu, G.X.; Qin, T.F. Genome-wide identification of cotton (Gossypium spp.) trehalose-6-phosphate phosphatase (TPP) gene family members and the role of GhTPP22 in the response to drought stress. Plants 2022, 11, 1079. [Google Scholar] [CrossRef]
- Krasensky, J.; Broyart, C.; Rabanal, F.A.; Jonak, C. The redox-sensitive chloroplast trehalose-6-phosphate phosphatase AtTPPD regulates salt stress tolerance. Antioxid. Redox Signal. 2014, 21, 1289–1304. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.C.M.; Hejazi, M.; Fettke, J.; Steup, M.; Feil, R.; Krause, U.; Arrivault, S.; Vosloh, D.; Figueroa, C.M.; Ivakov, A.; et al. Feedback inhibition of starch degradation in Arabidopsis leaves mediated by trehalose 6-phosphate. Plant Physiol. 2013, 163, 1142–1163. [Google Scholar] [CrossRef]
- Van Houtte, H.; López-Galvis, L.; Vandesteene, L.; Beeckman, T.; Van Dijck, P. Redundant and non-redundant roles of the trehalose-6-phosphate phosphatases in leaf growth, root hair specification and energy-responses in Arabidopsis. Plant Signal. Behav. 2013, 8, e23209. [Google Scholar] [CrossRef]
- Henry, C.; Bledsoe, S.W.; Griffiths, C.A.; Kollman, A.; Paul, M.J.; Sakr, S.; Lagrimini, L.M. Differential role for trehalose metabolism in salt-stressed maize. Plant Physiol. 2015, 169, 1072–1089. [Google Scholar] [CrossRef]
- Williams, B.; Njaci, I.; Moghaddam, L.; Long, H.; Dickman, M.B.; Zhang, X.R.; Mundree, S. Trehalose accumulation triggers autophagy during plant desiccation. PLoS Genet. 2015, 11, e1005705. [Google Scholar] [CrossRef]
- Lin, Q.F.; Yang, J.; Wang, Q.L.; Zhu, H.; Chen, Z.Y.; Dao, Y.H.; Wang, K. Overexpression of the trehalose-6-phosphate phosphatase family gene AtTPPF improves the drought tolerance of Arabidopsis thaliana. BMC Plant Biol. 2019, 19, 381. [Google Scholar] [CrossRef]
- Joshi, R.; Sahoo, K.K.; Singh, A.K.; Anwar, K.; Pundir, P.; Gauta, R.K.; Krishnamurthy, S.L.; Sopory, S.K.; Pareek, A.; Singla-Pareek, S.L. Enhancing trehalose biosynthesis improves yield potential in marker-free transgenic rice under drought, saline, and sodic conditions. J. Exp. Bot. 2020, 71, 653–668. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, L.; Liu, Y.; Yao, R.N.; He, D.L.; Yan, L.Y.; Chen, Y.N.; Huai, D.X.; Wang, Z.H.; Yu, B.L.; Kang, Y.P.; et al. Genome-wide analysis of UDP-glycosyltransferase gene family and identification of a flavonoid 7-O-UGT (AhUGT75A) enhancing abiotic stress in peanut (Arachis hypogaea L.). BMC Plant Biol. 2023, 23, 626. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Yi Chuan = Hered. 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Tang, H.B.; Debarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, D.P.; Zhang, Y.B.; Zhang, Z.; Zhu, J.; Yu, J. KaKs Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Clevenger, J.; Chu, Y.; Scheffler, B.; Ozias-Akins, P. A developmental transcriptome map for allotetraploid Arachis hypogaea. Front. Plant Sci. 2016, 7, 1446. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sun, B.; Zhang, F.; Xiao, N.; Jiang, M.; Yuan, Q.; Xue, S.L.; Miao, H.Y.; Chen, Q.; Li, M.Y.; Wang, X.R.; et al. An efficient mesophyll protoplast isolation, purification and PEG-mediated transient gene expression for subcellular localization in Chinese kale. Sci. Hortic. 2018, 241, 187–193. [Google Scholar] [CrossRef]
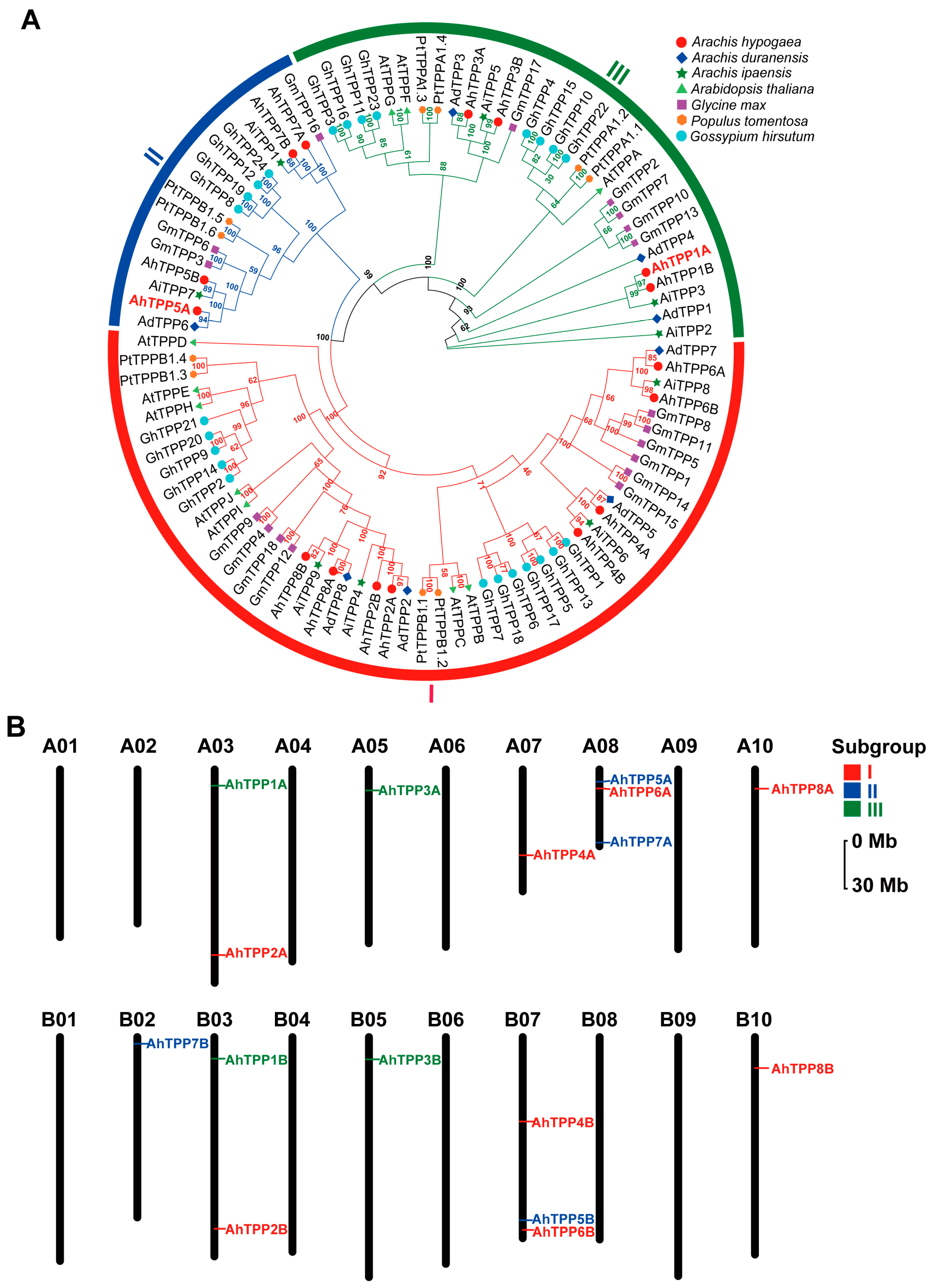
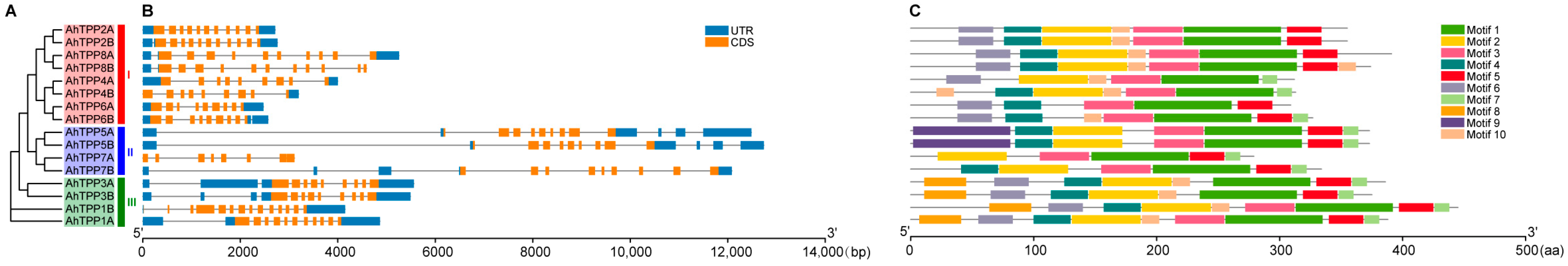

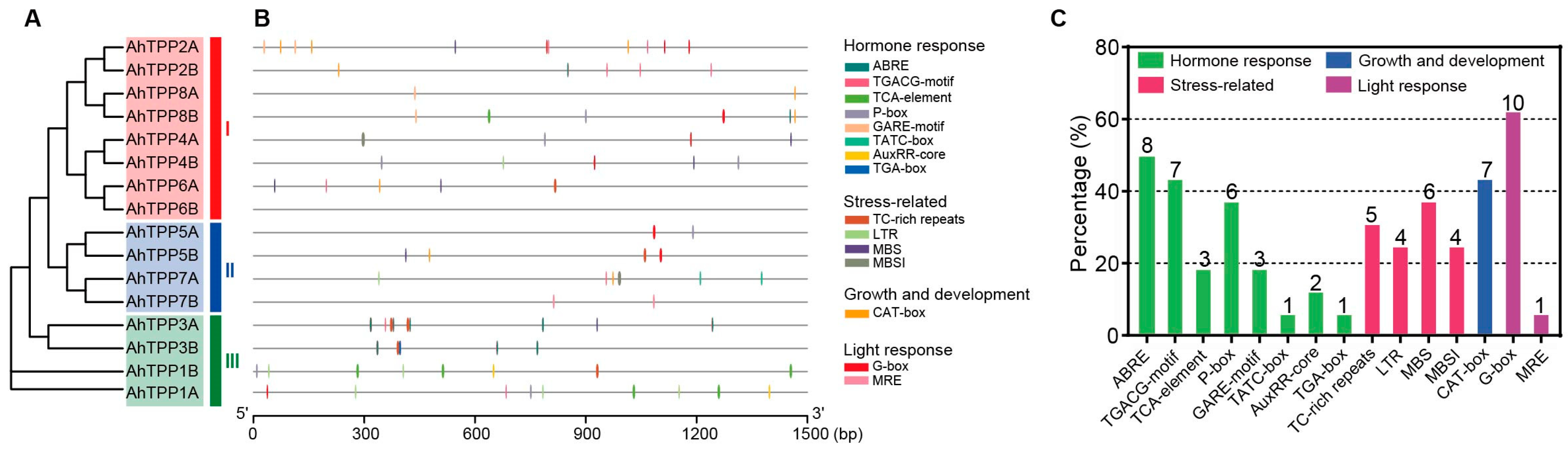



| Gene Name | Gene ID | Chr | Protein Length (aa) | pI | Molecular Weight (kDa) | GRAVY | Subcellular Location |
|---|---|---|---|---|---|---|---|
| AhTPP1A | Arahy.FIK9JS.1 | A03 | 388 | 6.57 | 43.21 | −0.275 | cytoplasm, nuclear |
| AhTPP2A | Arahy.P5P8C7.1 | A03 | 355 | 9.08 | 39.93 | −0.392 | apoplasm |
| AhTPP3A | Arahy.51Y4LP.1 | A05 | 386 | 6.22 | 43.30 | −0.381 | cytoplasm |
| AhTPP4A | Arahy.J3QXZL.1 | A07 | 312 | 9.31 | 35.41 | −0.366 | cytoplasm |
| AhTPP5A | Arahy.0FY2NM.1 | A08 | 373 | 9.22 | 42.41 | −0.332 | cytoplasm, nuclear |
| AhTPP6A | Arahy.B1753N.1 | A08 | 309 | 9.07 | 34.90 | −0.348 | apoplasm |
| AhTPP7A | Arahy.B2J19D.1 | A08 | 279 | 7.79 | 31.69 | −0.295 | cytoplasm |
| AhTPP8A | Arahy.SS1VDK.1 | A10 | 391 | 9.04 | 43.92 | −0.353 | apoplasm |
| AhTPP1B | Arahy.CK7SVG.1 | B03 | 445 | 7.59 | 49.87 | −0.256 | apoplasm |
| AhTPP2B | Arahy.FJ2B1F.1 | B03 | 355 | 9.22 | 39.95 | −0.393 | apoplasm |
| AhTPP3B | Arahy.5Q8BGE.1 | B05 | 375 | 6.03 | 42.05 | −0.410 | cytoplasm |
| AhTPP4B | Arahy.DP0G5T.1 | B07 | 313 | 9.48 | 35.21 | −0.296 | apoplasm |
| AhTPP5B | Arahy.G2WE73.1 | B07 | 373 | 9.33 | 42.31 | −0.343 | cytoplasm |
| AhTPP6B | Arahy.KW1U5A.1 | B07 | 327 | 9.18 | 36.74 | −0.378 | apoplasm |
| AhTPP7B | Arahy.U769C2.1 | B02 | 334 | 5.49 | 38.13 | −0.375 | cytoplasm |
| AhTPP8B | Arahy.S15BQI.1 | B10 | 374 | 9.12 | 41.95 | −0.472 | apoplasm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Wang, X.; Ouyang, L.; Yao, R.; Wang, Z.; Kang, Y.; Yan, L.; Chen, Y.; Huai, D.; Wang, Q.; et al. Genome-Wide Analysis of Trehalose-6-Phosphate Phosphatase Gene Family and Their Expression Profiles in Response to Abiotic Stress in Groundnut. Plants 2024, 13, 1056. https://doi.org/10.3390/plants13081056
Liu Y, Wang X, Ouyang L, Yao R, Wang Z, Kang Y, Yan L, Chen Y, Huai D, Wang Q, et al. Genome-Wide Analysis of Trehalose-6-Phosphate Phosphatase Gene Family and Their Expression Profiles in Response to Abiotic Stress in Groundnut. Plants. 2024; 13(8):1056. https://doi.org/10.3390/plants13081056
Chicago/Turabian StyleLiu, Yue, Xin Wang, Lei Ouyang, Ruonan Yao, Zhihui Wang, Yanping Kang, Liying Yan, Yuning Chen, Dongxin Huai, Qianqian Wang, and et al. 2024. "Genome-Wide Analysis of Trehalose-6-Phosphate Phosphatase Gene Family and Their Expression Profiles in Response to Abiotic Stress in Groundnut" Plants 13, no. 8: 1056. https://doi.org/10.3390/plants13081056





