GWAS in a Collection of Bulgarian Old and Modern Bread Wheat Accessions Uncovers Novel Genomic Loci for Grain Protein Content and Thousand Kernel Weight
Abstract
1. Introduction
2. Results
2.1. Phenotypic Variation
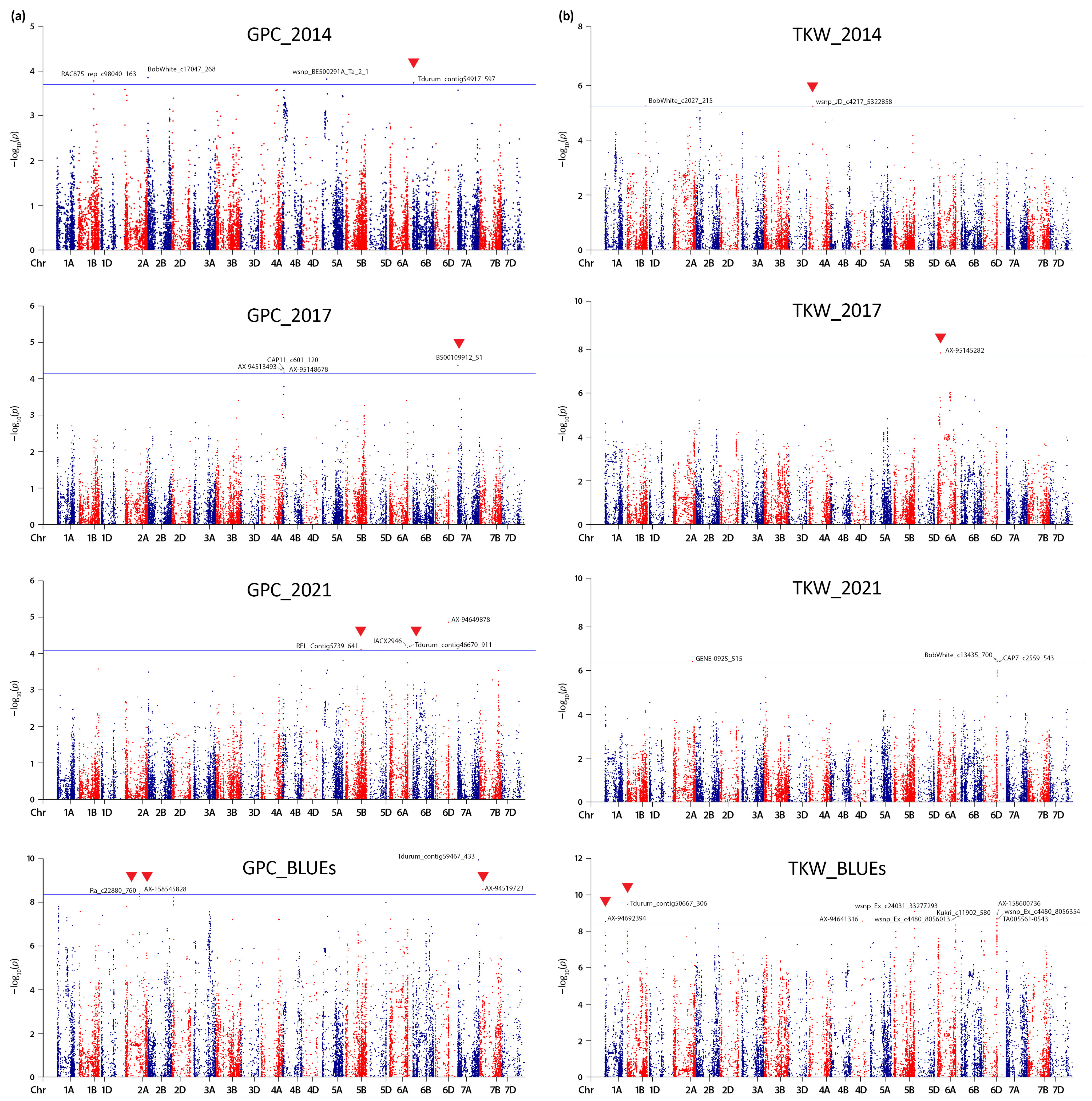
2.2. Linkage Disequilibrium (LD) Estimation, Genotype Relatedness, and Significant Quantitative Trait Nucleotides (QTNs)
2.3. Potential Candidate Genes
3. Discussion
3.1. Phenotypic Variation
3.2. Genomic Regions Associated with Grain Protein Content (GPC) and Thousand Kernel Weight (TKW)
3.3. Putative Candidate Genes Related to Grain Protein Content (GPC) and Thousand Kernel Weight (TKW)
4. Materials and Methods
4.1. Plant Material
4.2. Phenotyping
4.3. Statistical Analyses
4.4. Association Mapping and Candidate Gene Search
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Veraverbeke, W.S.; Delcour, J.A. Wheat protein composition and properties of wheat glutenin in relation to breadmaking functionality. Crit. Rev. Food Sci. Nutr. 2002, 42, 179–208. [Google Scholar] [CrossRef] [PubMed]
- Peña, R.J. Wheat for Bread and Other Foods. In Bread Wheat. Improvement and Production; FAO Plant Production and Protection Series No. 30; Curtis, B.S., Rajaram, S., Gómez Macpherson, H., Eds.; Food and Agriculture Organization of the United Nations: Rome, Italy, 2002; Available online: https://www.fao.org/3/y4011e/y4011e0w.htm#bm32 (accessed on 10 December 2023).
- Barneix, A.J. Physiology and biochemistry of source-regulated protein accumulation in the wheat grain. J. Plant Physiol. 2007, 164, 581–590. [Google Scholar] [CrossRef]
- Shewry, P.R.; Hassall, K.L.; Grausgruber, H.; Andersson, A.A.M.; Lampi, A.-M.; Piironen, V.; Rakszegi, M.; Ward, J.L.; Lovegrove, A. Do modern types of wheat have lower quality for human health? Nutr. Bull. 2020, 45, 362–373. [Google Scholar] [CrossRef]
- Cooper, M.; Woodruff, D.R.; Philips, I.G.; Basford, K.E.; Gilmour, A.R. Genotype-by-management interactions for grain yield and grain protein concentration of wheat. Field Crops Res. 2001, 70, 87–88. [Google Scholar] [CrossRef]
- Oury, F.X.; Godin, C. Yield and grain protein concentration in bread wheat: How to use the negative relationship between the two characters to identify favourable genotypes? Euphytica 2007, 157, 45–57. [Google Scholar] [CrossRef]
- Laidig, F.; Piepho, H.P.; Rentel, D.; Drobek, T.; Meyer, U.; Huesken, A. Breeding progress, environmental variation and correlation of winter wheat yield and quality traits in german official variety trials and on-farm during 1983–2014. Theor. Appl. Genet. 2017, 130, 223–245. [Google Scholar] [CrossRef] [PubMed]
- Geyer, M.; Mohler, V.; Hartl, L. Genetics of the inverse relationship between grain yield and grain protein content in common wheat. Plants 2022, 11, 2146. [Google Scholar] [CrossRef]
- Poudel, R.; Bhinderwala, F.; Morton, M.; Powers, R.; Rose, D.J. Metabolic profiling of historical and modern wheat cultivars using proton nuclear magnetic resonance spectroscopy. Sci. Rep. 2021, 11, 3080. [Google Scholar] [CrossRef]
- Majdrakov, P. For wheat from Pavlikeni. Seed Prod. 1945, 4, 132–141. (In Bulgarian) [Google Scholar]
- Ganeva, G.; Korzun, V.; Landjeva, S.; Tsenov, N.; Atanasova, M. Identification, distribution and effects on agronomic traits of the semi-dwarfing Rht alleles in Bulgarian bread wheat cultivars. Euphytica 2005, 145, 305–315. [Google Scholar] [CrossRef]
- Börner, A.; Landjeva, S.; Nagel, M.; Rehman Arif, M.A.; Allam, M.; Agacka, M.; Doroszewska, T.; Lohwasser, U. Plant genetic resources for food and agriculture (PGRFA)—Maintenance and research. Genet. Plant Physiol. 2014, 4, 13–21. [Google Scholar]
- Garcia, M.; Eckermann, P.; Haefele, S.; Satija, S.; Sznajder, B.; Timmins, A.; Baumann, U.; Wolters, P.; Mather, D.E.; Fleury, D. Genome-wide association mapping of grain yield in a diverse collection of spring wheat (Triticum aestivum L.) evaluated in southern Australia. PLoS ONE 2019, 14, e0211730. [Google Scholar] [CrossRef]
- Paina, C.; Gregersen, P.L. Recent advances in the genetics underlying wheat grain protein content and grain protein deviation in hexaploid wheat. Plant Biol. 2023, 25, 661–670. [Google Scholar] [CrossRef]
- Robert, N.; Hennequet, C.; Berard, P. Dry matter and nitrogen accumulation in wheat kernel: Genetic variation in rate and duration of grain filling. J. Genet. Breed. 2001, 55, 297–306. [Google Scholar]
- Alqudah, A.M.; Sallam, A.; Baenziger, P.S.; Börner, A. GWAS: Fast-forwarding gene identification and characterization in temperate cereals: Lessons from barley—A review. J. Adv. Res. 2020, 22, 119–135. [Google Scholar] [CrossRef]
- Daba, S.D.; Tyagi, P.; Brown-Guedira, G.; Mohammadi, M. Genome-wide association studies to identify loci and candidate genes controlling kernel weight and length in a historical United States wheat population. Front. Plant Sci. 2018, 9, 1045. [Google Scholar] [CrossRef]
- Muhammad, A.; Hu, W.; Li, Z.; Li, J.; Xie, G.; Wang, J.; Wang, L. Appraising the genetic architecture of kernel traits in hexaploid wheat using GWAS. Int. J. Mol. Sci. 2020, 21, 5649. [Google Scholar] [CrossRef]
- Muqaddasi, Q.H.; Brassac, J.; Ebmeyer, E.; Kollers, S.; Korzun, V.; Argillier, O.; Stiewe, G.; Plieske, J.; Ganal, M.W.; Röder, M.S. Prospects of GWAS and predictive breeding for European winter wheat’s grain protein content, grain starch content, and grain hardness. Sci. Rep. 2020, 10, 12541. [Google Scholar] [CrossRef]
- Alemu, A.; El Baouchi, A.; El Hanafi, S.; Kehel, Z.; Eddakhir, K.; Tadesse, W. Genetic analysis of grain protein content and dough quality traits in elite spring bread wheat (Triticum aestivum) lines through association study. J. Cereal Sci. 2021, 100, 103214. [Google Scholar] [CrossRef]
- Rufo, R.; López, A.; Lopes, M.S.; Bellvert, J.; Soriano, J.M. Identification of quantitative trait loci hotspots affecting agronomic traits and high-throughput vegetation indices in rainfed wheat. Front. Plant Sci. 2021, 12, 735192. [Google Scholar] [CrossRef]
- Suliman, S.; Alemu, A.; Abdelmula, A.A.; Badawi, G.H.; Al-Abdallat, A.; Tadesse, W. Genome-wide association analysis uncovers stable QTLs for yield and quality traits of spring bread wheat (Triticum aestivum) across contrasting environments. Plant Gene 2021, 25, 100269. [Google Scholar] [CrossRef]
- Leonova, I.N.; Kiseleva, A.A.; Berezhnaya, A.A.; Stasyuk, A.I.; Likhenko, I.E.; Salina, E.A. Identification of QTLs for grain protein content in Russian spring wheat varieties. Plants 2022, 11, 437. [Google Scholar] [CrossRef]
- White, J.; Sharma, R.; Balding, D.; Cockram, J.; Mackay, I.J. Genome-wide association mapping of Hagberg falling number, protein content, test weight, and grain yield in U.K. wheat. Crop Sci. 2022, 62, 965–981. [Google Scholar] [CrossRef]
- Alomari, D.Z.; Schierenbeck, M.; Alqudah, A.M.; Alqahtani, M.D.; Wagner, S.; Rolletschek, H.; Borisjuk, L.; Röder, M.S. Wheat grains as a sustainable source of protein for health. Nutrients 2023, 15, 4398. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, G.; Guo, X.; Chi, S.; Yu, H.; Jin, K.; Huang, H.; Wang, D.; Wu, C.; Tian, J.; et al. Genetic dissection of protein and starch during wheat grain development using QTL mapping and GWAS. Front. Plant Sci. 2023, 14, 1189887. [Google Scholar] [CrossRef]
- Kartseva, T.; Alqudah, A.M.; Aleksandrov, V.; Alomari, D.Z.; Doneva, D.; Arif, M.A.R.; Börner, A.; Misheva, S. Nutritional genomic approach for improving grain protein content in wheat. Foods 2023, 12, 1399. [Google Scholar] [CrossRef]
- Krishnappa, G.; Khan, H.; Krishna, H.; Devate, N.B.; Kumar, S.; Mishra, C.N.; Parkash, O.; Kumar, S.; Kumar, M.; Mamrutha, H.M.; et al. Genome-wide association study for grain protein, thousand kernel weight, and normalized difference vegetation index in bread wheat (Triticum aestivum L.). Genes 2023, 14, 637. [Google Scholar] [CrossRef]
- Sharma, A.; Arif, M.A.R.; Shamshad, M.; Rawale, K.S.; Brar, A.; Burgueño, J.; Shokat, S.; Ravinder Kaur, R.; Vikram, P.; Srivastava, P.; et al. Preliminary dissection of grain yield and related traits at differential nitrogen levels in diverse pre-breeding wheat germplasm through association mapping. Mol. Biotechnol. 2023, 65, 116–130. [Google Scholar] [CrossRef]
- Tyrka, M.; Krajewski, P.; Bednarek, P.T.; Rączka, K.; Drzazga, T.; Matysik, P.; Martofel, R.; Woźna-Pawlak, U.; Jasińska, D.; Niewińska, M.; et al. Genome-wide association mapping in elite winter wheat breeding for yield improvement. J. Appl. Genet. 2023, 64, 377–391. [Google Scholar] [CrossRef]
- Yannam, V.R.R.; Lopes, M.; Guzman, C.; Soriano, J.M. Uncovering the genetic basis for quality traits in the Mediterranean old wheat germplasm and phenotypic and genomic prediction assessment by cross-validation test. Front. Plant Sci. 2023, 4, 1127357. [Google Scholar] [CrossRef]
- Aleksandrov, V.; Kartseva, T.; Alqudah, A.M.; Kocheva, K.; Tasheva, K.; Börner, A.; Misheva, S. Genetic diversity, linkage disequilibrium and population structure of Bulgarian bread wheat assessed by genome-wide distributed SNP markers: From old germplasm to semi-dwarf cultivars. Plants 2021, 10, 1116. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chai, Y.; Zhang, X.; Lu, S.; Zhao, Z.; Wei, D.; Chen, L.; Hu, Y.-G. Multi-locus GWAS of quality traits in bread wheat: Mining more candidate genes and possible regulatory Network. Front. Plant Sci. 2020, 11, 1091. [Google Scholar] [CrossRef] [PubMed]
- Rathan, N.D.; Krishna, H.; Ellur, R.K.; Sehgal, D.; Govindan, V.; Ahlawat, A.K.; Krishnappa, G.; Jaiswal, J.P.; Singh, J.B.; Saiprasad, S.V.; et al. Genome-wide association study identifies loci and candidate genes for grain micronutrients and quality traits in wheat (Triticum aestivum L.). Sci. Rep. 2022, 12, 7037. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Xu, D.; Hanif, M.; Xia, X.; He, Z. Genetic architecture underpinning yield component traits in wheat. Theor. Appl. Genet. 2020, 33, 1811–1823. [Google Scholar] [CrossRef] [PubMed]
- Eltaher, S.; Sallam, A.; Emara, H.A.; Nower, A.A.; Salem, K.F.M.; Börner, A.; Baenziger, P.S.; Mourad, A.M.I. Genome-wide association mapping revealed SNP alleles associated with spike traits in wheat. Agronomy 2022, 12, 1469. [Google Scholar] [CrossRef]
- Yang, Y.; Amo, A.; Wei, D.; Chai, Y.; Zheng, J.; Qiao, P.; Cul, C.; Lu, S.; Chen, L.; Hu, Y. Large-scale integration of meta-QTL and genome-wide association study discovers the genomic regions and candidate genes for yield and yield-related traits in bread wheat. Theor. Appl. Genet. 2021, 134, 3083–3109. [Google Scholar] [CrossRef] [PubMed]
- Chidzanga, C.; Daniel Mullan, D.; Roy, S.; Baumann, U.; Garcia, M. Nested association mapping-based GWAS for grain yield and related traits in wheat grown under diverse Australian environments. Theor. Appl. Genet. 2022, 135, 4437–4456. [Google Scholar] [CrossRef] [PubMed]
- Arif, M.A.R.; Waheed, M.Q.; Lohwasser, U.; Shokat, S.; Alquddah, A.M.; Volkmar, C.; Börner, A. Genetic insight into the insect resistance in bread wheat exploiting the untapped natural diversity. Front. Genet. 2022, 13, 898905. [Google Scholar] [CrossRef]
- Rathan, N.D.; Krishnappa, G.; Singh, A.-M.; Govindan, V. Mapping QTL for phenological and grain-related traits in a mapping population derived from high-zinc-biofortified wheat. Plants 2023, 12, 220. [Google Scholar] [CrossRef]
- Mulugeta, B.; Tesfaye, K.; Ortiz, R.; Johansson, E.; Hailesilassie, T.; Hammenhag, C.; Hailu, F.; Geleta, M. Marker-trait association analyses revealed major novel QTLs for grain yield and related traits in durum wheat. Front. Plant Sci. 2023, 13, 1009244. [Google Scholar] [CrossRef]
- Akram, S.; Arif, M.A.R.; Hameed, A. A GBS-based GWAS analysis of adaptability and yield traits in bread wheat (Triticum aestivum L.). J. Appl. Genet. 2020, 62, 27–41. [Google Scholar] [CrossRef]
- Wan, C.; Dang, P.; Gao, L.; Wang, J.; Tao, J.; Qin, X.; Feng, B.; Gao, J. How does the environment affect wheat yield and protein content response to drought? A meta-analysis. Front. Plant Sci. 2022, 13, 896985. [Google Scholar] [CrossRef]
- Teng, Z.; Chen, Y.; Meng, S.; Duan, M.; Ye, N.; Zhang, J. Environmental stimuli: A major challenge during grain filling in cereals. Int. J. Mol. Sci. 2023, 24, 2255. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Hou, J.; Wang, Y.; Wang, M.; Zhang, W.; Fan, Y.; Huang, Z. Post-flowering soil waterlogging curtails grain yield formation by restricting assimilates supplies to developing grains. Front. Plant Sci. 2022, 13, 944308. [Google Scholar] [CrossRef] [PubMed]
- Herzog, M.; Striker, G.G.; Colmer, T.D.; Pedersen, O. Mechanisms of waterlogging tolerance in wheat—A review of root and shoot physiology. Plant Cell Environ. 2016, 39, 1068–1086. [Google Scholar] [CrossRef]
- Gooding, M.J.; Ellis, R.H.; Shewry, P.R.; Schofield, J.D. Effects of restricted water availability and increased temperature on the grain filling, drying and quality of winter wheat. J. Cereal Sci. 2003, 37, 295–309. [Google Scholar] [CrossRef]
- Sehgal, D.; Mondal, S.; Guzman, C.; Garcia Barrios, G.; Franco, C.; Singh, R.; Dreisigacker, S. Validation of candidate gene-based markers and identification of novel loci for thousand-grain weight in spring bread wheat. Front. Plant Sci. 2019, 10, 1189. [Google Scholar] [CrossRef]
- Han, F.; Liu, B.; Fedak, G.; Liu, Z. Genomic constitution and variation in five partial amphiploids of wheat–Thinopyrum intermedium as revealed by GISH, multicolor GISH and seed storage protein analysis. Theor. Appl. Genet. 2004, 109, 1070–1076. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, M.; Sepsi, A.; Tyankova, N.; Molnár-Láng, M. Molecular cytogenetic characterization of two high protein wheat-Thinopyrum intermedium partial amphiploids. J. Appl. Gen. 2011, 52, 269–277. [Google Scholar] [CrossRef]
- Fatiukha, A.; Filler, N.; Lupo, I.; Lidzbarsky, G.; Klymiuk, V.; Korol, A.B.; Pozniak, C.; Fahima, T.; Krugman, T. Grain protein content and thousand kernel weight QTLs identified in a durum × wild emmer wheat mapping population tested in five environments. Theor. Appl. Genet. 2020, 133, 119–131. [Google Scholar] [CrossRef]
- Liu, J.; Huang, L.; Wang, C.; Liu, Y.; Yan, Z.; Wang, Z.; Xiang, L.; Zhong, X.; Gong, F.; Zheng, Y.; et al. Genome-wide association study reveals novel genomic regions associated with high grain protein content in wheat lines derived from wild emmer wheat. Front. Plant Sci. 2019, 10, 464. [Google Scholar] [CrossRef] [PubMed]
- Lindeque, R.C.; van Biljon, A.; Labuschagne, M.T. Defining associations between grain yield and protein quantity and quality in wheat from the three primary production regions of South Africa. J. Cereal Sci. 2018, 79, 294e302. [Google Scholar] [CrossRef]
- Cseh, A.; Poczai, P.; Kiss, T.; Balla, K.; Berki, Z.; Horváth, A.; Kuti, C.; Karsai, I. Exploring the legacy of Central European historical winter wheat landraces. Sci. Rep. 2021, 11, 23915. [Google Scholar] [CrossRef] [PubMed]
- López-Fernández, M.; Pascual, L.; Faci, I.; Fernández, M.; Ruiz, M.; Benavente, E.; Giraldo, P. Exploring the end-use quality potential of a collection of spanish bread wheat landraces. Plants 2021, 10, 620. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Mullan, D.; Zhang, C.; Zhao, S.; Li, X.; Zhang, A.; Lu, Z.; Wang, Y.; Yan, G. Major genomic regions responsible for wheat yield and its components as revealed by meta-QTL and genotype–phenotype association analyses. Planta 2020, 252, 65. [Google Scholar] [CrossRef] [PubMed]
- Saini, P.; Sheikh, I.; Saini, D.K.; Mir, R.R.; Dhaliwal, H.S.; Tyagi, V. Consensus genomic regions associated with grain protein content in hexaploid and tetraploid wheat. Front. Genet. 2022, 13, 1021180. [Google Scholar] [CrossRef] [PubMed]
- Landjeva, S.; Korzun, V.; Ganeva, G. Evaluation of genetic diversity among Bulgarian winter wheat (Triticum aestivum L.) varieties during the period 1925–2003 using microsatellites. Genet. Res. Crop Evol. 2006, 53, 1605–1614. [Google Scholar] [CrossRef]
- Borrill, P.; Harrington, S.A.; Simmonds, J.; Uauy, C. Identification of transcription factors regulating senescence in wheat through gene regulatory network modelling. Plant Physiol. 2019, 180, 1740–1755. [Google Scholar] [CrossRef] [PubMed]
- Sultana, N.; Islam, S.; Juhasz, A.; Ma, W. Wheat leaf senescence and its regulatory gene network. Crop J. 2021, 9, 703–717. [Google Scholar] [CrossRef]
- Whan, A.; Dielen, A.-S.; Mieog, J.; Bowerman, A.F.; Robinson, H.M.; Byrne, K.; Colgrave, M.; Larkin, P.J.; Howitt, C.A.; Morell, M.K.; et al. Engineering α-amylase levels in wheat grain suggests a highly sophisticated level of carbohydrate regulation during development. J. Exp. Bot. 2014, 65, 5443–5457. [Google Scholar] [CrossRef]
- Kolli, R.; Soll, J.; Carrie, C. Plant mitochondrial inner membrane protein insertion. Int. J. Mol. Sci. 2018, 19, 641. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hu, Y.; Wang, Z.; Lin, X.; Li, Z.; Ren, Y.; Zhao, J. The translocase of the inner mitochondrial membrane 22-2 is required for mitochondrial membrane function during Arabidopsis seed development. J. Exp. Bot. 2023, 74, 4427–4448. [Google Scholar] [CrossRef] [PubMed]
- Zourelidou, M.; De Torres-Zabala, M.; Smith, C.; Bevan, M.W. Storekeeper defines a new class of plant-specific DNA-binding proteins and is a putative regulator of patatin expression. Plant J. 2002, 30, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Nietzsche, M.; Guerra, T.; Alseekh, S.; Wiermer, M.; Sonnewald, S.; Fernie, A.R.; Börnke, F. STOREKEEPER RELATED1/G-element binding protein (STKR1) interacts with protein kinase SnRK1. Plant Physiol. 2018, 176, 1773–1792. [Google Scholar] [CrossRef]
- Hajibarat, Z.; Saidi, A. Senescence-associated proteins and nitrogen remobilization in grain filling under drought stress condition. J. Genet. Eng. Biotechnol. 2022, 20, 101. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Lin, Z.; Chen, W.; Jiang, H.; Zhang, S.; Lin, H. Chemogenomic approach identified yeast YLR143W as diphthamide synthetase. Proc. Natl. Acad. Sci. USA 2012, 109, 19983–19987. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, L.; Liu, Y.; Sod, B.; Li, M.; Yang, T.; Gao, T.; Yang, Q.; Long, R. Overexpression of the elongation factor MtEF1A1 promotes salt stress tolerance in Arabidopsis thaliana and Medicago truncatula. BMC Plant Biol. 2023, 23, 138. [Google Scholar] [CrossRef] [PubMed]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef]
- Andleeb, T.; Borrill, P. Wheat NAM genes regulate the majority of early monocarpic senescence transcriptional changes including nitrogen remobilisation genes. G3 Genes Genomes Genet. 2023, 13, jkac275. [Google Scholar] [CrossRef]
- Konishi, M.; Yanagisawa, S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 2013, 4, 1617. [Google Scholar] [CrossRef]
- Mahmoud, D.; Pandey, R.; Sathee, L.; Dalal, M.; Singh, M.P.; Chinnusamy, V. Regulation of expression of genes associated with nitrate response by osmotic stress and combined osmotic and nitrogen deficiency stress in bread wheat (Triticum aestivum L.). Plant Physiol. Rep. 2020, 25, 200–215. [Google Scholar] [CrossRef]
- Schlaich, N.L. Flavin-containing monooxygenases in plants: Looking beyond detox. Trends Plant Sci. 2007, 12, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Wang, Z.; Liu, S.; Kong, X.; Wang, F.; Sun, G.; Geng, S.; Mao, L.; Zhou, P.; Li, A. Transcriptome analysis of developing wheat grains at rapid expanding phase reveals dynamic gene expression patterns. Biology 2022, 11, 281. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Mendoza, M.; Diaz, I.; Martinez, M. Insights on the proteases involved in barley and wheat grain germination. Int. J. Mol. Sci. 2019, 20, 2087. [Google Scholar] [CrossRef] [PubMed]
- Arif, M.A.R.; Börner, A. An SNP based GWAS analysis of seed longevity in wheat. Cereal Res. Comm. 2020, 48, 149–156. [Google Scholar] [CrossRef]
- Vierstra, R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xu, Y.; Luo, W.; Li, W.; Chen, N.; Zhang, D.; Chong, K. The F-box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice. Plant Physiol. 2013, 163, 1673–1685. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.J.; Kim, D.Y.; Choi, H.I.; Seo, Y.W.; Kim, J.B. Isolation and characterization of kelch repeat-containing F-box proteins from colored wheat. Mol. Boil. Rep. 2020, 47, 1129–1141. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.B.; Bellini, M. The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res. 2008, 36, 6482–6493. [Google Scholar] [CrossRef]
- Lopato, S.; Borisjuk, L.; Milligan, A.S.; Shirley, N.; Bazanova, N.; Langridge, P. Systematic identification of factors involved in post-transcriptional processes in wheat grain. Plant Mol. Biol. 2006, 62, 637–653. [Google Scholar] [CrossRef]
- Shao, A.; Ma, W.; Zhao, X.; Hu, M.; He, X.; Teng, W.; Li, H.; Tong, Y. The auxin biosynthetic TRYPTOPHAN AMINOTRANSFERASE RELATED TaTAR2. 1-3A increases grain yield of wheat. Plant Physiol. 2017, 174, 2274–2288. [Google Scholar] [CrossRef]
- Dufayard, J.F.; Bettembourg, M.; Fischer, I.; Droc, G.; Guiderdoni, E.; Périn, C.; Chantret, N.; Diévart, A. New insights on leucine-rich repeats receptor-like kinase orthologous relationships in angiosperms. Front. Plant Sci. 2017, 8, 243806. [Google Scholar] [CrossRef]
- Zha, X.; Luo, X.; Qian, X.; He, G.; Yang, M.; Li, Y.; Yang, J. Over-expression of the rice LRK1 gene improves quantitative yield components. Plant Biotechnol. J. 2009, 7, 611–620. [Google Scholar] [CrossRef]
- Si, L.; Chen, J.; Huang, X.; Gong, H.; Luo, J.; Hou, Q.; Zhou, T.; Lu, T.; Zhu, J.; Shangguan, Y.; et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016, 48, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Giancaspro, A.; Giove, S.L.; Zacheo, S.A.; Blanco, A.; Gadaleta, A. Genetic variation for protein content and yield-related traits in a durum population derived from an inter-specific cross between hexaploid and tetraploid wheat cultivars. Front. Plant Sci. 2019, 10, 1509. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Z.; Liu, D.; Zhang, K.; Li, A.; Mao, L. SQUAMOSA promoter-binding protein-like transcription factors: Star players for plant growth and development. J. Integr. Plant Biol. 2010, 52, 946–951. [Google Scholar] [CrossRef] [PubMed]
- Kuzay, S.; Xu, Y.; Zhang, J.; Katz, A.; Pearce, S.; Su, Z.; Fraser, M.; Anderson, J.A.; Brown-Guedira, G.; DeWitt, N.; et al. Identification of a candidate gene for a QTL for spikelet number per spike on wheat chromosome arm 7AL by high-resolution genetic mapping. Theor. Appl. Genet. 2019, 132, 2689–2705. [Google Scholar] [CrossRef] [PubMed]
- Voss-Fels, K.P.; Keeble-Gagnère, G.; Hickey, L.T.; Tibbits, J.; Nagornyy, S.; Hayden, M.J.; Wittkop, B. High-resolution mapping of rachis nodes per rachis, a critical determinant of grain yield components in wheat. Theor. Appl. Genet. 2019, 132, 2707–2719. [Google Scholar] [CrossRef]
- Xu, R.; Li, N.; Li, Y. Control of grain size by G protein signaling in rice. J. Integr. Plant Biol 2019, 61, 533–540. [Google Scholar] [CrossRef]
- Li, S.; Liu, Y.; Zheng, L.; Chen, L.; Li, N.; Corke, F.; Lu, Y.; Fu, X.; Zhu, Z.; Bevan, M.W.; et al. The plant-specific G protein γ subunit AGG3 influences organ size and shape in Arabidopsis thaliana. New Phytol. 2012, 194, 690–703. [Google Scholar] [CrossRef]
- Sun, S.; Wang, L.; Mao, H.; Shao, L.; Li, X.; Xiao, J.; Ouyang, Y.; Zhang, Q. A G-protein pathway determines grain size in rice. Nat. Commun. 2018, 9, 851. [Google Scholar] [CrossRef]
- Wendt, T.; Holme, I.; Dockter, C.; Preuß, A.; Thomas, W.; Druka, A.; Waugh, R.; Hansson, M.; Braumann, I. HvDep1 is a positive regulator of culm elongation and grain size in barley and impacts yield in an environment-dependent manner. PLoS ONE 2016, 11, e0168924. [Google Scholar] [CrossRef]
- Sun, H.; Qian, Q.; Wu, K.; Luo, J.; Wang, S.; Zhang, C.; Ma, Y.; Liu, Q.; Huang, X.; Yuan, Q.; et al. Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Gen. 2014, 46, 652–656. [Google Scholar] [CrossRef] [PubMed]
- Schierenbeck, M.; Alqudah, A.M.; Lohwasser, U.; Tarawneh, R.A.; Simón, M.R.; Börner, A. Genetic dissection of grain architecture related traits in a winter wheat population. BMC Plant Biol. 2021, 21, 417. [Google Scholar] [CrossRef]
- Kaur, S.; Dhugga, K.S.; Beech, R.; Singh, J. Genome-wide analysis of the cellulose synthase-like (Csl) gene family in bread wheat (Triticum aestivum L.). BMC Plant Biol. 2017, 17, 193. [Google Scholar] [CrossRef]
- Hyles, J.; Vautrin, S.; Pettolino, F.; MacMillan, C.; Stachurski, Z.; Breen, J.; Berges, H.; Spielmeyer, W. Repeat-length variation in a wheat cellulose synthase-like gene is associated with altered tiller number and stem cell wall composition. J. Exp. Bot. 2017, 68, 1519–1529. [Google Scholar] [CrossRef] [PubMed]
- Mossé, J.; Huet, J.C.; Baudet, J. The amino acid composition of wheat grain as a function of nitrogen content. J. Cereal Sci. 1985, 3, 115–130. [Google Scholar] [CrossRef]
- STATISTICA, Version 14; StatSoft Inc.: Tulsa, OK, USA, 2020.
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Yu, J.; Buckler, E.S. Genetic association mapping and genome organization of maize. Curr. Opin. Biotechnol. 2006, 17, 155–160. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Turner, S. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- Alaux, M.; Rogers, G.; Letellier, T.; Flores, R.; Alfama, F.; Pommier, C.; Mohellibi, N.; Durand, S.; Kimmel, E.; Michotey, C.; et al. Linking the International Wheat Genome Sequencing Consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol. 2018, 19, 111. [Google Scholar] [CrossRef] [PubMed]

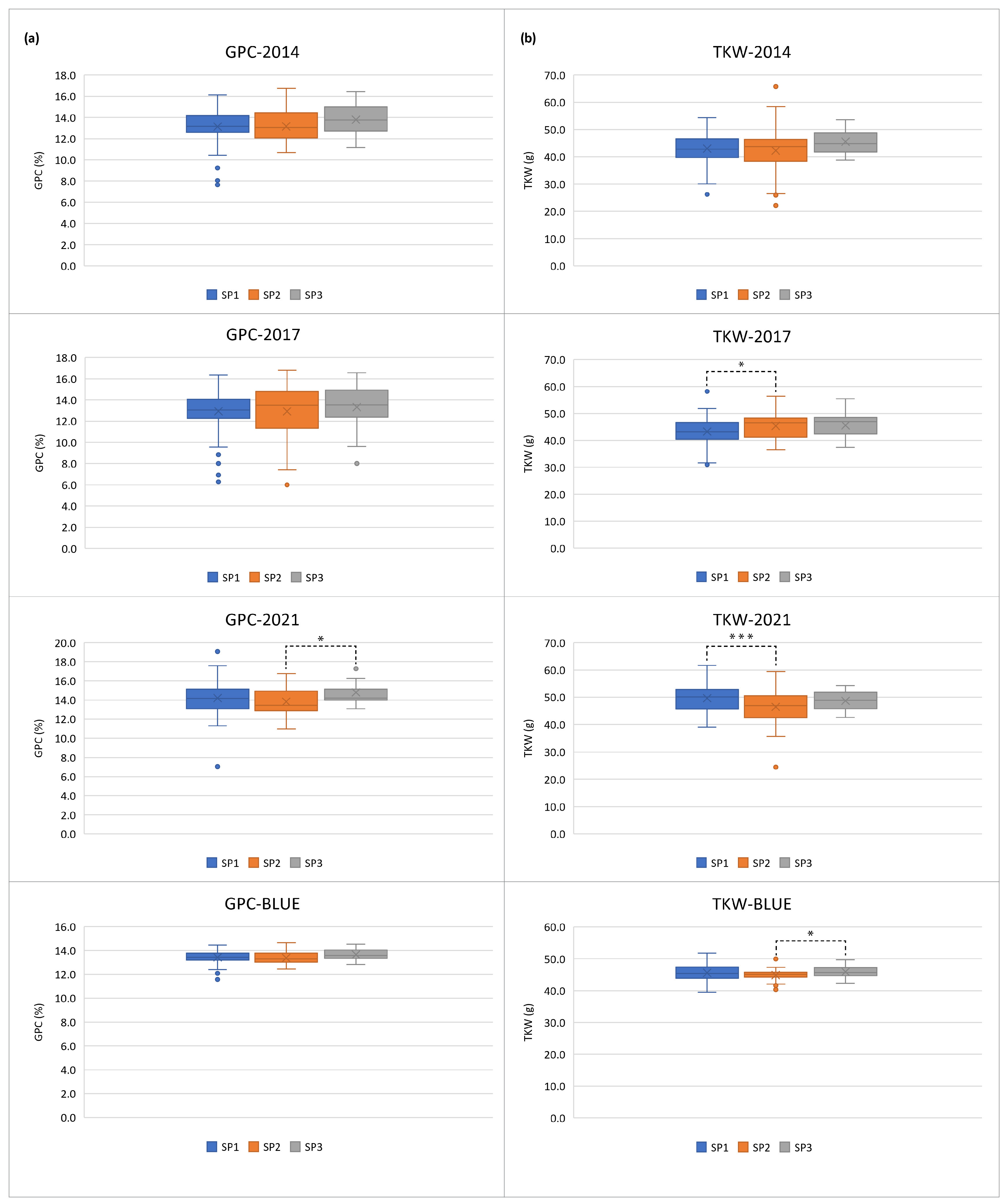
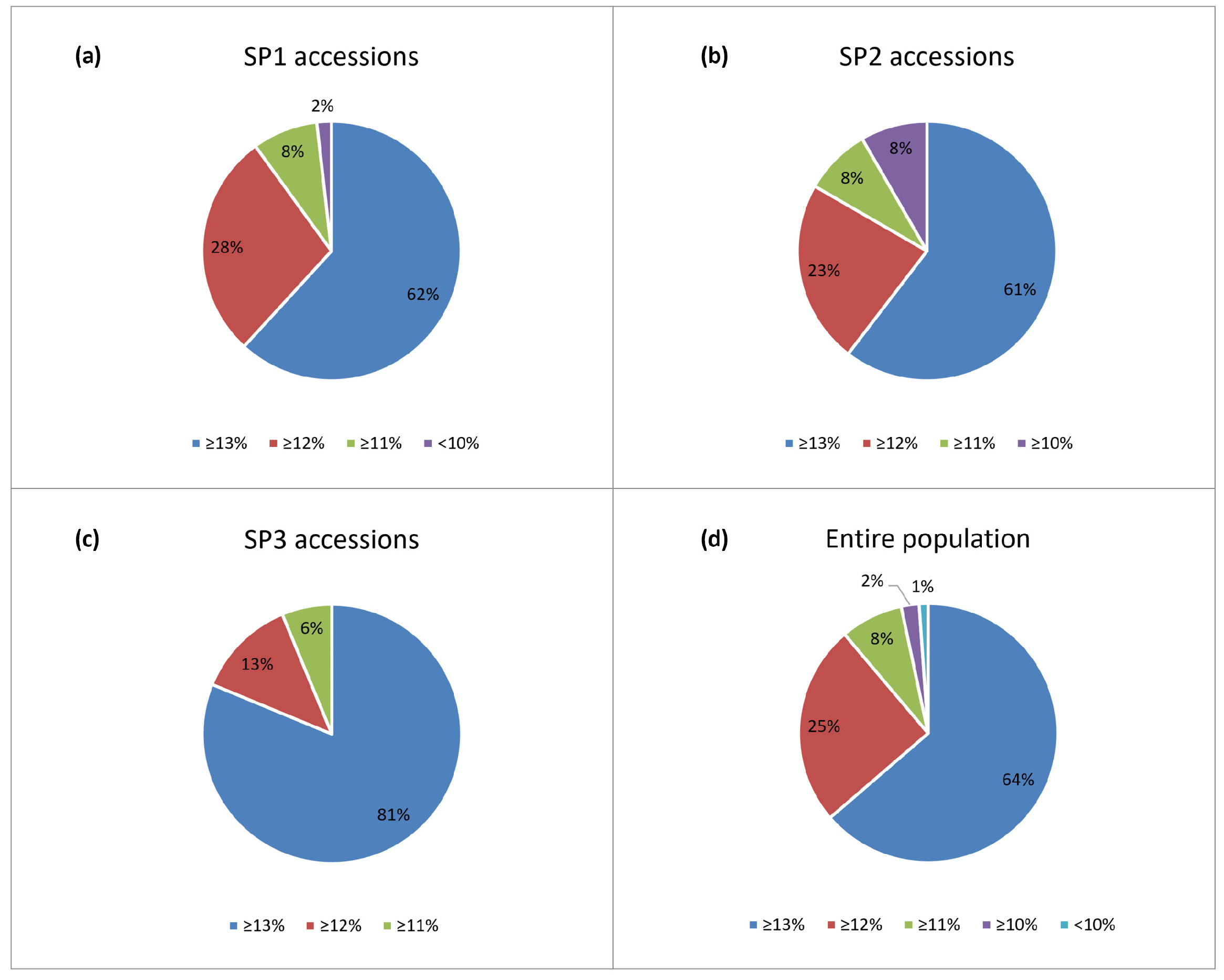
| Trait | Env. | Mean * | Std. Dev. | Min. | Max. | CV % | h2 |
|---|---|---|---|---|---|---|---|
| GPC (%) | Sofia 2014 | 13.2 b | 1.46 | 7.6 | 16.8 | 11.09 | 0.64 |
| Sofia 2017 | 12.9 b | 2.06 | 6.0 | 16.8 | 15.90 | 0.78 | |
| Sofia 2021 | 14.2 a | 1.62 | 7.1 | 19.4 | 11.43 | 0.69 | |
| Average | 13.4 | 1.34 | 9.4 | 16.8 | 10.00 | 0.82 | |
| BLUE | 13.4 | 0.51 | 11.6 | 14.7 | 3.81 | ||
| TKW (g) | Sofia 2014 | 43.1 b | 6.43 | 22.2 | 65.8 | 14.92 | 0.77 |
| Sofia 2017 | 44.0 b | 4.72 | 31.0 | 58.2 | 10.73 | 0.64 | |
| Sofia 2021 | 48.6 a | 5.42 | 24.4 | 61.6 | 11.14 | 0.70 | |
| Average | 45.2 | 4.28 | 33.7 | 55.0 | 9.46 | 0.81 | |
| BLUE | 45.4 | 2.25 | 39.6 | 51.8 | 4.94 |
| (a) | ||||||
| Source of Variation | SS | df | MS | F | p-Value | F Crit |
| Genotype (G) | 962.685 | 178 | 5.408 | 3.010 | 0.0000 | 1.233 |
| Environment (E) | 145.483 | 2 | 72.741 | 40.485 | 0.0000 | 3.021 |
| G × E | 639.638 | 356 | 1.797 | 7.210 | 0.0000 | 3.320 |
| Total | 1747.805 | 536 | ||||
| (b) | ||||||
| Source of Variation | SS | df | MS | F | p-Value | F Crit |
| Genotype (G) | 9792.257 | 178 | 55.013 | 2.897 | 0.0000 | 1.233 |
| Environment (E) | 3128.650 | 2 | 1564.325 | 82.375 | 0.0000 | 3.021 |
| G × E | 6760.582 | 356 | 18.990 | 4.3643 | 0.0000 | 4.092 |
| Total | 19681.49 | 536 | ||||
| GPC-2017 | GPC-2021 | GPC-BLUE | TKW-2014 | TKW-2017 | TKW-2021 | TKW-BLUE | |
|---|---|---|---|---|---|---|---|
| GPC-2014 | 0.69 *** | 0.50 *** | 0.93 *** | 0.00 | −0.09 | −0.08 | −0.05 |
| GPC-2017 | 0.10 | 0.61 *** | 0.04 | 0.01 | −0.01 | −0.02 | |
| GPC-2021 | 0.47 *** | 0.16 * | −0.01 | 0.12 | 0.06 | ||
| GPC-BLUE | −0.01 | −0.10 | −0.07 | 0.02 | |||
| TKW-2014 | 0.39 *** | 0.30 *** | 0.60 *** | ||||
| TKW-2017 | 0.53 *** | 0.42 *** | |||||
| TKW-2021 | 0.38 *** |
| Marker | Chr | Position (bp) | −log10 (p) | Effect | R2 | Gene ID | Annotation | Co-Located Loci a |
|---|---|---|---|---|---|---|---|---|
| GPC_2014 | ||||||||
| RAC875_rep_c98040_163 | 1B | 512,632,139 | 3.78 | −1.054 | 0.084 | TraesCS1B01G294600 | GTP cyclohydrolase II/3,4-dihydroxy-2-butanone 4-phosphate synthase | |
| BobWhite_c17047_268 | 2B | 11,077,394 | 3.85 | −1.011 | 0.089 | TraesCS2B01G023800 | Ethylene receptor | |
| wsnp_BE500291A_Ta_2_1 | 5A | 148,056,504 | 3.82 | −0.889 | 0.087 | TraesCS5A01G101200 | 1-acyl-sn-glycerol-3-phosphate acyltransferase | |
| Tdurum_contig54917_597 | 6B | 32,324,229 | 3.73 | 1.357 | 0.083 | TraesCS6B01G052900 | Kelch repeat–containing protein | [33] |
| GPC_2017 | ||||||||
| CAP11_c601_120 | 4B | 38,791,353 | 4.21 | −2.456 | 0.096 | TraesCS4B01G050200 | Histidine kinase | |
| AX-95148678 | 4B | 39,015,957 | 4.15 | −2.446 | 0.095 | TraesCS4B01G050400 | UDP-glucose 6-dehydrogenase | |
| AX-94513493 | 4B | 39,045,208 | 4.21 | −2.456 | 0.096 | TraesCS4B01G050500 | UV-B-induced protein, chloroplastic | |
| BS00109912_51 | 7A | 653,925 | 4.36 | 2.874 | 0.101 | NA | ||
| GPC_2021 | ||||||||
| RFL_Contig5739_641 | 5B | 531,538,634 | 4.11 | 1.412 | 0.092 | TraesCS5B01G350900 | Plant regulator RWP-RK family protein, putative | [34] |
| IACX2946 | 6A | 599,046,570 | 4.16 | −2.172 | 0.092 | TraesCS6A01G378000 | Protein kinase family protein | [23,33] |
| Tdurum_contig46670_911 | 6A | 599,050,921 | 4.16 | 2.172 | 0.092 | TraesCS6A01G378100 | Diphthine-ammonia ligase | [23,33] |
| AX-94649878 | 6D | 453,347,731 | 4.86 | −1.954 | 0.111 | TraesCS6D01G362900 | NAC domain protein | |
| GPC_BLUE | ||||||||
| Ra_c22880_760 | 2A | 498,105,752 | 8.38 | 7.918 | 0.217 | TraesCS2A01G289800 | Alpha-amylase | |
| AX-158545828 | 2A | 501,850,514 | 8.46 | 8.069 | 0.219 | TraesCS2A01G291200 | Mitochondrial import inner membrane translocase subunit TIM22 | |
| Tdurum_contig59467_433 | 7A | 715,692,069 | 9.94 | −9.349 | 0.267 | NA | ||
| AX-94519723 | 7B | 121,890,360 | 8.57 | 7.372 | 0.222 | TraesCS7B01G105700 | DNA-binding storekeeper protein-related transcriptional regulator | |
| Marker | Chr | Position (bp) | −log10 (p) | Effect | R2 | Gene ID | Annotation | Co-Located Loci a |
|---|---|---|---|---|---|---|---|---|
| TKW_2014 | ||||||||
| BobWhite_c2027_215 | 1B | 640,555,504 | 5.27 | −10.268 | 0.124 | TraesCS1B01G415400 | TSA: Wollemia nobilis Ref_Wollemi_Transcript_14910_3291 transcribed RNA sequence | [21,35] |
| wsnp_JD_c4217_5322858 | 4A | 140,291,399 | 5.25 | 8.388 | 0.123 | TraesCS4A01G115300 | Small nuclear ribonucleoprotein | [36] |
| TKW_2017 | ||||||||
| AX-95145282 | 6A | 100,766,008 | 7.84 | 4.780 | 0.199 | NA | ||
| TKW_2021 | ||||||||
| GENE-0925_515 | 2A | 656,820,630 | 6.42 | −4.526 | 0.143 | NA | ||
| BobWhite_c13435_700 | 6D | 471,008,534 | 6.43 | −4.534 | 0.144 | NA | [37] | |
| CAP7_c2559_543 | 6D | 471,017,962 | 6.40 | −4.519 | 0.143 | TraesCS6D01G402800 | Zinc finger CCCH domain-containing protein 19 | [37] |
| TKW_BLUEs | ||||||||
| AX-94692394 | 1A | 7,186,246 | 8.54 | 1.927 | 0.209 | TraesCS1A01G012600 | Leucine-rich repeat receptor-like protein kinase family protein | [21,35] |
| Tdurum_contig50667_306 | 1B | 20,588,032 | 9.49 | 2.705 | 0.238 | TraesCS1B01G041200 | F-box protein-like | |
| AX-94641316 | 4D | 374,832,658 | 8.55 | −2.485 | 0.210 | TraesCS4D01G219400 | Regulator of Vps4 activity in the MVB pathway protein, putative | |
| wsnp_Ex_c24031_33277293 | 5B | 707,131,836 | 9.10 | −3.002 | 0.226 | TraesCS5B01G560400 | PGR5-like protein 1A, chloroplastic | [35,37] |
| wsnp_Ex_c4480_8056013 | 6A | 611,487,998 | 8.76 | −2.084 | 0.216 | TraesCS6A01G405500 | Lysine-specific histone demethylase 1-3-like protein | [35,37,38] |
| Kukri_c11902_580 | 6A | 611,491,408 | 8.76 | −2.085 | 0.216 | TraesCS6A01G405500 | Lysine-specific histone demethylase 1-3-like protein | [35,37,38] |
| wsnp_Ex_c4480_8056354 | 6D | 464,946,552 | 8.67 | −2.078 | 0.214 | TraesCS6D01G389200 | Lysine-specific histone demethylase 1-3-like protein | [37] |
| AX-158600736 | 6D | 464,948,739 | 8.70 | −2.078 | 0.215 | TraesCS6D01G389200 | Lysine-specific histone demethylase 1-3-like protein | [37] |
| TA005561-0543 | 6D | 465,050,368 | 8.48 | −2.065 | 0.211 | NA | [37] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kartseva, T.; Aleksandrov, V.; Alqudah, A.M.; Arif, M.A.R.; Kocheva, K.; Doneva, D.; Prokopova, K.; Börner, A.; Misheva, S. GWAS in a Collection of Bulgarian Old and Modern Bread Wheat Accessions Uncovers Novel Genomic Loci for Grain Protein Content and Thousand Kernel Weight. Plants 2024, 13, 1084. https://doi.org/10.3390/plants13081084
Kartseva T, Aleksandrov V, Alqudah AM, Arif MAR, Kocheva K, Doneva D, Prokopova K, Börner A, Misheva S. GWAS in a Collection of Bulgarian Old and Modern Bread Wheat Accessions Uncovers Novel Genomic Loci for Grain Protein Content and Thousand Kernel Weight. Plants. 2024; 13(8):1084. https://doi.org/10.3390/plants13081084
Chicago/Turabian StyleKartseva, Tania, Vladimir Aleksandrov, Ahmad M. Alqudah, Mian Abdur Rehman Arif, Konstantina Kocheva, Dilyana Doneva, Katelina Prokopova, Andreas Börner, and Svetlana Misheva. 2024. "GWAS in a Collection of Bulgarian Old and Modern Bread Wheat Accessions Uncovers Novel Genomic Loci for Grain Protein Content and Thousand Kernel Weight" Plants 13, no. 8: 1084. https://doi.org/10.3390/plants13081084
APA StyleKartseva, T., Aleksandrov, V., Alqudah, A. M., Arif, M. A. R., Kocheva, K., Doneva, D., Prokopova, K., Börner, A., & Misheva, S. (2024). GWAS in a Collection of Bulgarian Old and Modern Bread Wheat Accessions Uncovers Novel Genomic Loci for Grain Protein Content and Thousand Kernel Weight. Plants, 13(8), 1084. https://doi.org/10.3390/plants13081084








