OsSCYL2 is Involved in Regulating ABA Signaling-Mediated Seed Germination in Rice
Abstract
1. Introduction
2. Results
2.1. Bioinformatics Analysis of OsSCYL2
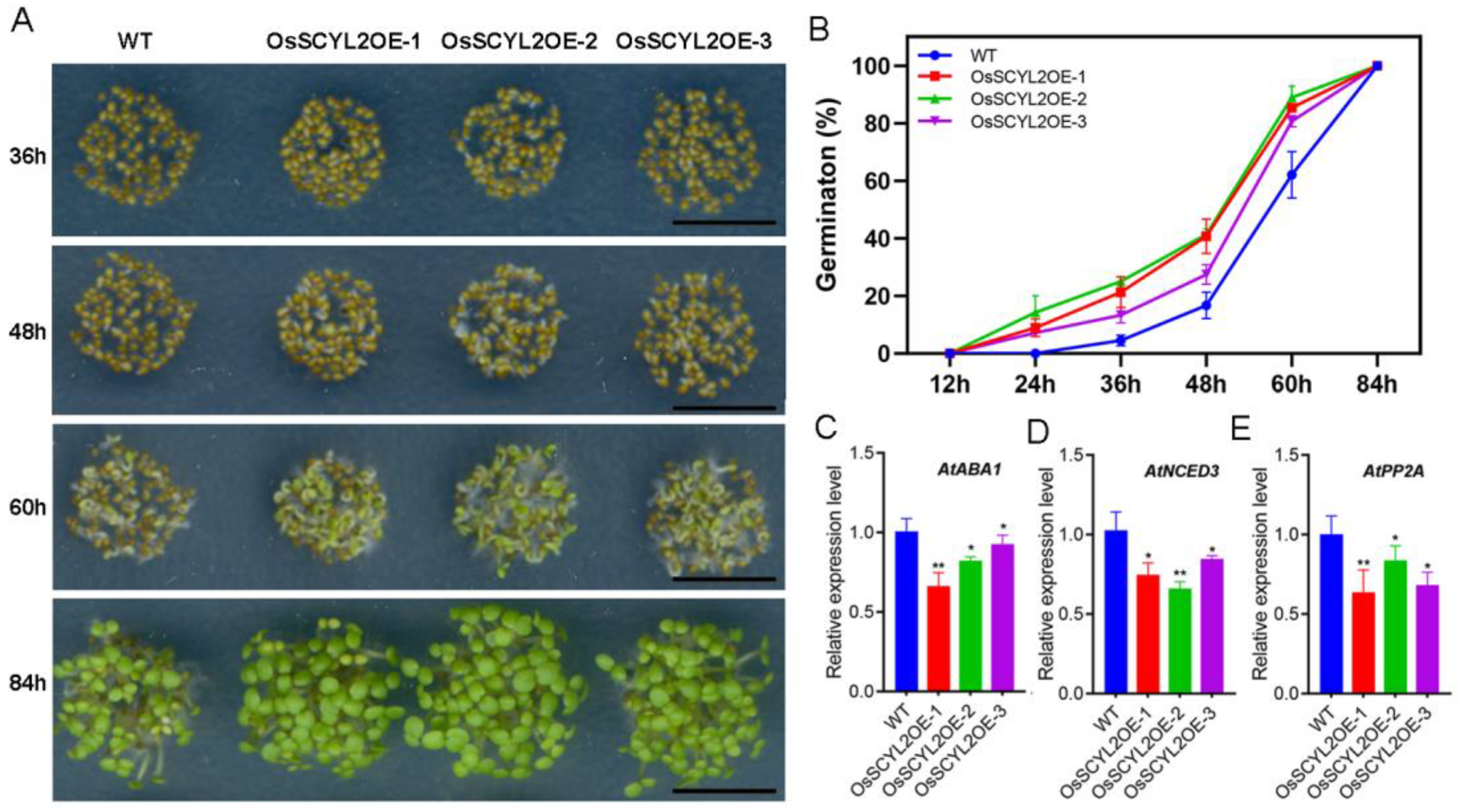
2.2. OsSCYL2 Affects the Speed of Seed Germination
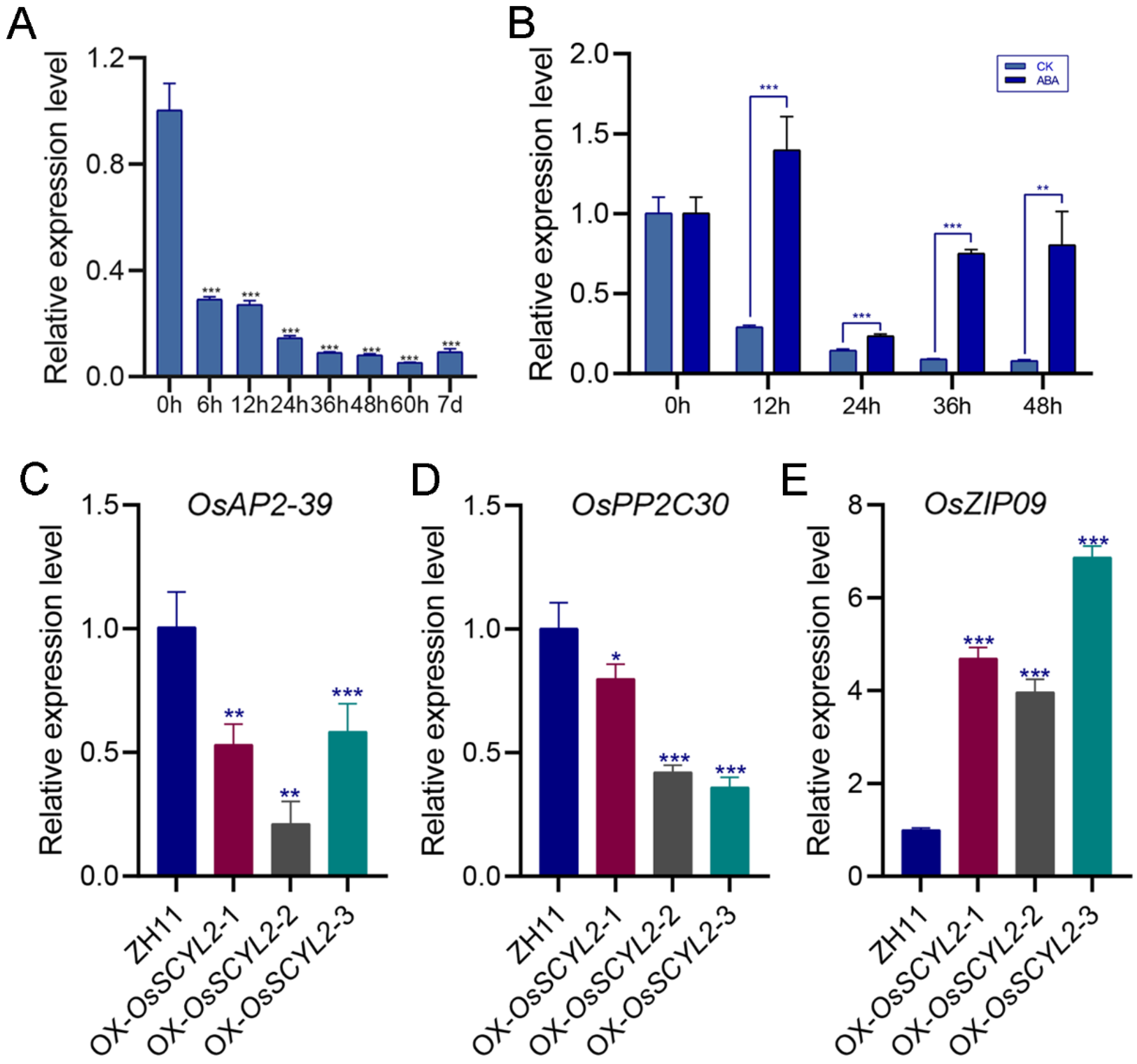
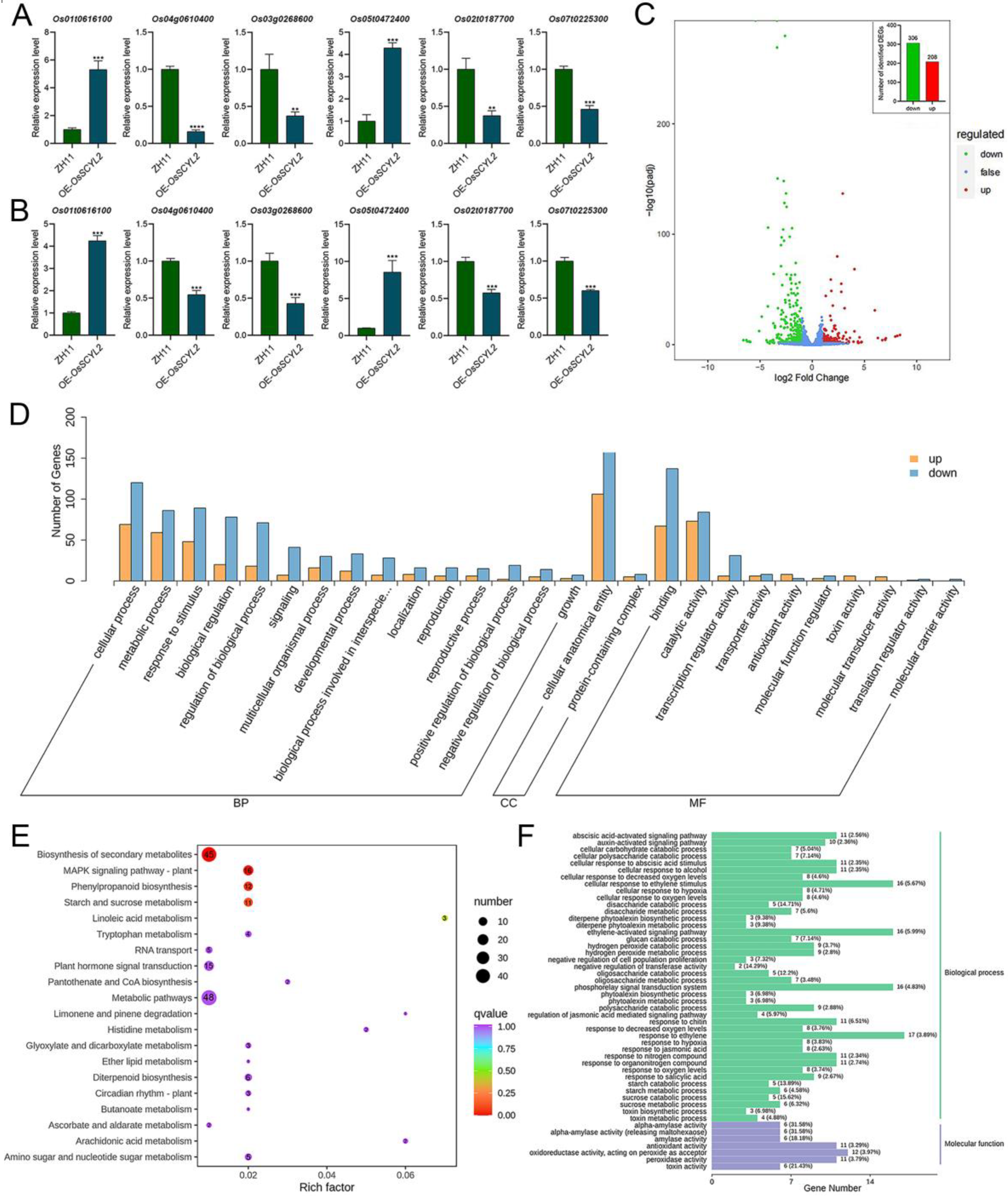
2.3. Overexpression of OsSCYL2 Resulted in Changes in ABA-Regulated Genes
2.4. OsSCYL2 Regulates MAPK and Plant Hormones Signaling Pathways
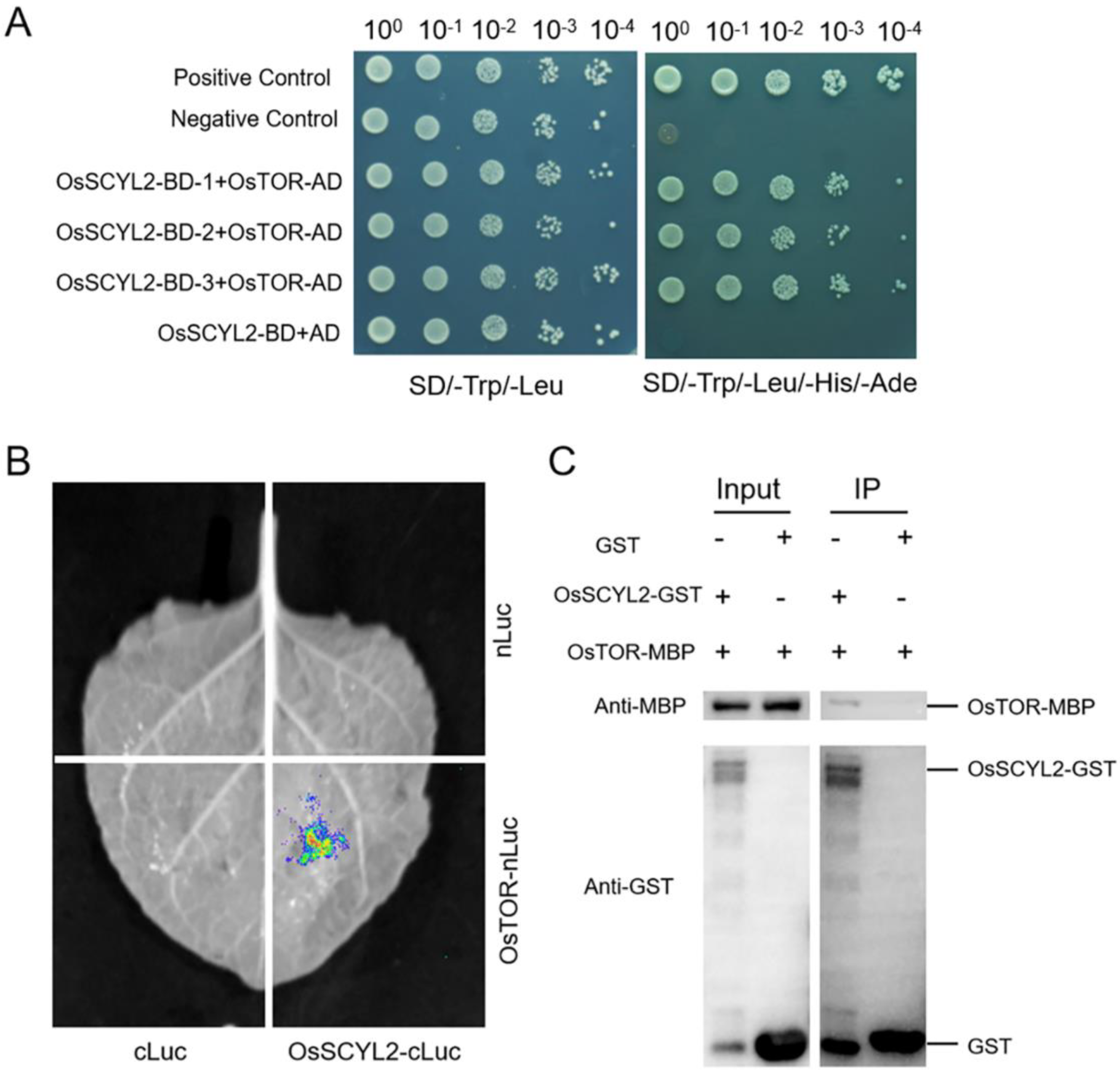
2.5. OsSCYL2 Interacted with OsTOR in Rice
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Bioinformatics and Expression Analysis of OsSCYL2
4.3. Seed Germination Assay and ABA Treatment Assay
4.4. RNA Extractions, RNA Library Construction, and Sequencing
4.5. KEGG and GO Analyses
4.6. Yeast Two-Hybrid Assay
4.7. Split-Luciferase Complementation Assay (Split-LUC)
4.8. Glutathione Stransferase (GST) Pull-Down Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gommers, C.M.M.; Monte, E. Seedling establishment: A dimmer Switch-regulated process between dark and light signaling. Plant Physiol. 2017, 176, 1061–1074. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Baek, W.; Jung, J.; Kim, J.H.; Lee, S.C. Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 2015, 16, 15251–15270. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Zhou, W.; Chen, F.; Luo, X.; Yang, W. Abscisic acid and gibberellins antagonistically mediate plant development and abiotic stress responses. Front. Plant Sci. 2018, 9, 416. [Google Scholar] [CrossRef]
- Shu, K.; Zhou, W.; Yang, W. APETALA 2-domain-containing transcription factors: Focusing on abscisic acid and gibberellins antagonism. New Phytol. 2018, 217, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Liu, X.D.; Xie, Q.; He, Z.H. Two faces of one seed: Hormonal regulation of dormancy and germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Dai, X.; Zhang, W.H. A rice F-box gene, OsFbx352, is involved in glucose-delayed seed germination in rice. J. Exp. Bot. 2012, 63, 5559–5568. [Google Scholar] [CrossRef]
- Hu, Q.; Lin, C.; Guan, Y.; Sheteiwy, M.S.; Hu, W.; Hu, J. Inhibitory effect of eugenol on seed germination and pre-harvest sprouting of hybrid rice (Oryza sativa L.). Sci. Rep. 2017, 7, 5295. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hasanuzzaman, M.; Wen, H.; Zhang, J.; Peng, T.; Sun, H.; Zhao, Q. High temperature and drought stress cause abscisic acid and reactive oxygen species accumulation and suppress seed germination growth in rice. Protoplasma 2019, 256, 1217–1227. [Google Scholar] [CrossRef]
- Zhou, C.; Lin, Q.; Lan, J.; Zhang, T.; Liu, X.; Miao, R.; Mou, C.; Nguyen, T.; Wang, J.; Zhang, X.; et al. WRKY transcription factor OsWRKY29 represses seed dormancy in rice by weakening abscisic acid response. Front. Plant Sci. 2020, 11, 691. [Google Scholar] [CrossRef]
- He, Y.; Chen, S.; Liu, K.; Chen, Y.; Cheng, Y.; Zeng, P.; Zhu, P.; Xie, T.; Chen, S.; Zhang, H.; et al. OsHIPL1, a hedgehog-interacting protein-like 1 protein, increases seed vigor in rice. Plant Biotechnol. J. 2022, 20, 1346–1362. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Andújar, C.; Ordiz, M.I.; Huang, Z.; Nonogaki, M.; Beachy, R.N.; Nonogaki, H. Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc. Natl. Acad. Sci. USA 2011, 108, 17225–17229. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Ye, N.; Zhang, J. Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiol. 2009, 50, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.; Li, H.; Zhu, G.; Liu, Y.; Liu, R.; Xu, W.; Jing, Y.; Peng, X.; Zhang, J. Copper suppresses abscisic acid catabolism and catalase activity, and inhibits seed germination of rice. Plant Cell Physiol. 2014, 55, 2008–2016. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Yan, A.; Chen, Z. The pivotal role of abscisic acid signaling during transition from seed maturation to germination. Plant Cell Rep. 2017, 36, 689–703. [Google Scholar] [CrossRef] [PubMed]
- Miyakawa, T.; Fujita, Y.; Yamaguchi-Shinozaki, K.; Tanokura, M. Structure and function of abscisic acid receptors. Trends Plant Sci. 2013, 18, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Nie, K.; Zhou, H.; Yan, X.; Zhan, Q.; Zheng, Y.; Song, C.P. ABI5 modulates seed germination via feedback regulation of the expression of the PYR/PYL/RCAR ABA receptor genes. New Phytol. 2020, 228, 596–608. [Google Scholar] [CrossRef]
- Lopez-Molina, L.; Mongrand, S.; McLachlin, D.T.; Chait, B.T.; Chua, N.H. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002, 32, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Giraudat, J.; Hauge, B.M.; Valon, C.; Smalle, J.; Parcy, F.; Goodman, H.M. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 1992, 4, 1251–1261. [Google Scholar]
- Zhao, L.; Hu, Y.; Chong, K.; Wang, T. ARAG1, an ABA-responsive DREB gene, plays a role in seed germination and drought tolerance of rice. Ann. Bot. 2010, 105, 401–409. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhao, J.; Feng, D.; Huang, Z.; Liang, J.; Zhang, Y.; Cheng, J.; Ying, J.; Wang, Z. RNA-Seq study reveals AP2-domain-containing signalling regulators involved in initial imbibition of seed germination in rice. Rice Sci. 2020, 27, 302–314. [Google Scholar]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family—An update. Curr. Opin. Plant Biol. 2018, 45 Pt A, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Dröge-Laser, W.; Weiste, C. The C/S (1) bZIP network: A regulatory nub orchestrating plant energy homeostasis. Trends Plant Sci. 2018, 23, 422–433. [Google Scholar] [CrossRef]
- Alves, M.S.; Dadalto, S.P.; Gonçalves, A.B.; De Souza, G.B.; Barros, V.A.; Fietto, L.G. Plant bZIP transcription factors responsive to pathogens: A review. Int. J. Mol. Sci. 2013, 14, 7815–7828. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Cho, J.I.; Han, M.; Ahn, C.H.; Jeon, J.S.; An, G.; Park, P.B. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 2010, 167, 1512–1520. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Guan, Y.; Ren, H.; Zhang, F.; Chen, F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 2008, 66, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lin, Q.; Wu, T.; Duan, E.; Huang, Y.; Yang, C.; Mou, C.; Lan, J.; Zhou, C.; Xie, K.; et al. OsDOG1L-3 regulates seed dormancy through the abscisic acid pathway in rice. Plant Sci. 2020, 298, 110570. [Google Scholar] [CrossRef]
- Zhu, C.C.; Wang, C.X.; Lu, C.Y.; Wang, J.D.; Zhou, Y.; Xiong, M.; Zhang, C.Q.; Liu, Q.Q.; Li, Q.F. Genome-wide identification and expression analysis of OsbZIP09 target genes in rice reveal its mechanism of controlling seed germination. Int. J. Mol. Sci. 2021, 22, 1661. [Google Scholar] [CrossRef]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J.; et al. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 2012, 10, 2–11. [Google Scholar] [CrossRef]
- Xie, W.; Li, X.; Wang, S.; Yuan, M. OsWRKY53 promotes abscisic acid accumulation to accelerate leaf senescence and inhibit seed germination by downregulating abscisic acid catabolic genes in rice. Front. Plant Sci. 2021, 12, 816156. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Mao, C.; Zhong, Q.; Yao, X.; Li, P.; Liu, C.; Ming, F. OsNAC2 is involved in multiple hormonal pathways to mediate germination of rice seeds and establishment of seedling. Front. Plant Sci. 2021, 12, 699303. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Dobrenel, T.; Caldana, C.; Hanson, J.; Robaglia, C.; Vincentz, M.; Veit, B.; Meyer, C. TOR signaling and nutrient sensing. Annu. Rev. Plant Biol. 2016, 67, 261–285. [Google Scholar] [CrossRef]
- Bosotti, R.; Isacchi, A.; Sonnhammer, E.L. FAT: A novel domain in PIK-related kinases. Trends Biochem. Sci. 2000, 25, 225–227. [Google Scholar] [CrossRef] [PubMed]
- Kunz, J.; Schneider, U.; Howald, I.; Schmidt, A.; Hall, M.N. HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J. Biol. Chem. 2000, 275, 37011–37020. [Google Scholar] [CrossRef] [PubMed]
- Schmelzle, T.; Hall, M.N. TOR, a central controller of cell growth. Cell 2000, 103, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Sarbassov, D.D.; Ali, S.M.; King, J.E.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002, 110, 163–175. [Google Scholar] [CrossRef]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 2002, 10, 457–468. [Google Scholar] [CrossRef]
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR signaling in growth and metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef]
- Moreau, M.; Azzopardi, M.; Clément, G.; Dobrenel, T.; Marchive, C.; Renne, C.; Martin-Magniette, M.L.; Taconnat, L.; Renou, J.P.; Robaglia, C.; et al. Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 2012, 24, 463–481. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.H.; Veit, B.; Hanson, M.R. The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol. 2005, 3, 12. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.A.; Giavalisco, P. Regulatory-associated protein of TOR 1B (RAPTOR1B) regulates hormonal switches during seed germination in Arabidopsis thaliana. Plant Signal. Behav. 2019, 14, 1613130. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.C.; Liu, X.; Fu, L.; Hou, Y.J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell 2018, 69, 100–112.e6. [Google Scholar] [CrossRef] [PubMed]
- Boudeau, J.; Miranda-Saavedra, D.; Barton, G.J.; Alessi, D.R. Emerging roles of pseudokinases. Trends Cell Biol. 2006, 16, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, S. SCYL pseudokinases in neuronal function and survival. Neural Regen. Res. 2016, 11, 42–44. [Google Scholar] [CrossRef] [PubMed]
- Conner, S.D.; Schmid, S.L. CVAK104 is a novel poly-l-lysine-stimulated kinase that targets the beta2-subunit of AP2. J. Biol. Chem. 2005, 280, 21539–21544. [Google Scholar] [CrossRef]
- Borner, G.H.; Rana, A.A.; Forster, R.; Harbour, M.; Smith, J.C.; Robinson, M.S. CVAK104 is a novel regulator of clathrin-mediated SNARE sorting. Traffic 2007, 8, 893–903. [Google Scholar] [CrossRef]
- Gingras, S.; Earls, L.R.; Howell, S.; Smeyne, R.J.; Zakharenko, S.S.; Pelletier, S. SCYL2 protects CA3 pyramidal neurons from excitotoxicity during functional maturation of the mouse hippocampus. J. Neurosci. 2015, 35, 10510–10522. [Google Scholar] [CrossRef]
- Jung, J.Y.; Lee, D.W.; Ryu, S.B.; Hwang, I.; Schachtman, D.P. SCYL2 genes are involved in clathrin-mediated vesicle trafficking and essential for plant growth. Plant Physiol. 2017, 175, 194–209. [Google Scholar] [CrossRef]
- Yao, Y.; Zhou, J.; Cheng, C.; Niu, F.; Zhang, A.; Sun, B.; Tu, R.; Wan, J.; Li, Y.; Huang, Y.; et al. A conserved clathrin-coated vesicle component, OsSCYL2, regulates plant innate immunity in rice. Plant Cell Environ. 2022, 45, 542–555. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, K.M.; Petersen, J. Fission yeast SCYL1/2 homologue Ppk32: A novel regulator of TOR signalling that governs survival during Brefeldin A induced stress to protein trafficking. PLoS Genet. 2016, 12, e1006041. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Ni, Y.; Tu, Y.; Wang, Y.; Zhang, Z.; Jiao, Y.; Zhang, X. A SCYL2 gene from Oryza sativa is involved in phytosterol accumulation and regulates plant growth and salt stress. Plant Sci. 2024, 343, 112062. [Google Scholar] [CrossRef] [PubMed]
- Yaish, M.W.; El-Kereamy, A.; Zhu, T.; Beatty, P.H.; Good, A.G.; Bi, Y.M.; Rothstein, S.J. The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 2010, 6, e1001098. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Hwang, H.; Hong, J.-W.; Lee, Y.-N.; Ahn, I.P.; Yoon, I.S.; Yoo, S.-D.; Lee, S.; Lee, S.C.; Kim, B.-G. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 2011, 63, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhu, C.; Zhou, Y.; Xiong, M.; Wang, J.; Bai, H.; Lu, C.; Zhang, C.; Liu, Q.; Li, Q. OsbZIP09, a unique OsbZIP transcription factor of rice, promotes rather than suppresses seed germination by attenuating abscisic acid pathway. Rice Sci. 2021, 28, 358–367. [Google Scholar]
- Finkelstein, R. Abscisic acid synthesis and response. Arab. Book 2013, 11, e0166. [Google Scholar] [CrossRef]
- Cafferkey, R.; Young, P.R.; McLaughlin, M.M.; Bergsma, D.J.; Koltin, Y.; Sathe, G.M.; Faucette, L.; Eng, W.K.; Johnson, R.K.; Livi, G.P. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol. Cell. Biol. 1993, 13, 6012–6023. [Google Scholar] [PubMed]
- Ali, I.; Sher, H.; Ali, A.; Hussain, S.; Ullah, Z. Simplified floral dip transformation method of Arabidopsis thaliana. J. Microbiol. Methods 2022, 197, 106492. [Google Scholar] [CrossRef]
- Wei, J.; Choi, H.; Jin, P.; Wu, Y.; Yoon, J.; Lee, Y.-S.; Quan, T.; An, G. GL2-type homeobox gene Roc4 in rice promotes flowering time preferentially under long days by repressing Ghd7. Plant Sci. 2016, 252, 133–143. [Google Scholar] [CrossRef]
- Kim, S.L.; Choi, M.; Jung, K.-H.; An, G. Analysis of the early-flowering mechanisms and generation of T-DNA tagging lines in Kitaake, a model rice cultivar. J. Exp. Bot. 2013, 64, 4169–4182. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, H.; Lian, X.; Converse, R.; Zhu, L. Identification of interacting motifs between armadillo repeat containing 1 (ARC1) and Exocyst 70 A1 (Exo70A1) proteins in Brassica oleracea. Protein J. 2016, 35, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zou, Y.; Shang, Y.; Lin, H.; Wang, Y.; Cai, R.; Tang, X.; Zhou, J.-M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2007, 146, 323–324. [Google Scholar] [CrossRef]
- Li, Z.; Wang, X.; Cao, X.; Chen, B.; Ma, C.; Lv, J.; Sun, Z.; Qiao, K.; Zhu, L.; Zhang, C.; et al. GhTULP34, a member of tubby-like proteins, interacts with GhSKP1A to negatively regulate plant osmotic stress. Genomics 2021, 113, 462–474. [Google Scholar] [CrossRef]


| ID | Description | KEGG | p Value | Regulated |
|---|---|---|---|---|
| Os01g0511000-01 | Predicted: LOB domain-containing protein 29 | K13945 | 2.87563 × 10−48 | down |
| Os04g0208200-01 | Predicted: senescence-specific cysteine protease SAG39 | K16292 | 0.001279091 | up |
| Os05g0545400-01 | Mitogen-activated protein kinase kinase kinase 19 | K20716 | 2.04441 × 10−28 | down |
| Os02g0187700-00 | Myb-like DNA-binding domain containing protein | K09422 | 9.00346 × 10−19 | down |
| Os06g0637500-02 | Predicted: transcription factor MYB | K09422 | 1.52605 × 10−18 | down |
| Os01g0699600-01 | Mitogen-activated protein kinase kinase kinase | K20716 | 1.02578 × 10−16 | down |
| Os04g0586500-01 | Predicted: receptor-like protein kinase FERONIA | -- | 7.77913 × 10−12 | down |
| Os03g0268600-01 | Protein phosphatase 2C 30 | K14497 | 2.91137 × 10−7 | down |
| Os05g0109825-00 | Predicted: E3 ubiquitin-protein ligase RING1 | K11982 | 2.08451 × 10−5 | down |
| Os09g0243200-01 | Predicted: E3 ubiquitin-protein ligase CIP8 | K22378 | 0.00023672 | up |
| Os05g0545300-01 | Predicted: mitogen-activated protein kinase kinase kinase 17/18 | K20716 | 0.001312045 | down |
| Os05g0494600-01 | Predicted: EID1-like F-box protein 3 | -- | 0.002511623 | down |
| Os11g0521900-01 | Predicted: RNA polymerase II C-terminal domain phosphatase-like 3/4 | K18999 | 0.002660793 | down |
| Os04g0610400-01 | APETALA-2-Like transcription factor gene; OsAP2-39 | K09286 | 1.9587 × 10−132 | down |
| Os05g0472400-01 | Zinc transporter 9 isoform X1 | K14709 | 9.95893 × 10−18 | up |
| Os01g0963600-01 | Abscisic acid-stress-ripening-inducible 6 protei | -- | 3.81499 × 10−50 | down |
| Os02g0657000-01 | AP2/EREBP-type transcription factor; ABA-responsive DREB gene | K09286 | 5.08219 × 10−12 | down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, M.; Zhang, W.; Jiao, Y.; Yang, Q.; Chen, M.; Cheng, H.; Cheng, B.; Zhang, X. OsSCYL2 is Involved in Regulating ABA Signaling-Mediated Seed Germination in Rice. Plants 2024, 13, 1088. https://doi.org/10.3390/plants13081088
Xu M, Zhang W, Jiao Y, Yang Q, Chen M, Cheng H, Cheng B, Zhang X. OsSCYL2 is Involved in Regulating ABA Signaling-Mediated Seed Germination in Rice. Plants. 2024; 13(8):1088. https://doi.org/10.3390/plants13081088
Chicago/Turabian StyleXu, Minyan, Wei Zhang, Yuhuan Jiao, Qing Yang, Meng Chen, Hu Cheng, Beijiu Cheng, and Xin Zhang. 2024. "OsSCYL2 is Involved in Regulating ABA Signaling-Mediated Seed Germination in Rice" Plants 13, no. 8: 1088. https://doi.org/10.3390/plants13081088
APA StyleXu, M., Zhang, W., Jiao, Y., Yang, Q., Chen, M., Cheng, H., Cheng, B., & Zhang, X. (2024). OsSCYL2 is Involved in Regulating ABA Signaling-Mediated Seed Germination in Rice. Plants, 13(8), 1088. https://doi.org/10.3390/plants13081088





