Transcriptional Regulation and Gene Mapping of Internode Elongation and Late Budding in the Chinese Cabbage Mutant lcc
Abstract
:1. Introduction
2. Results
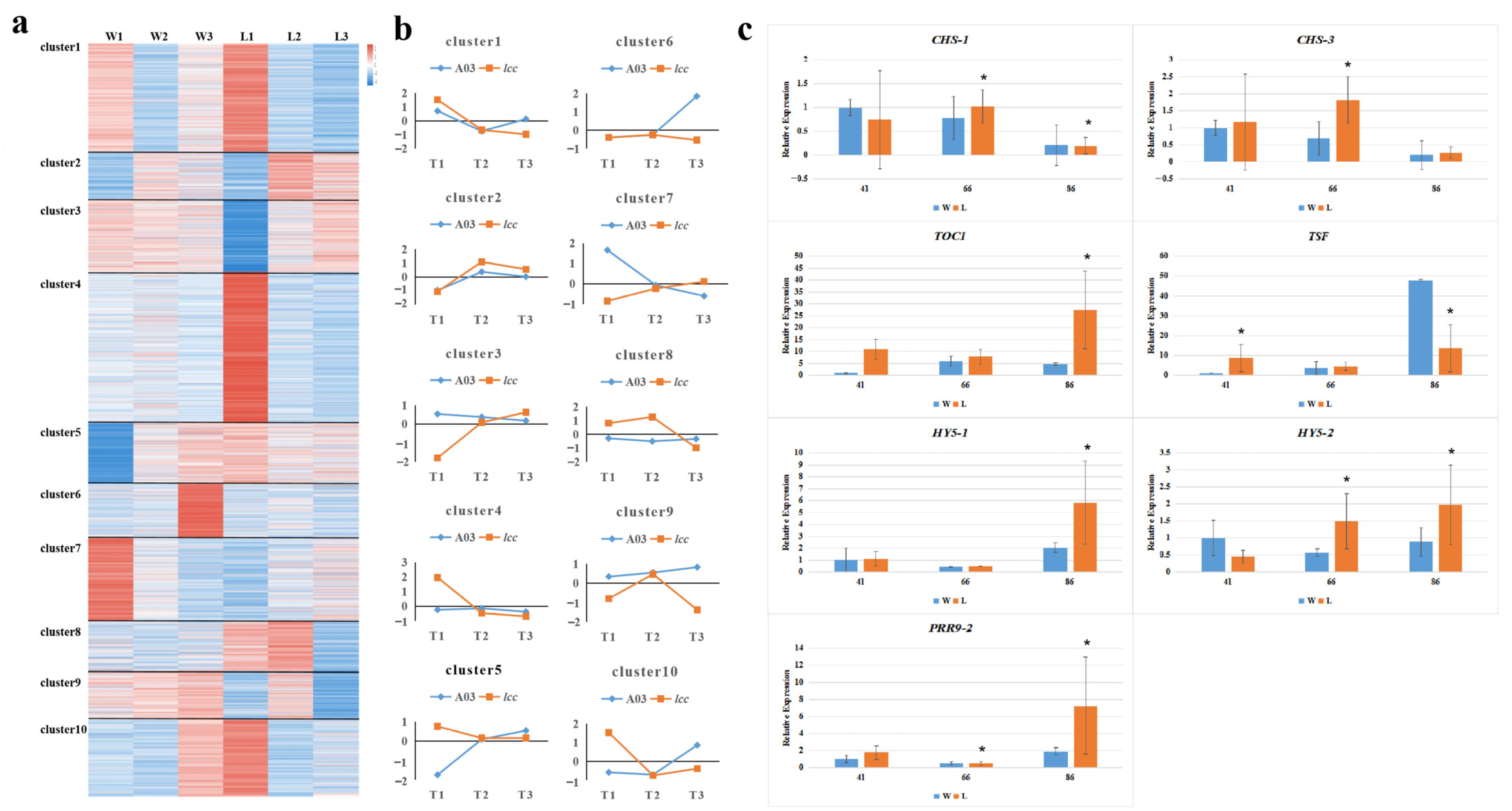
2.1. Transcriptome Analysis Reveals Genetic Factors Underlying Internode Length and Budding Time Regulation in Chinese Cabbage
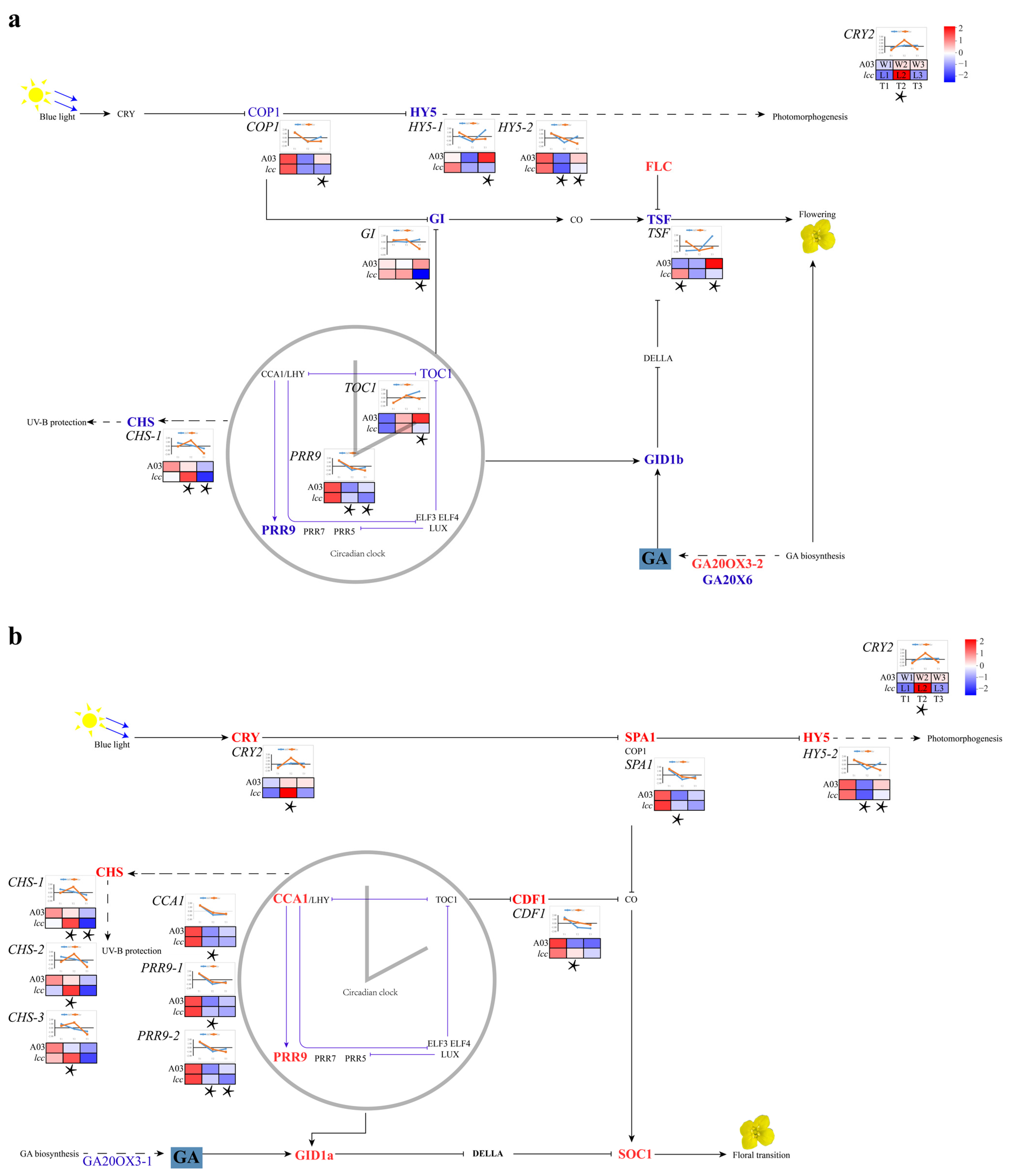
2.1.1. Plant Hormone Gene Transcripts Modulated Internode Growth
2.1.2. Expression Analysis of Genes Influencing Budding Time in Chinese Cabbage
2.2. QTL Analysis of Internode Length and Budding Time Traits
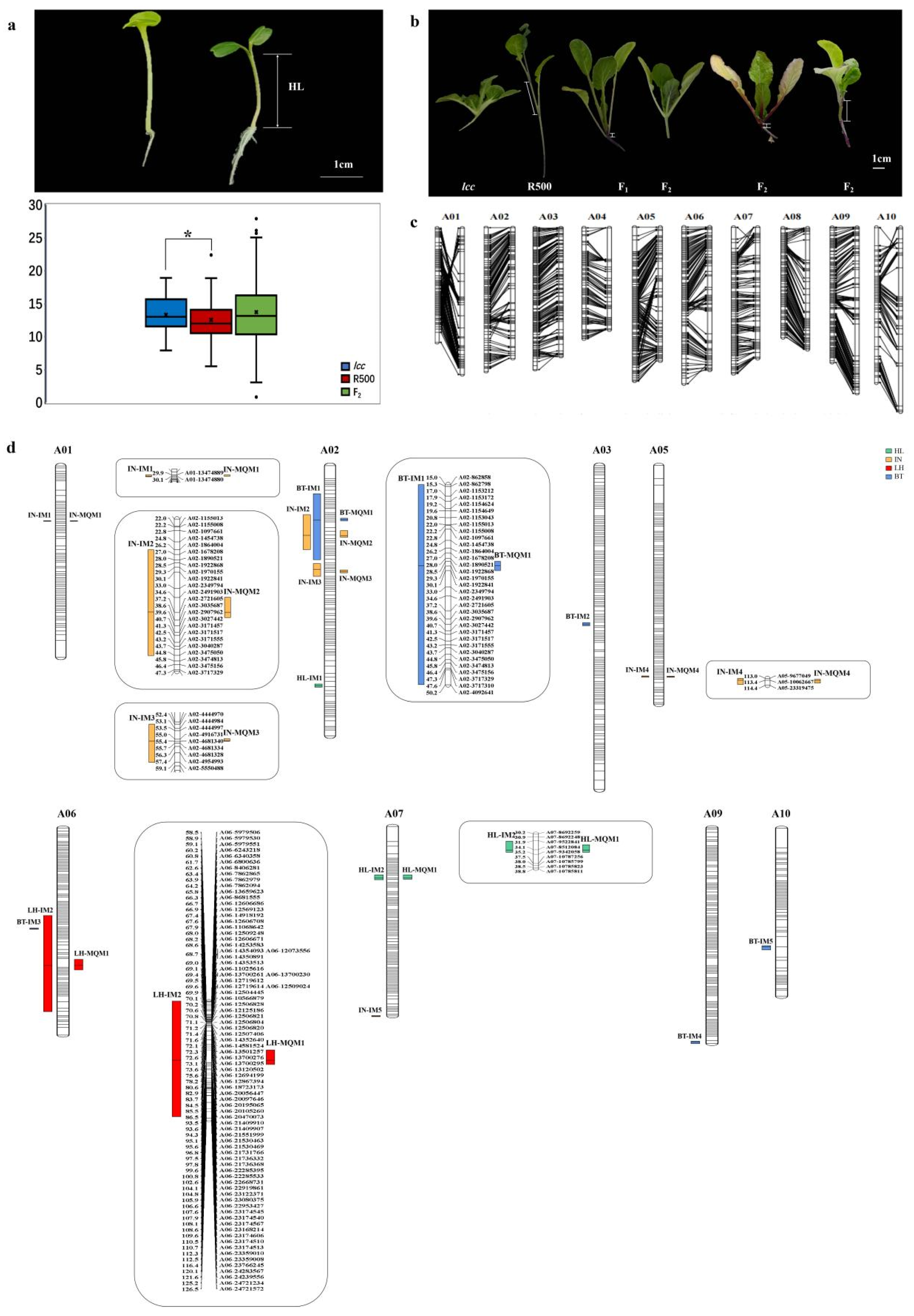
2.2.1. Phenotyping Data Analysis of Parental Lines and the F2 Population
2.2.2. A High-Density Genetic Linkage Map for the F2 Population
2.2.3. QTL Mapping
2.2.4. Differentially Expressed Genes within QTLs
3. Discussion
3.1. Hormone Genes Related to Internode Elongation
3.2. Link between Budding Time and the Biological Clock
4. Material and Methods
4.1. Plant Material and Growth Conditions for RNA-Seq
4.2. RNA Isolation and Sequencing
4.3. Bioinformatics Analysis of RNA-Seq Data
4.4. RT–qPCR
4.5. Plant Material, Growth Conditions, and Trait Measurements for QTLs
4.6. DNA Isolation and Resequencing
4.7. Sequencing and Alignment with the Reference Genome
4.8. Identification and Annotation of SNPs
4.9. Construction of a Genetic Linkage Map and QTL Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Franzke, A.; Lysak, M.A.; Al-Shehbaz, I.A.; Koch, M.A.; Mummenhoff, K. Cabbage family affairs: The evolutionary history of Brassicaceae. Trends Plant. Sci. 2011, 16, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Bahadur, B.; Pullaiah, T.; Krishnamurthy, K. Plant diversity, organization, function and improvement. In Plant Biology and Biotechnology; Springer: Berlin/Heidelberg, Germany, 2015; pp. 361–383. [Google Scholar]
- Wang, Y.; Li, J. Molecular basis of plant architecture. Annu. Rev. Plant. Biol. 2008, 59, 253–279. [Google Scholar] [CrossRef] [PubMed]
- Taiz, L.; Zeiger, E.; Moller, I.M.; Murphy, A. Plant Physiology and Development, 6th ed.; Sinauer Associates Incorporated: Sunderland, MA, USA, 2014. [Google Scholar]
- Sablowski, R.; Carnier Dornelas, M. Interplay between cell growth and cell cycle in plants. J. Exp. Bot. 2014, 65, 2703–2714. [Google Scholar] [CrossRef]
- Gray, W.M. Hormonal regulation of plant growth and development. PLoS Biol. 2004, 2, E311. [Google Scholar] [CrossRef] [PubMed]
- Nagai, K.; Mori, Y.; Ishikawa, S.; Furuta, T.; Gamuyao, R.; Niimi, Y.; Hobo, T.; Fukuda, M.; Kojima, M.; Takebayashi, Y.; et al. Antagonistic regulation of the gibberellic acid response during stem growth in rice. Nature 2020, 584, 109–114. [Google Scholar] [CrossRef]
- Lee, J.; Moon, S.; Jang, S.; Lee, S.; An, G.; Jung, K.H.; Park, S.K. OsbHLH073 negatively regulates internode elongation and plant height by modulating GA homeostasis in rice. Plants 2020, 9, 547. [Google Scholar] [CrossRef]
- Ikram, A.U.; Ding, Y.; Su, Y. OsARP6 is involved in internode elongation by regulating cell-cycle-related genes. Biomolecules 2021, 11, 1100. [Google Scholar] [CrossRef] [PubMed]
- Yamamuro, C.; Ihara, Y.; Wu, X.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 2000, 12, 1591–1606. [Google Scholar] [CrossRef]
- Ejaz, M.; Bencivenga, S.; Tavares, R.; Bush, M.; Sablowski, R. ARABIDOPSIS THALIANA HOMEOBOX GENE 1 controls plant architecture by locally restricting environmental responses. Proc. Natl. Acad. Sci. USA 2021, 118, e2018615118. [Google Scholar] [CrossRef]
- Devlin, P.F.; Patel, S.R.; Whitelam, G.C. Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 1998, 10, 1479–1487. [Google Scholar] [CrossRef] [PubMed]
- Patil, V.; McDermott, H.I.; McAllister, T.; Cummins, M.; Silva, J.C.; Mollison, E.; Meikle, R.; Morris, J.; Hedley, P.E.; Waugh, R.; et al. APETALA2 control of barley internode elongation. Development 2019, 146, dev170373. [Google Scholar] [CrossRef]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef]
- Denay, G.; Chahtane, H.; Tichtinsky, G.; Parcy, F. A flower is born: An update on Arabidopsis floral meristem formation. Curr. Opin. Plant. Biol. 2017, 35, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, H.; Zhou, D.; Li, C.; Ding, Q.; Yang, X.; Wang, F.; Zheng, H.; Gao, J. Genetic and transcriptome analysis of leaf trichome development in Chinese Cabbage (Brassica rapa L. subsp. pekinensis) and molecular marker development. Int. J. Mol. Sci. 2022, 23, 12721. [Google Scholar] [CrossRef] [PubMed]
- Schaller, G.E.; Street, I.H.; Kieber, J.J. Cytokinin and the cell cycle. Curr. Opin. Plant. Biol. 2014, 21, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Qin, L.; Lee, S.; Fu, X.; Richards, D.E.; Cao, D.; Luo, D.; Harberd, N.P.; Peng, J. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 2004, 131, 1055–1064. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Minami, A.; Takahashi, K.; Inoue, S.I.; Tada, Y.; Kinoshita, T. Brassinosteroid induces phosphorylation of the Plasma membrane H+-ATPase during hypocotyl elongation in Arabidopsis thaliana. Plant Cell Physiol. 2019, 60, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Bai, M.Y.; Oh, E.; Zhu, J.Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 2012, 46, 1701–1724. [Google Scholar] [CrossRef]
- Gomes, G.L.B.; Scortecci, K.C. Auxin and its role in plant development: Structure, signalling, regulation and response mechanisms. Plant Biol. 2021, 23, 894–904. [Google Scholar] [CrossRef]
- Petrásek, J.; Mravec, J.; Bouchard, R.; Blakeslee, J.J.; Abas, M.; Seifertová, D.; Wisniewska, J.; Tadele, Z.; Kubes, M.; Covanová, M.; et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 2006, 312, 914–918. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.X.; Webb, C.J.; Knowles, S.M.; Kim, S.H.; Wang, Z.; Tobin, E.M. CCA1 and ELF3 interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 2012, 158, 1079–1088. [Google Scholar] [CrossRef]
- Wang, H.; Lu, Y.; Gu, A.; Wang, Y.; Zhao, J.; Shen, S.; Feng, D. Expression analysis of key clock genes of lcc-1 mutant from Chinese cabbage under different photoperiods. J. Hebei Agric. Univ. 2019, 42, 21–28, (Chinese with English abstract). [Google Scholar]
- Khanna, R.; Shen, Y.; Toledo-Ortiz, G.; Kikis, E.A.; Johannesson, H.; Hwang, Y.S.; Quail, P.H. Functional profiling reveals that only a small number of phytochrome-regulated early-response genes in Arabidopsis are necessary for optimal deetiolation. Plant Cell 2006, 18, 2157–2171. [Google Scholar] [CrossRef] [PubMed]
- Burko, Y.; Seluzicki, A.; Zander, M.; Pedmale, U.V.; Ecker, J.R.; Chory, J. Chimeric Activators and Repressors Define HY5 Activity and Reveal a Light-Regulated Feedback Mechanism. Plant Cell 2020, 32, 967–983. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.; Peer, W.A.; Taiz, L. Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 2000, 211, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Xu, X.; Xu, J. Effects of vernalization temperature on flower bud differentiation and bolting of Chinese cabbage (Brassica campestris L. ssp. pekinensis). Acta Agric. Boreali-Sin. 2003, 24, 75–78, (Chinese with English abstract). [Google Scholar]
- Yu, R.; Su, T.; Yu, S.; Zhang, F.; Yu, Y.; Zhang, D.; Zhao, X.; Wang, W.; Lu, G. Effects of Plumule-vernalization and Seeding-vernalization on Bolting and Flowering Times of Different Chinese Cabbage Varieties. China Veg. 2016, 5, 27–32, (Chinese with English abstract). [Google Scholar]
- Chen, T.; Chen, X.; Zhang, S.; Zhu, J.; Tang, B.; Wang, A.; Dong, L.; Zhang, Z.; Yu, C.; Sun, Y.; et al. The Genome Sequence Archive Family: Toward Explosive Data Growth and Diverse Data Types. Genom. Proteom. Bioinform. 2021, 19, 578–583. [Google Scholar] [CrossRef]
- CNCB-NGDC Members and Partners. Database Resources of the National Genomics Data Center, China Natonal Center for Bioinformation in 2022. Nucleic Acids Res. 2022, 50, D27–D38. [Google Scholar] [CrossRef]
- Ma, Q.; Chen, C.; Zeng, Z.; Zou, Z.; Li, H.; Zhou, Q.; Chen, X.; Sun, K.; Li, X. Transcriptomic analysis between self- and cross-pollinated pistils of tea plants (Camellia sinensis). BMC Genom. 2018, 19, 289. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Li, X.; Lu, Y.; Wang, S.; Zhang, X.; Zhang, K.; Su, X.; Liu, M.; Feng, D.; Luo, S.; et al. Construction of a high-density mutant population of Chinese cabbage facilitates the genetic dissection of agronomic traits. Mol Plant. 2022, 15, 913–924. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.; Doyle, J. A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Ooijen, J.W.; Ooijen, J.W.; Verlaat, J.V.; Ooijen, J.W.; Tol, J.; Dalén, J.; Buren, J.B.; Meer, J.V.; Krieken, J.H.; Ooijen, J.W.; et al. JoinMap® 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma BV: Wageningen, The Netherlands, 2006. [Google Scholar]
- Shi, P.; Xu, Z.; Zhang, S.; Wang, X.; Ma, X.; Zheng, J.; Xing, L.; Zhang, D.; Ma, J.; Han, M.; et al. Construction of a high-density SNP-based genetic map and identification of fruit-related QTLs and candidate genes in peach [Prunus persica (L.) Batsch. BMC Plant. Biol. 2020, 20, 438. [Google Scholar] [CrossRef]

| Gene Name | Gene ID | Forward Primer | Reverse Primer |
|---|---|---|---|
| Actin | BraA02g003190.3C | GGAGCTGAGAGATTCCGTTG | GAACCACCACTGAGGACGAT |
| CHS-3 | BraA10g024990.3C | CGCGTGTGTTCTCTTCATATTGG | CAAGACCACTGTCTCTACGGTAAG |
| CHS-1 | BraA02g005190.3C | GAGGAAGTCTAAGGAAGATGGTGTG | TTAGACAGGAACGCTGTGTAGG |
| PRR9-2 | BraA05g001070.3C | GAGAAGCAAGATCAAACCACCAAG | GCTGCCTGGCTGTTCTCATA |
| HY5-1 | BraA02g003870.3C | AAGAGACCAAGCGGCTAAAGAG | CTCTAAGTCTTTCACTCTGGTCTCC |
| HY5-2 | BraA05g029990.3C | CGAGGGAGAGGAAGAAAGTGTATG | TTAGTGATTGTCGTCAGCTTTAGGC |
| TOC1 | BraA03g044360.3C | GTCATGTGCCTTTACAGAATGGTC | GCTTAGTCACTCTCACCTCGTT |
| TSF | BraA07g031650.3C | AACCCGCACCTTCGAGAATATC | GAACAATACCAGCACGAGACGA |
| Trait Name | Abbreviation | Measurement |
|---|---|---|
| Hypocotyl length | HL | Length of the hypocotyl (the part of a seedling below the cotyledon, above the root), measured using a ruler at the seedling stage (cm). |
| Internode length | IN | The distance between the base of the cotyledon petioles to the base of second true leaf petioles, scored as 1, looks like that of lcc or F1; 2, longer than that of F1; or 3, looks like that of R500. Measured by eye at the seedling stage. |
| Leaf hairs | LH | Determined with visual assessment. |
| Budding time | BT | Days after the first bud of the F2 population appeared (days). |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Xuan, S.; Zhao, J.; Li, H.; Lu, Y.; Li, R.; Wang, Y.; Shen, S.; Sun, X.; Feng, D. Transcriptional Regulation and Gene Mapping of Internode Elongation and Late Budding in the Chinese Cabbage Mutant lcc. Plants 2024, 13, 1083. https://doi.org/10.3390/plants13081083
Zhang Y, Xuan S, Zhao J, Li H, Lu Y, Li R, Wang Y, Shen S, Sun X, Feng D. Transcriptional Regulation and Gene Mapping of Internode Elongation and Late Budding in the Chinese Cabbage Mutant lcc. Plants. 2024; 13(8):1083. https://doi.org/10.3390/plants13081083
Chicago/Turabian StyleZhang, Yunqin, Shuxin Xuan, Jiaojiao Zhao, Hui Li, Yin Lu, Rui Li, Yanhua Wang, Shuxing Shen, Xiaoxue Sun, and Daling Feng. 2024. "Transcriptional Regulation and Gene Mapping of Internode Elongation and Late Budding in the Chinese Cabbage Mutant lcc" Plants 13, no. 8: 1083. https://doi.org/10.3390/plants13081083
APA StyleZhang, Y., Xuan, S., Zhao, J., Li, H., Lu, Y., Li, R., Wang, Y., Shen, S., Sun, X., & Feng, D. (2024). Transcriptional Regulation and Gene Mapping of Internode Elongation and Late Budding in the Chinese Cabbage Mutant lcc. Plants, 13(8), 1083. https://doi.org/10.3390/plants13081083





