From Phenotypes to Genotypes: Enhancing the Identification of Cymbidium Species with DNA Barcoding
Abstract
1. Introduction
2. Results
2.1. Analysis of Amplification Success Rate and Sequence Characteristics
2.2. Analysis of Barcoding Gap
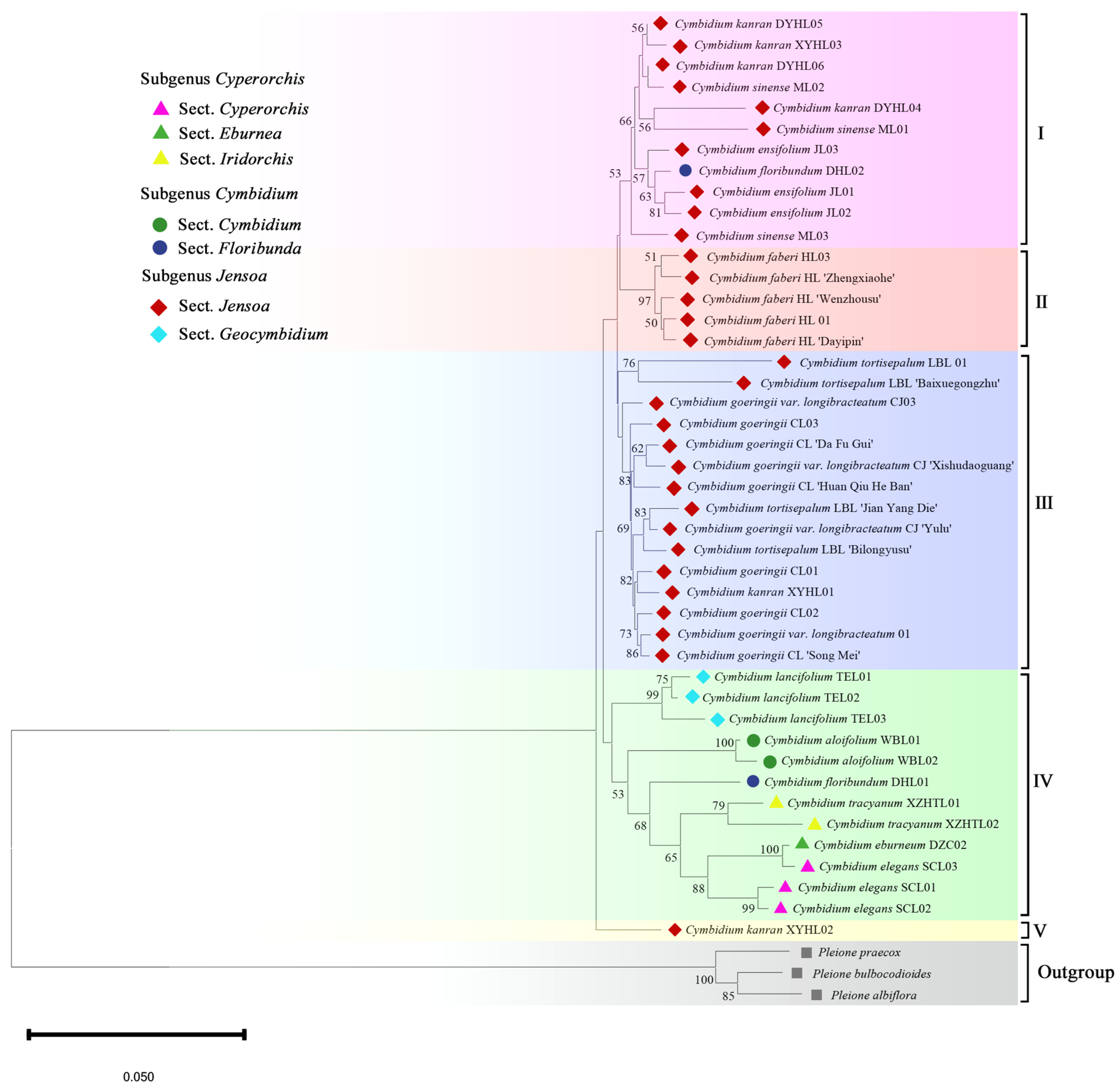
2.3. Phylogenetic Analysis
2.4. Genetic Diversity Analysis of Phenotypic Characters in Orchid Germplasm Resources
2.4.1. Diversity Analysis of Quantitative Traits
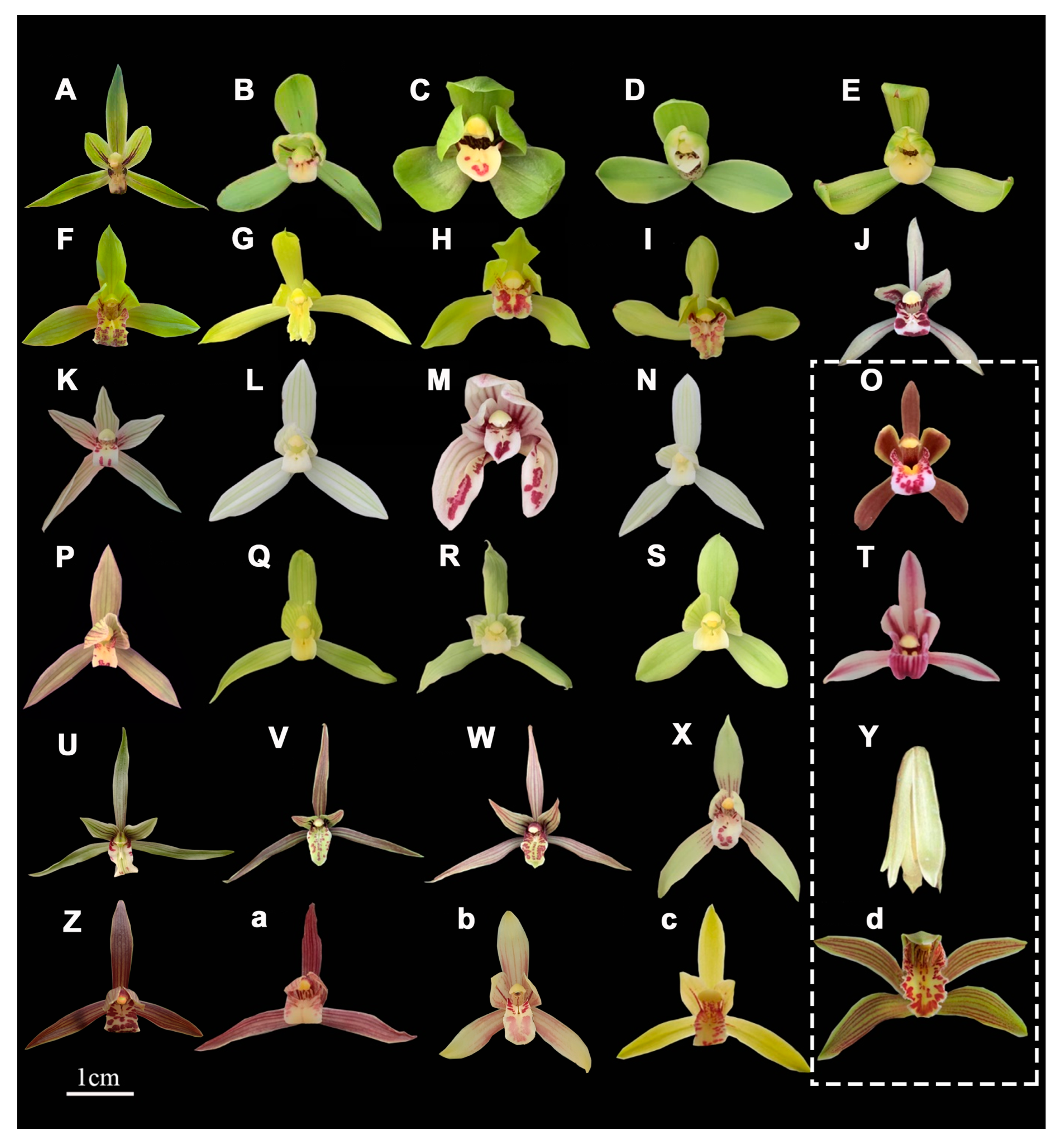
2.4.2. Diversity Evaluation of Quality Traits
2.4.3. Principal Component Analysis of Phenotypic Traits
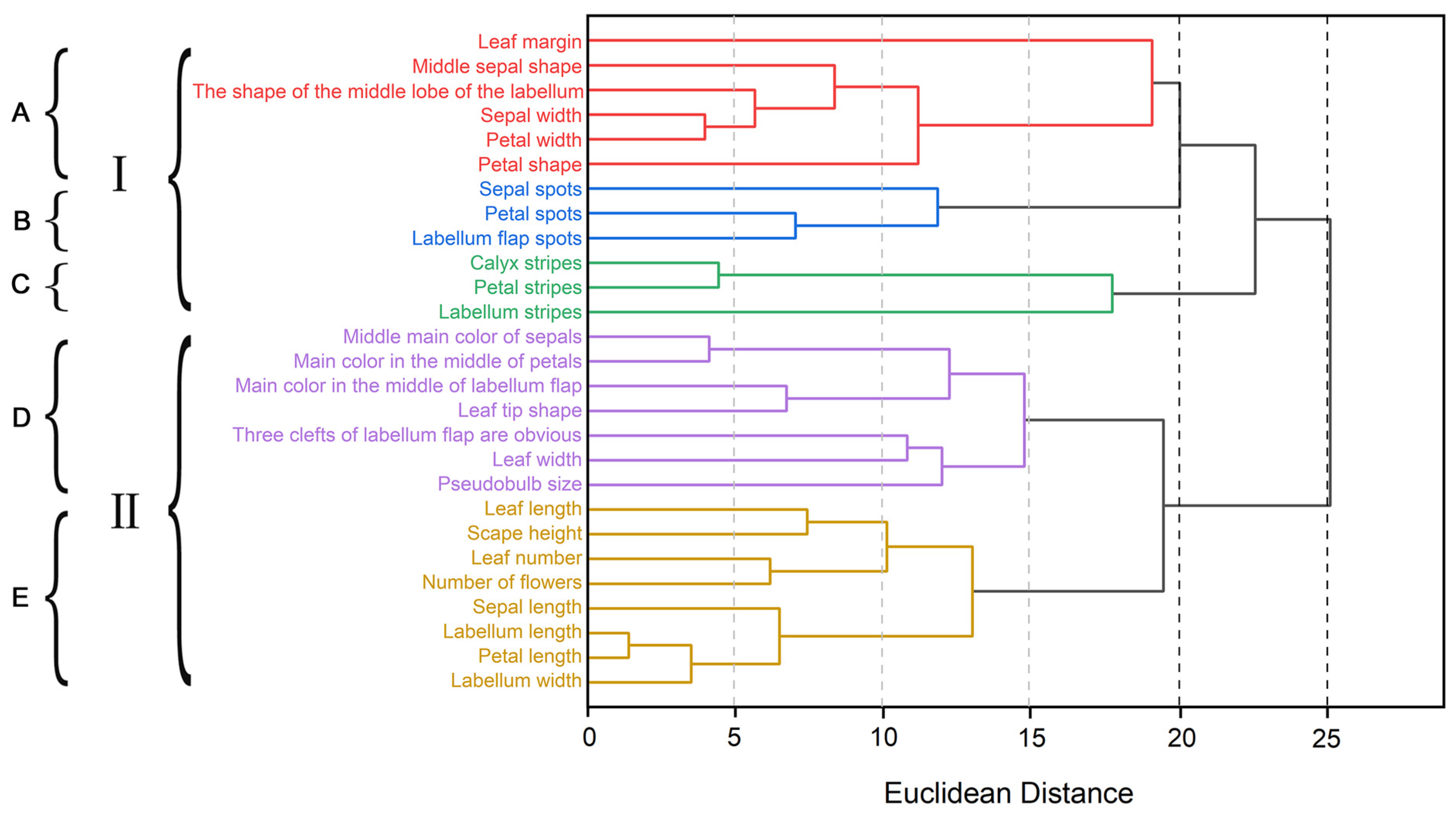
2.5. Cluster Analysis of Phenotypic Traits
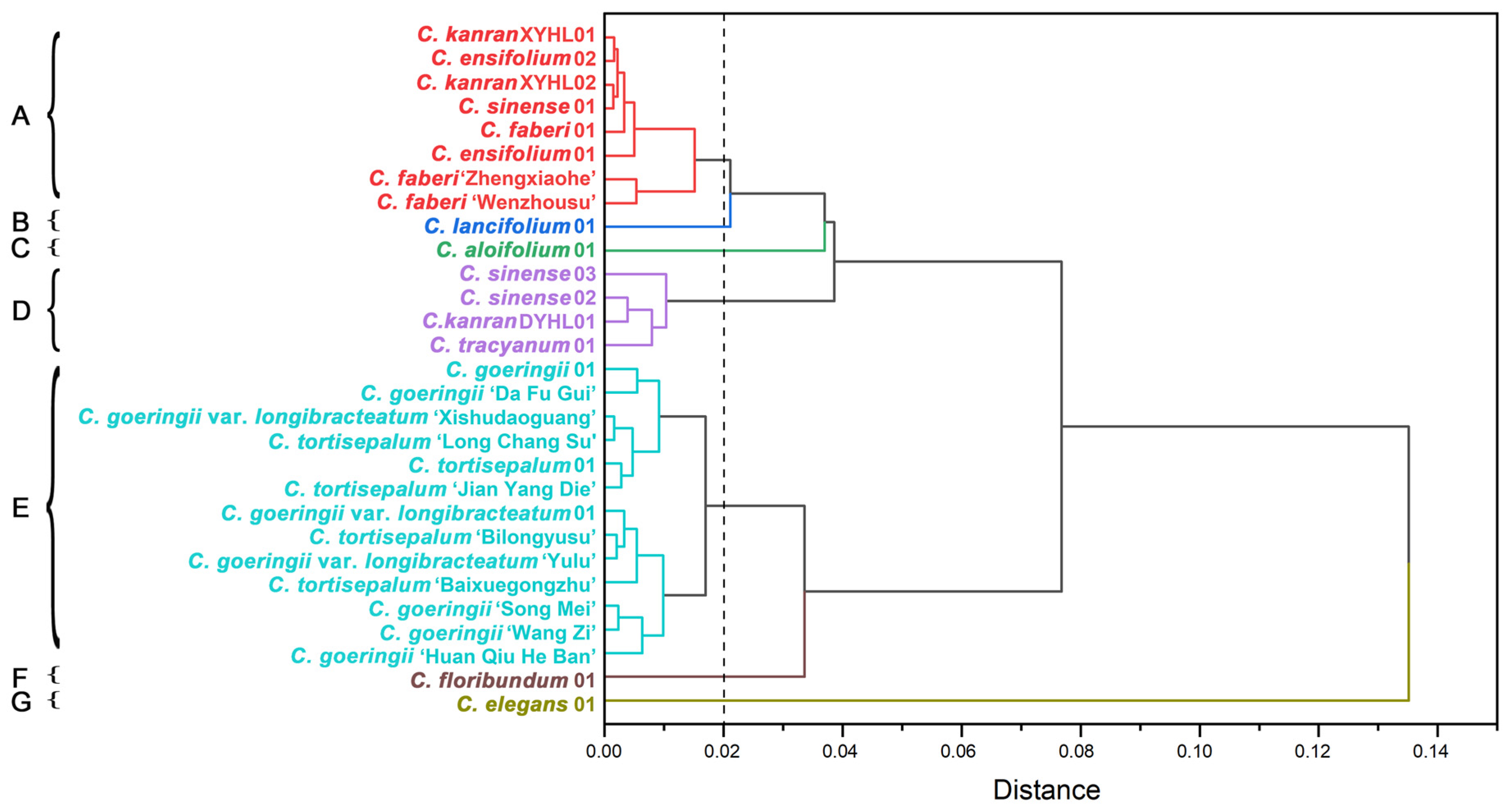
2.6. The Support of Molecular Characters for Morphological Features
3. Discussion
3.1. The Universality and Applicability of Sequences
3.2. Assessment of the Discriminating Power of Single and a Combination of Barcodes
3.3. The Phylogeny of the Cymbidium
3.4. Identification and Classification of Cymbidium Species Through the Combination of Morphological Analysis and DNA Barcoding Technology
3.5. Cymbidium Species Diversity
4. Materials and Method
4.1. Plant Materials
4.2. DNA Extraction, Amplification, and Sequencing
4.3. Data Analysis
4.4. Selection and Determination Methods of Phenotypic Traits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, X.Q.; Ji, Z.H. The Complete Book of Chinese Orchids; China Forestry Publishing House: Beijing, China, 1998; Volume 3, pp. 1–282. [Google Scholar]
- Elazazi, E.; Ziems, L.; Mahmood, T. Genotypic Selection Using Quantitative Trait Loci for Better Productivity under High Temperature Stress in Tomato (Solanum lycopersicum L.). Horticulturae 2024, 10, 874. [Google Scholar] [CrossRef]
- Yan, H. Morphological and ISSR Molecular Identification of Several Cymbidium Species. Master’s Thesis, Sichuan Agricultural University, Ya’an, China, 2012. [Google Scholar]
- Freudenstein, J.V. Orchid phylogenetics and evolution: History, current status and prospects. Ann. Bot. 2024, mcae202. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Cao, B.; Zhang, Y.Q.; Chen, Q.X.; Chen, N.C. Genetic Diversity Analysis of Chinese Orchid Germplasm Resources Based on SRAP Markers. Chin. J. Trop. Crops 2020, 41, 929–938. [Google Scholar]
- Schlechter, R. Die Gattungen Cymbidium Sw. und Cyperorchis Bl. Repert. Nov. Specierum Regni Veg. 1924, 20, e96–e110. [Google Scholar] [CrossRef]
- Seth, C.J.; Cribb, P.J. A Reassessment of the Sectional Limits in the Genus Cymbidium Swartz. In Orchid Biology, Reviews and Prospectives 3; Cornell University Press: Ithaca, NY, USA; London, UK, 1984. [Google Scholar]
- Puy, D.D.; Cribb, P. The Genus Cymbidium; Helm, C., Ed.; The University of Chicago Press: Chicago, IL, USA, 1988; Volume 10. [Google Scholar]
- Liu, Z.J.; Chen, X.Q.; Ru, Z.Z. The Genus Cymbidium in China; Science Press: Beijing, China, 2006. [Google Scholar]
- Hollingsworth, P.M. Refining the DNA Barcode for Land Plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19451–19452. [Google Scholar] [CrossRef] [PubMed]
- Li, D.Z.; Gao, L.M.; Li, H.T.; Wang, H.; Ge, X.J.; Liu, J.Q.; Chen, Z.D.; Zhou, S.L.; Chen, S.L.; Yang, J.B.; et al. Comparative Analysis of a Large Dataset Indicates that Internal Transcribed Spacer (ITS) should be Incorporated into the Core Barcode for Seed Plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [PubMed]
- Schindel, D.E.; Miller, S.E. DNA Barcoding a Useful Tool for Taxonomists. Nature 2005, 435, 17. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Jin, Y.Y.; Gao, Y.D.; Zhang, Y.; Ying, Q.C.; Shen, C.J.; Lu, J.J.; Zhan, X.R.; Wang, H.Z.; Feng, S.G. The Complete chloroplast Genomes of Two Physalis Species, Physalis macrophysa and P. ixocarpa: Comparative Genomics, Evolutionary Dynamics and Phylogenetic Relationships. Agronomy 2023, 13, 135. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; Dewaard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. Lond. B 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. Bold: The Barcode of Life Data System. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Ren, B.Q.; Chen, Z.R. Plant DNA Barcode Technology. J. Bot. 2010, 45, 1–12. [Google Scholar]
- Yang, J.B.; Tang, M.; Li, H.T.; Zhang, Z.R.; Li, D.Z. Complete Chloroplast Genome of the Genus Cymbidium: Lights into the Species Identification, Phylogenetic Implications and Population Genetic Analyses. BMC Evol. Biol. 2013, 13, 84. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Huang, Y.W.; Huang, J.L.; Ya, J.D.; Zhe, M.Q.; Zeng, C.X.; Zhang, Z.R.; Zhang, S.B.; Li, D.Z.; Li, H.T.; et al. DNA Barcoding of Cymbidium by Genome Skimming: Call for Next-Generation Nuclear Barcodes. Mol. Ecol. Resour. 2022, 23, 424–439. [Google Scholar] [CrossRef]
- Wang, X.M.; Cheng, L.J.; Li, Z.L.; Zhang, Z.C.; Wang, Y.Y. Characterization of the Complete Chloroplast Genome of a Chinese Endangered Species Cymbidium wenshanense Y. S. Wu et F. Y. Liu. Mitochondrial DNA B Resour. 2023, 8, 815–818. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.K.; Dkhar, J.; Kumaria, S.; Tandon, P.; Rao, S.R. Assessment of Phylogenetic Inter-Relationships in the Genus Cymbidium (Orchidaceae) Based on Internal Tanscribed Spacer Region of rDNA. Gene 2012, 495, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Kim, M.J.; Lee, J.S.; Ryu, K.H. Genetic Diversity and Phylogenetic Relationships among and within Species of Oriental Cymbidiums Based on RAPD Analysis. Sci. Hortic. 2006, 108, 79–85. [Google Scholar] [CrossRef]
- Zhang, G.Q.; Chen, G.Z.; Chen, L.J.; Zhai, J.W.; Huang, J.; Wu, X.Y.; Li, M.H.; Peng, D.H.; Rao, W.H.; Liu, Z.J.; et al. Phylogenetic Incongruence in Cymbidium Orchids. Plant Divers. 2021, 43, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.H.; Zeng, X.; Yang, D.; Ren, G.M.; Chu, G.Y.; Yuan, Z.R.; Luo, K.; Xiao, P.G.; Chen, S.L. Identification of Medicinal Vines by ITS2 using Complementary Discrimination Methods. J. Ethnopharmacol. 2012, 141, 242–249. [Google Scholar] [CrossRef]
- Dong, W.P.; Xu, C.; Li, C.H.; Sun, J.H.; Zuo, Y.J.; Shi, S.; Cheng, T.; Guo, J.J.; Zhou, S.L. Ycf1, the Most Promising Plastid DNA Barcode of Land Plants. Sci. Rep. 2015, 5, 8348. [Google Scholar] [CrossRef]
- Ning, S.P.; Yan, H.F.; Hao, G.; Ge, X.J. Research Progress on Plant DNA Barcode. Biodivers. Sci. 2008, 5, 417–425. [Google Scholar]
- Zhang, X.S. Evolution and Maintenance of the Environmental Component of the Phenotypic Variance: Benefit of Plastic Traits under Changing Environments. Am. Nat. 2005, 166, 569–580. [Google Scholar] [CrossRef]
- Kolb, A.; Ehrlén, J. Environmental Context Drives Seed Predator-Mediated Selection on a Floral Display Trait. Evol. Ecol. 2010, 24, 433–445. [Google Scholar] [CrossRef]
- Sletvold, N.; Grindeland, J.M.; Ågren, J. Vegetation Context Influences the Strength and Targets of Pollinator-Mediated Selection in a Deceptive Orchid. Ecology 2013, 94, 1236–1242. [Google Scholar] [CrossRef]
- Ning, H.J.; Ao, S.Y.; Fan, Y.Y.; Fu, J.X.; Xu, C.M. Correlation Analysis between the Karyotypes and Phenotypic Traits of Chinese Cymbidium Cultivars. Hortic. Environ. Biotechnol. 2018, 59, 93–103. [Google Scholar] [CrossRef]
- Li, W.; Wang, P.; Qi, Q.G.; Zhang, Q.C.; Gao, X.; Lin, M.F.; Cui, Y.T. Phenotypic Diversity and Variation of Lonicera Caerulea Populations in the Changbai Mountain Alongside the Elevation Gradient. Pol. J. Environ. Stud. 2021, 30, 705–716. [Google Scholar] [CrossRef]
- Ren, J.F.; Yang, J.; Li, H.W.; Yun, Y.; Zhang, L. Research Progress and Prospects of DNA Barcoding in Orchids. Mod. Hortic. 2015, 9, 11–12. [Google Scholar]
- Hollingsworth, P.; Graham, S.; Little, D. Choosing and using a Plant DNA Barcode. PLoS ONE 2011, 6, 236–254. [Google Scholar] [CrossRef] [PubMed]
- Mehraj, S.; Parihar, T.J.; Murtaza, D.; Hurrah, A.A.; Wani, I.A.; Lone, F.; Mufti, S.; Zargar, S.M.; Khan, I.; Sheikh, P.A. Newly emerging aquatic macrophytes in Northern Himalayan, Dal Lake of Kashmir Valley identified through DNA Barcoding and Its Antioxidant Profiling. Ecol. Genet. Genom. 2023, 27, 100162. [Google Scholar]
- Cai, H.Y.; Xing, S.T.; He, R.R.; Zhou, B.; Zhao, F. Phylogenetic Analysis of Rhododendron in Mount Taishan Mountain based on ITS2 Sequence and Morphological Characteristics. Plant Physiol. J. 2017, 8, 1489–1498. [Google Scholar]
- Cai, Y.M.; Dai, J.P.; Zheng, Y.X.; Ren, Y.Y.; Chen, H.M.; Feng, T.T.; Gao, X.X.; Zhu, S. DNA Barcode Screening for Molecular Identification of Uncaria Plants. Chin. Tradit. Herb. Drugs 2022, 53, 1828–1837. [Google Scholar]
- Wang, D.X. ITS-1 Sequencing and Single Nucleotide Polymorphism Variation Site Analysis of Gastrodia elata. Biotechnology 2012, 22, 48–51. [Google Scholar]
- Wu, J.; Li, D.; Boyd, B.; Balan, R.K.; George, S.; Peacock, L.; Pal, C. Comparative Performance of a Multi-Locus Barcoding Approach to enhance Taxonomic Resolution of New Zealand Mosquitoes (Diptera: Culicidae). Aust. Entomol. 2023, 62, 77–95. [Google Scholar] [CrossRef]
- Sayed, H.A.; Mostafa, S.; Haggag, I.M.; Hassan, N.A. DNA Barcoding of Prunus Species Collection Conserved in the National Gene Bank of Egypt. Mol. Biotechnol. 2023, 65, 410–418. [Google Scholar] [CrossRef]
- Zhang, J.J. Phylogenetic Analysis of Dioscorea Plants Based on psbA-trnH Gene and 18S rDNA. Master’s Thesis, Inner Mongolia University, Hohhot, China, 2017. [Google Scholar]
- Zhou, H.; Ma, S.J.; Yang, P.; Yao, H. Research Progress on Identification of Dendrobium Medicinal Plants based on DNA Sequence. Mod. Tradit. Chin. Med. Mater. Med. World Sci. Technol. 2015, 17, 950–957. [Google Scholar]
- Kim, H.M.; Oh, S.H.; Bhandari, G.S.; Kim, C.S.; Park, C.W. DNA Barcoding of Orchidaceae in Korea. Mol. Ecol. Resour. 2014, 14, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.Q.; Xiang, X.G.; Chen, Z. Species Identification of Alnus (Betulaceae) using nrDNA and cpDNA Genetic Markers. Mol. Ecol. Resour. 2010, 10, 594–605. [Google Scholar] [CrossRef]
- Fazekas, A.; Burgess, K.; Kesanakurti, P.; Graham, S.W.; Newmaster, S.G.; Husband, B.C.; Percy, D.M.; Hajibabaei, M.; Barrett, S.C.H. Multiple Multilocus DNA Barcodes from the Plastid Genome Discriminate Plant Species Equally Well. PLoS ONE 2008, 3, e2802. [Google Scholar] [CrossRef]
- Zheng, Y.D. Molecular Phylogenetic Study of Fagopyrum Plants in Southwest China. Master’s Thesis, Sichuan Agricultural University, Ya’an, China, 2013. [Google Scholar]
- Yu, W.B.; Huang, P.H.; Li, D.Z.; Wang, H. Incongruence between nuclear and chloroplast DNA phylogenies in Pedicularis section Cyathophora (Orobanchaceae). PLoS ONE 2013, 8, e74828. [Google Scholar] [CrossRef]
- Guo, Y.Y.; Luo, Y.B.; Liu, Z.J.; Wang, X.Q. Reticulate evolution and sea-level fluctuations together drove species diversification of slipper orchids (Paphiopedilum) in South-East Asia. Mol. Ecol. 2015, 24, 2838–2855. [Google Scholar] [CrossRef]
- De Ré, F.C.; Robe, L.J.; Wallau, G.L.; Loreto, E.L.S. Inferring the phylogenetic position of the Drosophila flavopilosa group: Incongruence within and between mitochondrial and nuclear multilocus datasets. J. Zool. Syst. Evol. Res. 2017, 55, 208–221. [Google Scholar] [CrossRef]
- Kanzi, A.M.; Trollip, C.; Wingfield, M.J.; Barnes, I.; Van der Nest, M.A.; Wingfield, B.D. Phylogenomic incongruence in Ceratocystis: A clue to speciation. BMC Genom. 2020, 21, 362. [Google Scholar] [CrossRef]
- Tang, Y.; Yukawa, T.; Bateman, R.M.; Jiang, H.; Peng, H. Phylogeny and classification of the east Asian Amitostigma alliance (Orchidaceae: Orchideae) based on six DNA markers. BMC Evol. Biol. 2015, 15, 96. [Google Scholar] [CrossRef] [PubMed]
- Perkins, J.; Hayashi, T.; Peakall, R.; Flematti, G.R.; Bohman, B. The volatile chemistry of orchid pollination. Nat. Prod. Rep. 2023, 40, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.Q.; Liu, K.W.; Li, Z.; Lohaus, R.; Hsiao, Y.Y.; Niu, S.C.; Wang, J.Y.; Lin, Y.C.; Xu, Q.; Chen, L.J. The Apostasia genome and the evolution of orchids. Nature 2017, 549, 379–383. [Google Scholar] [CrossRef]
- Sugahara, M.; Minamoto, T.; Fuchikawa, T.; Michinomae, M.; Shimizu, I. Apis cerana japonica discriminates between floral color phases of the oriental orchid, Cymbidium floribundum. Zool. Sci. 2010, 27, 901–906. [Google Scholar] [CrossRef]
- Dressler, R.L.; Dodson, C.H. Classification and Phylogeny in the Orchidaceae. Ann. Mo. Bot. Gard. 1960, 47, 25–68. [Google Scholar] [CrossRef]
- Burns-Balogh, P.; Funk, V.A. A phylogenetic Analysis of the Orchidaceae. Smithson. Contrib. Bot. 1986, 61, 1–79. [Google Scholar] [CrossRef]
- Wang, H.C.; Moore, M.J.; Soltis, P.S.; Bell, C.D.; Brockington, S.F.; Alexandre, R.; Davis, C.C.; Latvis, M.; Manchester, S.R.; Soltis, D.E. Rosid Radiation and the Rapid Rise of Angiosperm-Dominated Forests. Proc. Natl. Acad. Sci. USA 2009, 106, 3853–3858. [Google Scholar] [CrossRef] [PubMed]
- Ao, S.Y. Analysis on Phenotypic Characters and Karyotype of Cymbidium; Zhejiang A&F University: Hangzhou, China, 2014. [Google Scholar]
- Ji, X.B.; Wang, G.D.; Kang, J.C.; Qiao, G.; Wen, X.P. Comparative Analysis of Morphology and RAPD Markers of 12 Orchid Plant Resources in Guizhou Province. Seed 2008, 2, 56–60. [Google Scholar]
- Wei, X.Y.; Liu, H.; Ma, H.; Bie, T.D.; Sun, Y. Analysis of Genetic Diversity and Fingerprint Map Construction of 96 Orchid Germplasm Resources Based on ISSR Markers. J. Plant Genet. Resour. 2024, 25, 586–599. [Google Scholar]
- Yang, Y.Y. Phenotypic Diversity and ITS Marker Analysis of konjac Plants. Master’s Thesis, Yunnan University, Kunming, China, 2020. [Google Scholar]
- Zhe, M.Q.; Zhang, L.; Liu, F.; Huang, Y.W.; Fan, W.S.; Yang, J.B.; Zhu, A.D. Plastid RNA Editing Reduction Accompanied with Genetic Variations in Cymbidium, a Genus with Diverse Lifestyle Modes. Plant Divers. 2022, 44, 316–321. [Google Scholar] [CrossRef]
- Chen, H.Y.; Zhang, Z.R.; Yao, X.; Ya, J.D.; Jin, X.H.; Wang, L.; Lu, L.; Li, D.Z.; Yang, J.B.; Yu, W.B. Plastid Phylogenomics Provides New Insights into the Systematics, Diversification, and Biogeography of Cymbidium (Orchidaceae). Plant Divers. 2024, 46, 448–461. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.M.; Gao, L.; Wang, H.Z.; Feng, S.G. Molecular Identification and Phylogenetic Analysis of Cymbidium Species (Orchidaceae) Based on the Potential DNA Barcodes matK, rbcL, psbA-trnH, and Internal Transcribed Spacer. Agronomy 2024, 14, 933. [Google Scholar] [CrossRef]
- Crayn, D.M.; Winter, K.; Smith, J.A.C. Multiple Origins of Crassulacean Acid Metabolism and the Epiphytic Habit in the Neotropical Family Bromeliaceae. Proc. Natl. Acad. Sci. USA 2004, 101, 3703–3708. [Google Scholar] [CrossRef] [PubMed]
- Schneider, H.; Schuetipelz, E.; Pryer, K.M.; Cranfill, R.; Magallón, S.; Lupia, R. Ferns Diversified in the Shadow of Angiosperms. Nature 2004, 428, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Schuettpelz, E.; Pryer, K.M. Evidence for a Cenozoic Radiation of Ferns in an Angiosperm–Dominated Canopy. Proc. Natl. Acad. Sci. USA 2009, 106, 11200–11205. [Google Scholar] [CrossRef] [PubMed]
- Zotz, G.; Weigelt, P.; Kessler, M.; Kreft, H.; Taylor, A. EpiList 1.0: A global Checklist of Vascular Epiphytes. Ecology 2021, 102, e03326. [Google Scholar] [CrossRef] [PubMed]
- Raskoti, B.B.; Ale, R. DNA Barcoding of Medicinal Orchids in Asia. Sci. Rep. 2021, 11, 23651. [Google Scholar] [CrossRef]
- Worthy, S.J.; Bucalo, K.; Perry, E.; Reynolds, A.; Cruse-Sanders, J.; Pérez, A.J.; Burgess, K.S. Ability of rbcL and matK DNA Barcodes to Discriminate between Montane Forest Orchids. Plant Syst. Evol. 2022, 308, 19. [Google Scholar] [CrossRef]
- Givnish, T.J.; Spalink, D.; Ames, M.; Lyon, S.P.; Hunter, S.J.; Zuluaga, A.; Iles, W.J.D.; Clements, M.A.; Arroyo, M.T.K.; Leebens-Mack, J. Orchid phylogenomics and Multiple Drivers of Their Extraordinary Diversification. Proc. R. Soc. B 2015, 282, 171–180. [Google Scholar] [CrossRef]
- Pérez-Escobar, O.A.; Dodsworth, S.; Bogarín, D.; Bellot, S.; Balbuena, J.A.; Schley, R.J.; Kikuchi, I.A.; Morris, S.K.; Epitawalage, N.; Cowan, R. Hundreds of Nuclear and Plastid Loci Yield Novel Insights into Orchid Relationships. Am. J. Bot. 2021, 108, 1166–1180. [Google Scholar] [CrossRef]
- Pérez-Escobar, O.A.; Bogarín, D.; Przelomska, N.A.S.; Ackerman, J.D.; Balbuena, J.A.; Bellot, S.; Bühlmann, R.P.; Cabrera, B.; Cano, J.A.; Charitonidou, M. The Origin and Speciation of Orchids. New Phytol. 2024, 242, 700–716. [Google Scholar] [CrossRef]
- Zhang, W.X.; Zhang, G.Q.; Zeng, P.; Zhang, Y.Q.; Hu, H.; Liu, Z.J.; Cai, J. Genome Sequence of Apostasia ramifera Provides Insights into the Adaptive Evolution in Orchids. BMC Genom. 2021, 22, 536. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.Q.; Xu, Q.; Bian, C.; Tsai, W.C.; Yeh, C.M.; Liu, K.W.; Yoshida, K.; Zhang, L.S.; Chang, S.B.; Chen, F. The Dendrobium Catenatum Lindl. Genome Sequence Provides Insights into Polysaccharide Synthase, Floral Development and Adaptive Evolution. Sci. Rep. 2016, 6, 19029. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Tan, K.; Ren, M.X. Effects of monsoon on distribution patterns of tropical plants in Asia. Chin. J. Plant Ecol. 2017, 41, 1103–1112, (In Chinese with English Abstract). [Google Scholar]
- Ding, W.N.; Ree, R.H.; Spicer, R.A.; Xing, Y.W. Ancient orogenic and monsoon-driven assembly of the world’s richest temperate alpine flora. Science 2020, 369, 578–581. [Google Scholar] [CrossRef]
- Ye, J.W.; Tian, B.; Li, D.Z. Monsoon intensification in East Asia triggered the evolution of its flora. Front. Plant Sci. 2022, 13, 1046538. [Google Scholar] [CrossRef]
- Ashokan, A.; Xavier, A.; Suksathan, P.; Ardiyani, M.; Leong-Skornicková, J.; Newman, M.; Kress, W.J.; Gowda, V. Himalayan orogeny and monsoon intensification explain species diversification in an endemic ginger (Hedychium: Zingiberaceae) from the Indo-Malayan Realm. Mol. Phylogenet. Evol. 2022, 170, 107440. [Google Scholar] [CrossRef]
- Chinese Academy of Sciences. Flora of China; Science Press: Beijing, China, 1999; Volume 18, p. 191. [Google Scholar]
- Sun, Y.; Skinner, D.; Liang, G.; Hulbert, S.H. Phylogenetic Analysis of Sorghum and Related Taxa using Internal Transcribed Spacers of Nuclear Ribosomal DNA. Theor. Appl. Genet. 1994, 89, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Cuenoud, P.; Savolainen, V.; Chatrou, L.W.; Powell, M.; Grayer, R.; Chase, M.W. Molecular Phylogenetics of Caryophyllales Based on Nuclear 18S rDNA and Plastid rbcL, atpB, and matK DNA Sequences. Am. J. Bot. 2002, 89, 132–144. [Google Scholar] [CrossRef]
- Sang, T.; Crawford, D.J.; Stuessy, T.F. Chloroplast DNA Phylogeny, Reticulate Evolution, and Biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 1997, 84, 1120–1132. [Google Scholar] [CrossRef]
- Simpson, B.B. Paraphyly of Tarasa (Malvaceae) and Diverse Origins of the Polyploid Species. Syst. Bot. 2003, 28, 723–737. [Google Scholar]
- Goldman, D.H.; Freudenstein, J.V.; Kores, P.J.; Molvray, M.; Jarrell, D.C.; Whitten, W.M.; Cameron, K.M.; Jansen, R.K.; Chase, M.W. Phylogenetics of Arethuseae (Orchidaceae) Based on Plastid matK and rbcL Sequences. Syst. Bot. 2001, 37, 670–695. [Google Scholar]
- Sun, G. Interspecific Polymorphism at Non-Coding Regions of Chloroplast, Mitochondrial DNA and rRNA IGS Region in Elymus Species. Hereditas 2002, 137, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Meier, R.; Zhang, G.Y.; Ali, F. The Use of Mean Instead of Smallest Interspecific Distances Exaggerates the Size of the “Barcoding Gap” and Leads to Misidentification. Syst. Biol. 2008, 57, 809–813. [Google Scholar] [CrossRef]
- Chen, S.L.; Yao, H.; Han, J.P.; Liu, C.; Song, J.Y.; Shi, L.C.; Zhu, Y.J.; Ma, X.Y.; Gao, T.; Pang, X.H.; et al. Validation of the ITS2 Region as a Novel DNA Barcode for Identifying Medicinal Plant Species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.G.; Jiang, M.Y.; Shi, Y.J.; Jiao, K.L.; Shen, C.J.; Lu, J.J.; Ying, Q.C.; Wang, H.Z. Application of the Ribosomal DNA ITS2 Region of Physalis (Solanaceae): DNA Barcoding and Phylogenetic Study. Front. Plant Sci. 2016, 7, 1047. [Google Scholar] [CrossRef]
- Carl, R. Woese Institute for Genomic Biology, University of Illinois at Urbana-Champaign. Pattern Analysis of Phylogenetic Trees Could Reveal Connections Between Evolution, Ecology. Available online: https://www.sciencedaily.com/releases/2020/06/200626125018.htm (accessed on 10 February 2025).
- Somrup, S.; Sangsawang, A.; Mcmillan, N.; Winitchai, S.; Inthoncharoen, J.; Liu, S.K.; Muangmai, N. Pinctada phuketensis sp. nov. (Bivalvia, Ostreida, Margaritidae), a New Pearl oyster Species from Phuket, Western Coast of Thailand. ZooKeys 2022, 1119, 181–195. [Google Scholar] [CrossRef]





| DNA Barcodes | Number of Samples | Number of Successful Amplifications | Amplification Success Rate | Number of Successful Sequences | Sequencing Success Rate |
|---|---|---|---|---|---|
| ITS | 48 | 48 | 100% | 46 | 95.8% |
| matK | 48 | 48 | 100% | 48 | 100% |
| rbcL | 48 | 48 | 100% | 48 | 100% |
| psbA-trnH | 48 | 48 | 100% | 46 | 95.8% |
| trnL-F | 48 | 48 | 100% | 46 | 95.8% |
| Sequence Information | ITS | matK | rbcL | trnL-F | psbA-trnH |
|---|---|---|---|---|---|
| Comparison length (bp) | 885 | 861 | 645 | 775 | 865 |
| GC content (%) | 63.6% | 31.1% | 41.1% | 34.62% | 33.36% |
| Conserved site | 672 (75.93%) | 784 (91.06%) | 627 (97.21%) | 721 (93.03%) | 732 (84.62%) |
| Total variation sites | 165 (18.64%) | 73 (8.48%) | 17 (2.64%) | 23 (2.97%) | 96 (11.10%) |
| Parsimony-informative site | 49 | 40 | 10 | 11 | 74 |
| Sequence Information | ITS + matK | matK + psbA-trnH | ITS + psbA-trnH | ITS + psbA-trnH + matK |
|---|---|---|---|---|
| Comparison length (bp) | 1716 | 1726 | 1720 | 2581 |
| GC content (%) | 48.1% | 32.5% | 50.3% | 43.6% |
| Conserved site | 1399 (81.53%) | 1516 (87.83%) | 1367 (79.48%) | 2162 (83.77%) |
| Total variation sites | 303 (17.66%) | 169 (9.79%) | 306 (17.79%) | 368 (14.26%) |
| Parsimony-informative site | 124 | 114 | 154 | 188 |
| Single polymorphic loci | 179 | 54 | 151 | 179 |
| DNA Barcodes | Intraspecific Genetic Distance | Interspecific Genetic Distance | ||||
|---|---|---|---|---|---|---|
| Minimum | Maximum | Mean | Minimum | Maximum | Mean | |
| ITS | 0 | 0.09657 | 0.01759 | 0 | 0.14136 | 0.02890 |
| matk | 0 | 0.02262 | 0.00335 | 0 | 0.03115 | 0.01208 |
| rbcL | 0 | 0.00782 | 0.00088 | 0 | 0.01731 | 0.00424 |
| trnL-F | 0 | 0.00980 | 0.00168 | 0 | 0.02004 | 0.00434 |
| psbA-trnH | 0 | 0.02128 | 0.00513 | 0 | 0.04525 | 0.01588 |
| DNA Barcodes | Intraspecific Genetic Distance | Interspecific Genetic Distance | ||||
|---|---|---|---|---|---|---|
| Minimum | Maximum | Mean | Minimum | Maximum | Mean | |
| ITS + matK | 0 | 0.04979 | 0.01041 | 0 | 0.07025 | 0.02025 |
| ITS + psbA-trnH | 0 | 0.04855 | 0.01147 | 0 | 0.09987 | 0.02258 |
| matK + psbA-trnH | 0 | 0.02387 | 0.00409 | 0 | 0.04290 | 0.01366 |
| ITS + matK + psbA-trnH | 0 | 0.03395 | 0.00824 | 0 | 0.06201 | 0.01841 |
| Barcodes | Primer Name | Primer Sequence (5′–3′) | Reaction Procedure | References |
|---|---|---|---|---|
| ITS | 17SE | ACGAATTCATGGTCCGGTGAAGTGTTCG | 95 °C 3 min, 35 cycle (95 °C 15 s, 62 °C 15 s, 72 °C 15 s), 72 °C 5 min | Sun et al., 1994 [79] |
| 26SE | TAGAATTCCCCGGTTCGCTCGCCGTTAC | Sun et al., 1994 | ||
| matK | 390F | CGATCTATTCATTCAATATTTC | 95 °C 3 min, 35 cycle (95 °C 15 s, 46.5 °C 15 s, 72 °C 15 s), 72 °C 5 min | Cuenoud et al., 2002 [80] |
| 1326R | TCTAGCACACGAAAGTCGAAGT | Cuenoud et al., 2002 | ||
| psbA-trnH | psbA | GTTATGCATGAACGTAATGCTC | 95 °C 3 min, 35 cycle (95 °C 15 s, 55 °C 15 s, 72 °C 15 s), 72 °C 5 min | Sang et al., 1997 [81] |
| trnH2 | CGCGCATGGTGGATTCACAATCC | Tate, 2002 [82] | ||
| rbcL | 1F | ATGTCACCACAAACAGAAAC | 95 °C 3 min, 35 cycle (95 °C 15 s, 56 °C 15 s, 72 °C 15 s), 72 °C 5 min | Goldman et al., 2001 [83] |
| 724R | TGCCATGTACCYGCAGTTGC | Goldman et al., 2001 | ||
| trnL-F | c | CGAAATCGGTAGACGCTACG | 95 °C 3 min, 35 cycle (95 °C 15 s, 53 °C 15 s, 72 °C 15 s), 72 °C 5 min | Taberlet et al., 1991 [84] |
| f | ATTTGAACTGGTGACACGAG | Taberlet et al., 1991 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Y.; Chen, Y.; Ding, H.; Liu, X.; Cao, F.; Xu, L. From Phenotypes to Genotypes: Enhancing the Identification of Cymbidium Species with DNA Barcoding. Plants 2025, 14, 619. https://doi.org/10.3390/plants14040619
Peng Y, Chen Y, Ding H, Liu X, Cao F, Xu L. From Phenotypes to Genotypes: Enhancing the Identification of Cymbidium Species with DNA Barcoding. Plants. 2025; 14(4):619. https://doi.org/10.3390/plants14040619
Chicago/Turabian StylePeng, Yaonan, Yao Chen, Hongfan Ding, Xiangdong Liu, Fuxiang Cao, and Lu Xu. 2025. "From Phenotypes to Genotypes: Enhancing the Identification of Cymbidium Species with DNA Barcoding" Plants 14, no. 4: 619. https://doi.org/10.3390/plants14040619
APA StylePeng, Y., Chen, Y., Ding, H., Liu, X., Cao, F., & Xu, L. (2025). From Phenotypes to Genotypes: Enhancing the Identification of Cymbidium Species with DNA Barcoding. Plants, 14(4), 619. https://doi.org/10.3390/plants14040619







