Transcriptome Analysis Reveals the Pivotal Genes and Regulation Pathways Under Cold Stress and Identifies SbERF027, an AP2/ERF Gene That Confers Cold Tolerance in Sorghum
Abstract
:1. Introduction
2. Results
2.1. Cold Tolerance Evaluation
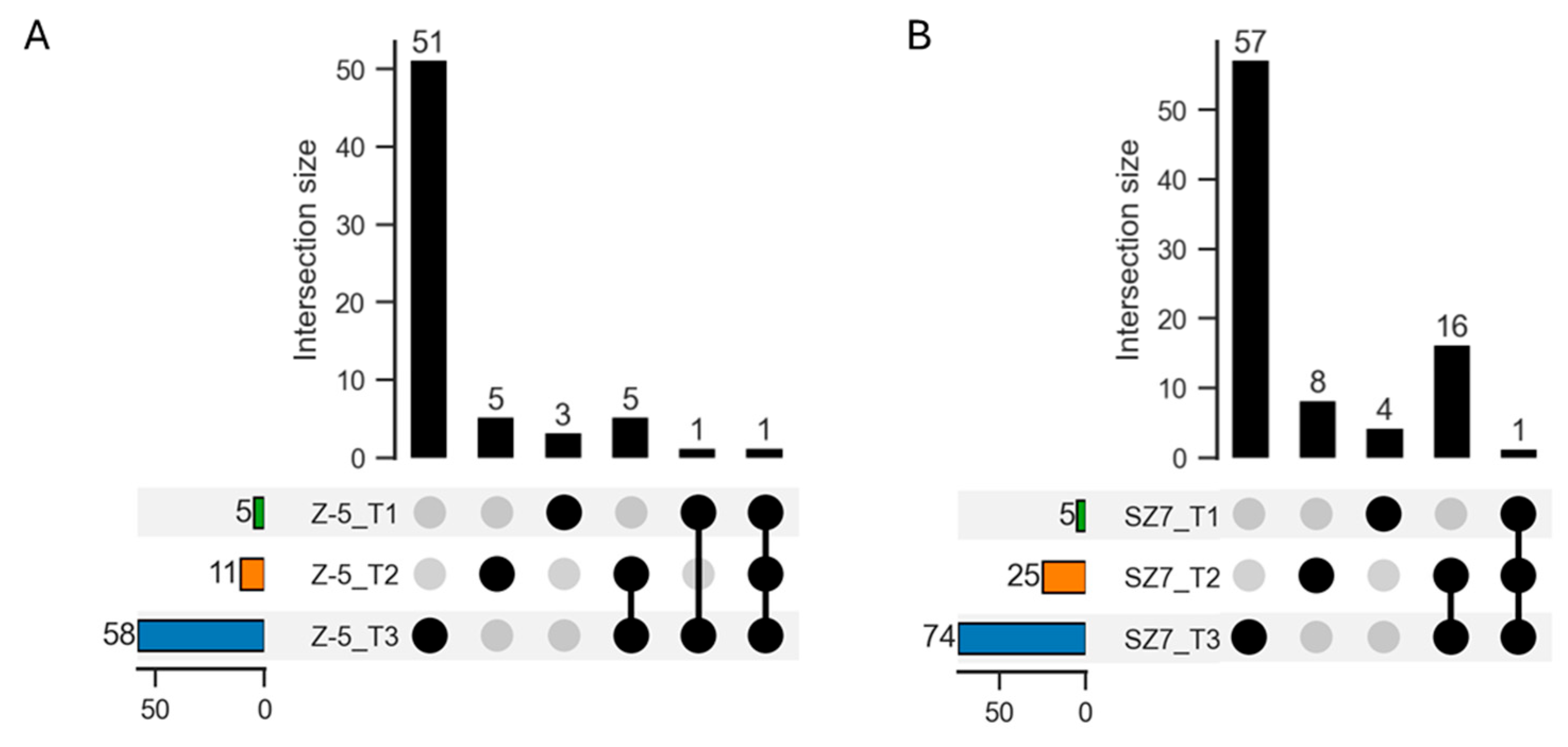
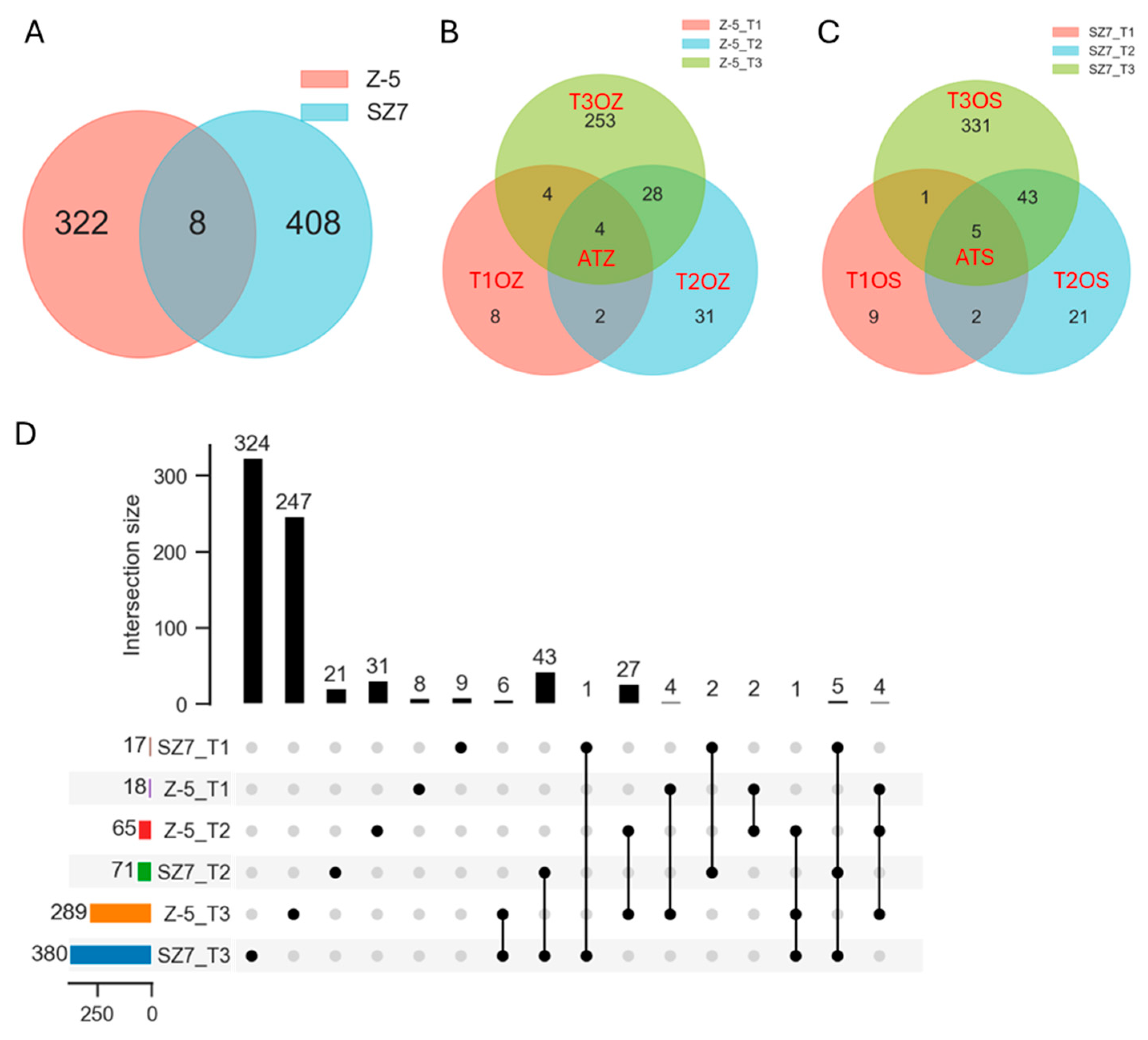
2.2. Transcriptome Sequencing Data
2.3. Functional Annotation of DEGs
2.4. Analysis of DEGs Enriched in GO Terms Related to “Cold”
2.5. KEGG Pathway Enrichment Analysis
2.6. TFs Related to Cold Tolerance
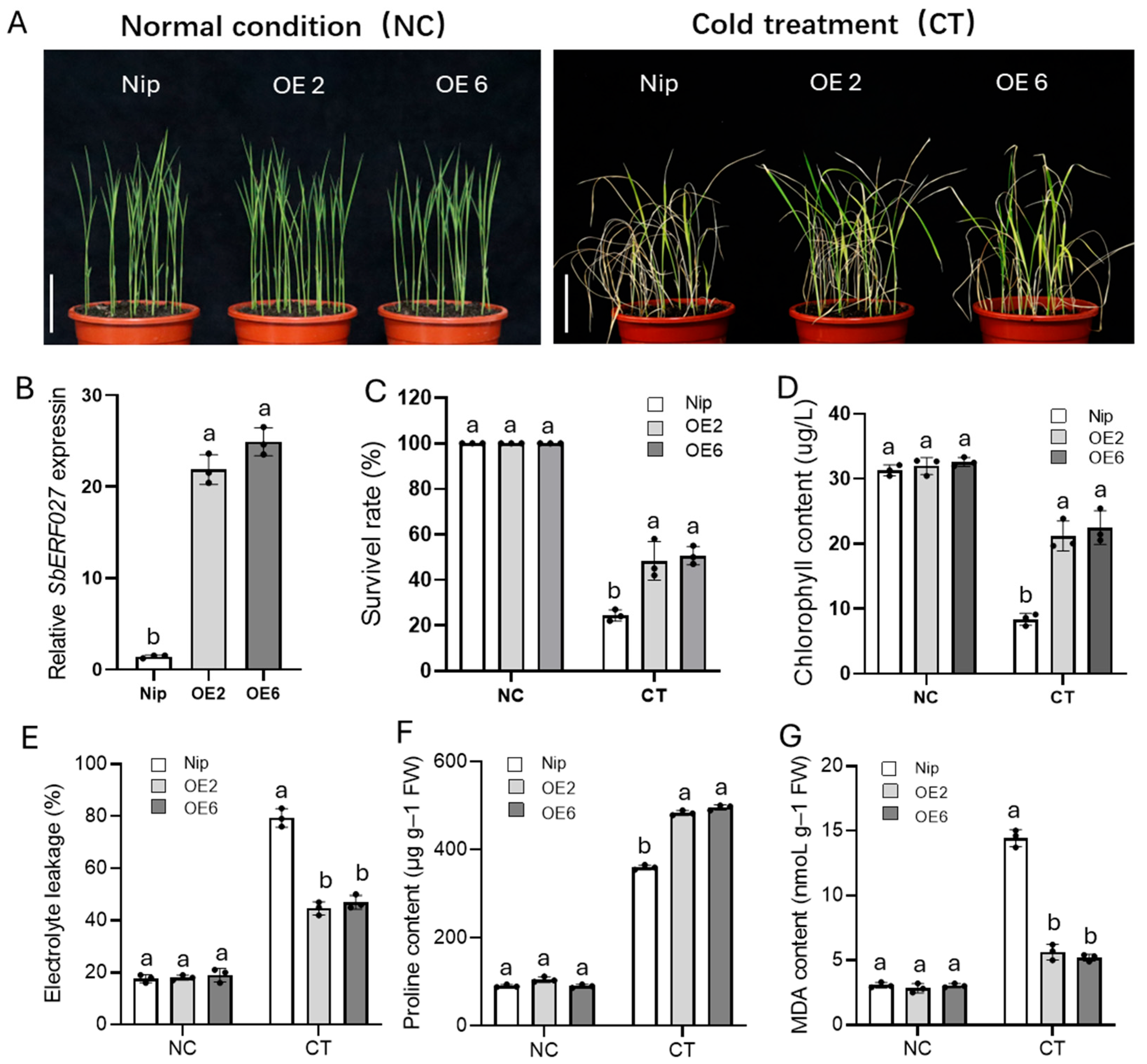
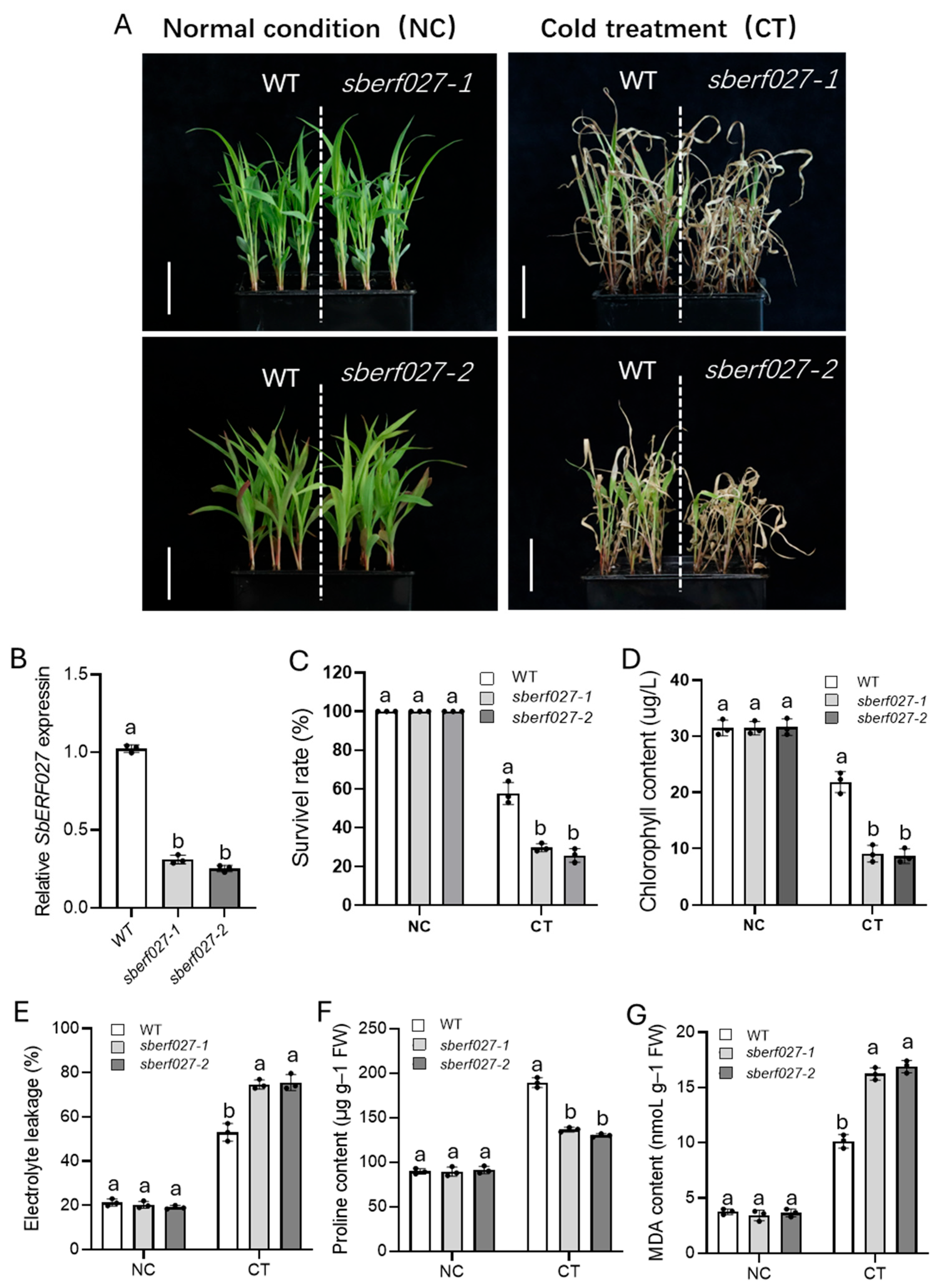
2.7. SbERF027 Positively Regulates Cold Tolerance in Sorghum Seedlings
2.8. Analysis of SbERF027 Expression Patterns
2.9. SNP-911 on the Promoter of SbERF027 Was Responsible for Transcriptional Differences
3. Discussion
3.1. Discovery of Genes for Cold Tolerance in Sorghum Seedlings
3.2. Gene Function and Regulatory Pathways Under Cold Stress
3.3. AP2/ERF TFs Play a Pivotal Role in the Cold Stress Response in Plants
3.4. Possible Involved Pathways of SbERF027 in Response to Cold Stress in Sorghum
4. Conclusions
5. Materials and Method
5.1. Plant Culture and Phenotypic Evaluation
5.2. Transcriptome Analysis
5.3. Plasmid Construction and Genetic Transformation
5.4. Expression Patterns and Subcellular Localization
5.5. Determination of Physiological Indices
5.6. Transcriptional Activity Assays
5.7. Transactivation Assays of SbERF027
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chopra, R.; Burow, G.; Burke, J.J.; Gladman, N.; Xin, Z. Genome-Wide Association Analysis of Seedling Traits in Diverse Sorghum Germplasm under Thermal Stress. BMC Plant Biol. 2017, 17, 12. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, S.; Kravcov, N.; Schaffasz, A.; Snowdon, R.J.; Wittkop, B.; Windpassinger, S. Genetic Architecture of Novel Sources for Reproductive Cold Tolerance in Sorghum. Front. Plant Sci. 2021, 12, 772177. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H. Genomics of Sorghum. Int. J. Plant Genom. 2008, 2008, 362451. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.-Y.; Guo, X.-S.; He, B.; Sun, L.-J.; Peng, Y.; Dong, S.-S.; Liu, T.-F.; Jiang, S.; Ramachandran, S.; Liu, C.-M.; et al. Genome-Wide Patterns of Genetic Variation in Sweet and Grain Sorghum (Sorghum bicolor). Genome Biol. 2011, 12, R114. [Google Scholar] [CrossRef]
- Lou, Q.; Guo, H.; Li, J.; Han, S.; Khan, N.U.; Gu, Y.; Zhao, W.; Zhang, Z.; Zhang, H.; Li, Z.; et al. Cold-Adaptive Evolution at the Reproductive Stage in Geng/Japonica Subspecies Reveals the Role of OsMAPK3 and OsLEA9. Plant J. 2022, 111, 1032–1051. [Google Scholar] [CrossRef]
- Yu, J.; Tuinstra, M.R.; Claassen, M.M.; Gordon, W.B.; Witt, M.D. Analysis of Cold Tolerance in Sorghum under Controlled Environment Conditions. Field Crops Res. 2004, 85, 21–30. [Google Scholar] [CrossRef]
- Xin, Z.; Wang, M.; Cuevas, H.E.; Chen, J.; Harrison, M.; Pugh, N.A.; Morris, G. Sorghum Genetic, Genomic, and Breeding Resources. Planta 2021, 254, 114. [Google Scholar] [CrossRef]
- Zhang, G.; Shi, H.; Zhang, H. Characterization and Evaluation for Cold Tolerance of Sorghum Germplasm. Chin. Agric. Sci. Bull. 2012, 28, 87–91. [Google Scholar] [CrossRef]
- Xu, J.; Ding, Y.; Cao, N.; Cheng, B.; Gao, X.; Zou, G.; Zhang, L. Analysis of Cold Tolerance of Grain Sorghum Germplasm Resources. Seed 2021, 40, 40–45. [Google Scholar] [CrossRef]
- Zhu, L.; Ding, Y.; Xu, J.; Cao, N.; Xu, Y.; Wen, X.; Zhang, L. Genetic Diversity Analysis and Core Germplasms Identification of Guizhou Sorghum Resources. Seed 2023, 42, 74–82. [Google Scholar] [CrossRef]
- Zhang, G.; Ge, Y.; Zhang, Z. Identification and Comprehensive Evaluation of Cold Tolerance of Sorghum Germplasms During Seed Germination. Seed 2021, 40, 28–40. [Google Scholar] [CrossRef]
- Revilla, P.; Rodríguez, V.M.; Ordás, A.; Rincent, R.; Charcosset, A.; Giauffret, C.; Melchinger, A.E.; Schön, C.-C.; Bauer, E.; Altmann, T.; et al. Association Mapping for Cold Tolerance in Two Large Maize Inbred Panels. BMC Plant Biol. 2016, 16, 127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Li, F.; Liu, H.; Yang, W.; Chong, K.; Xu, Y. OsMAPK3 Phosphorylates OsbHLH002/OsICE1 and Inhibits Its Ubiquitination to Activate OsTPP1 and Enhances Rice Chilling Tolerance. Dev. Cell 2017, 43, 731–743.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, S.; Dong, J.; Yang, T.; Mao, X.; Liu, Q.; Wang, X.; Liu, B. A Novel Functional Gene Associated with Cold Tolerance at the Seedling Stage in Rice. Plant Biotechnol. J. 2017, 15, 1141–1148. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Pan, Y.; Li, J.; Zhou, L.; Shi, H.; Zeng, Y.; Guo, H.; Yang, S.; Zheng, W.; et al. Natural Variation in CTB4a Enhances Rice Adaptation to Cold Habitats. Nat. Commun. 2017, 8, 14788. [Google Scholar] [CrossRef]
- Guo, H.; Wu, T.; Li, S.; He, Q.; Yang, Z.; Zhang, W.; Gan, Y.; Sun, P.; Xiang, G.; Zhang, H.; et al. The Methylation Patterns and Transcriptional Responses to Chilling Stress at the Seedling Stage in Rice. Int. J. Mol. Sci. 2019, 20, 5089. [Google Scholar] [CrossRef]
- Knoll, J.; Gunaratna, N.; Ejeta, G. QTL Analysis of Early-Season Cold Tolerance in Sorghum. Theor. Appl. Genet. 2008, 116, 577–587. [Google Scholar] [CrossRef]
- Burow, G.; Burke, J.J.; Xin, Z.; Franks, C.D. Genetic Dissection of Early-Season Cold Tolerance in Sorghum (Sorghum bicolor (L.) Moench). Mol. Breed. 2011, 28, 391–402. [Google Scholar] [CrossRef]
- Bekele, W.A.; Fiedler, K.; Shiringani, A.; Schnaubelt, D.; Windpassinger, S.; Uptmoor, R.; Friedt, W.; Snowdon, R.J. Unravelling the Genetic Complexity of Sorghum Seedling Development under Low-Temperature Conditions. Plant Cell Environ. 2014, 37, 707–723. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Wang, Y.-H.; Sastry, D.V.S.S.R.; Dwivedi, S.L.; Prasad, P.V.V.; Burrell, A.M.; Klein, R.R.; Morris, G.P.; Klein, P.E. Association Mapping of Germinability and Seedling Vigor in Sorghum under Controlled Low-Temperature Conditions. Genome 2016, 59, 137–145. [Google Scholar] [CrossRef]
- Parra-Londono, S.; Fiedler, K.; Kavka, M.; Samans, B.; Wieckhorst, S.; Zacharias, A.; Uptmoor, R. Genetic Dissection of Early-Season Cold Tolerance in Sorghum: Genome-Wide Association Studies for Seedling Emergence and Survival under Field and Controlled Environment Conditions. Theor. Appl. Genet. 2018, 131, 581–595. [Google Scholar] [CrossRef] [PubMed]
- Moghimi, N.; Desai, J.S.; Bheemanahalli, R.; Impa, S.M.; Vennapusa, A.R.; Sebela, D.; Perumal, R.; Doherty, C.J.; Jagadish, S.V.K. New Candidate Loci and Marker Genes on Chromosome 7 for Improved Chilling Tolerance in Sorghum. J. Exp. Bot. 2019, 70, 3357–3371. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, S.; Mufumbo, R.; Windpassinger, S.; Jordan, D.; Mace, E.; Snowdon, R.J.; Hathorn, A. Genetic and Genomic Diversity in the Sorghum Gene Bank Collection of Uganda. BMC Plant Biol. 2022, 22, 378. [Google Scholar] [CrossRef] [PubMed]
- La Borde, N.; Rajewski, J.; Dweikat, I. Novel QTL for Chilling Tolerance at Germination and Early Seedling Stages in Sorghum. Front. Genet. 2023, 14, 1129460. [Google Scholar] [CrossRef]
- Borde, N.L.; Dweikat, I. Identification of Genomic Regions Associated with Seedling Frost Tolerance in Sorghum. Genes 2023, 14, 2117. [Google Scholar] [CrossRef]
- Salas Fernandez, M.G.; Schoenbaum, G.R.; Goggi, A.S. Novel Germplasm and Screening Methods for Early Cold Tolerance in Sorghum. Crop Sci. 2014, 54, 2631–2638. [Google Scholar] [CrossRef]
- Maulana, F.; Weerasooriya, D.; Tesso, T. Sorghum Landrace Collections from Cooler Regions of the World Exhibit Magnificent Genetic Differentiation and Early Season Cold Tolerance. Front. Plant Sci. 2017, 8, 756. [Google Scholar] [CrossRef]
- Franks, C.D.; Burow, G.B.; Burke, J.J. A Comparison of US and Chinese Sorghum Germplasm for Early Season Cold Tolerance. Crop Sci. 2006, 46, 1371–1376. [Google Scholar] [CrossRef]
- Du, Q.; Fang, Y.; Jiang, J.; Chen, M.; Li, X.; Xie, X. Genome-Wide Identification and Characterization of the JAZ Gene Family and Its Expression Patterns under Various Abiotic Stresses in Sorghum bicolor. J. Integr. Agric. 2022, 21, 3540–3555. [Google Scholar] [CrossRef]
- Nagaraju, M.; Kumar, A.; Rajasheker, G.; Manohar Rao, D.; Kavi Kishor, P.B. DnaJs, the Critical Drivers of Hsp70s: Genome-Wide Screening, Characterization and Expression of DnaJ Family Genes in Sorghum bicolor. Mol. Biol. Rep. 2020, 47, 7379–7390. [Google Scholar] [CrossRef]
- Nagaraju, M.; Kumar, A.; Jalaja, N.; Rao, D.M.; Kishor, P.B.K. Functional Exploration of Chaperonin (HSP60/10) Family Genes and Their Abiotic Stress-Induced Expression Patterns in Sorghum bicolor. Curr. Genom. 2021, 22, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Ramegowda, V.; Basu, S.; Krishnan, A.; Pereira, A. Rice GROWTH UNDER DROUGHT KINASE Is Required for Drought Tolerance and Grain Yield under Normal and Drought Stress Conditions. Plant Physiol. 2014, 166, 1634–1645. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Chung, P.J.; Park, S.-H.; Redillas, M.C.F.R.; Kim, Y.S.; Suh, J.-W.; Kim, J.-K. Overexpression of OsERF48 Causes Regulation of OsCML16, a Calmodulin-like Protein Gene That Enhances Root Growth and Drought Tolerance. Plant Biotechnol. J. 2017, 15, 1295–1308. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Yu, J.; Miao, J.; Li, J.; Zhang, H.; Wang, X.; Liu, P.; Zhao, Y.; Jiang, C.; Yin, Z.; et al. Natural Variation in OsLG3 Increases Drought Tolerance in Rice by Inducing ROS Scavenging. Plant Physiol. 2018, 178, 451–467. [Google Scholar] [CrossRef]
- Wang, J.; Ren, Y.; Liu, X.; Luo, S.; Zhang, X.; Liu, X.; Lin, Q.; Zhu, S.; Wan, H.; Yang, Y.; et al. Transcriptional Activation and Phosphorylation of OsCNGC9 Confer Enhanced Chilling Tolerance in Rice. Mol. Plant 2021, 14, 315–329. [Google Scholar] [CrossRef]
- Chen, H.-C.; Chien, T.-C.; Chen, T.-Y.; Chiang, M.-H.; Lai, M.-H.; Chang, M.-C. Overexpression of a Novel ERF-X-Type Transcription Factor, OsERF106MZ, Reduces Shoot Growth and Tolerance to Salinity Stress in Rice. Rice 2021, 14, 82. [Google Scholar] [CrossRef]
- Jia, M.; Meng, X.; Song, X.; Zhang, D.; Kou, L.; Zhang, J.; Jing, Y.; Liu, G.; Liu, H.; Huang, X.; et al. Chilling-Induced Phosphorylation of IPA1 by OsSAPK6 Activates Chilling Tolerance Responses in Rice. Cell Discov. 2022, 8, 71. [Google Scholar] [CrossRef]
- Sun, M.; Shen, Y.; Chen, Y.; Wang, Y.; Cai, X.; Yang, J.; Jia, B.; Dong, W.; Chen, X.; Sun, X. Osa-miR1320 Targets the ERF Transcription Factor OsERF096 to Regulate Cold Tolerance via JA-Mediated Signaling. Plant Physiol. 2022, 189, 2500–2516. [Google Scholar] [CrossRef]
- Xu, L.; Yang, L.; Li, A.; Guo, J.; Wang, H.; Qi, H.; Li, M.; Yang, P.; Song, S. An AP2/ERF Transcription Factor Confers Chilling Tolerance in Rice. Sci. Adv. 2024, 10, eado4788. [Google Scholar] [CrossRef]
- Lata, C.; Mishra, A.K.; Muthamilarasan, M.; Bonthala, V.S.; Khan, Y.; Prasad, M. Genome-Wide Investigation and Expression Profiling of AP2/ERF Transcription Factor Superfamily in Foxtail Millet (Setaria italica L.). PLoS ONE 2014, 9, e113092. [Google Scholar] [CrossRef]
- Li, Z.; Wang, G.; Liu, X.; Wang, Z.; Zhang, M.; Zhang, J. Genome-Wide Identification and Expression Profiling of DREB Genes in Saccharum spontaneum. BMC Genom. 2021, 22, 456. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liao, J.; Ling, Q.; Xi, Y.; Qian, Y. Genome-Wide Identification and Expression Profiling Analysis of Maize AP2/ERF Superfamily Genes Reveal Essential Roles in Abiotic Stress Tolerance. BMC Genom. 2022, 23, 125. [Google Scholar] [CrossRef] [PubMed]
- Mathur, S.; Priyadarshini, S.S.; Singh, V.; Vashisht, I.; Jung, K.-H.; Sharma, R.; Sharma, M.K. Comprehensive Phylogenomic Analysis of ERF Genes in Sorghum Provides Clues to the Evolution of Gene Functions and Redundancy among Gene Family Members. 3 Biotech 2020, 10, 139. [Google Scholar] [CrossRef] [PubMed]
- Boonburapong, B.; Buaboocha, T. Genome-Wide Identification and Analyses of the Rice Calmodulin and Related Potential Calcium Sensor Proteins. BMC Plant Biol. 2007, 7, 4. [Google Scholar] [CrossRef]
- Hurry, V.; Druart, N.; Cavaco, A.; Gardeström, P.; Strand, Å. Photosynthesis at Low Temperatures. In Plant Cold Hardiness; Li, P.H., Palva, E.T., Eds.; Springer: Boston, MA, USA, 2002; pp. 161–179. ISBN 978-1-4613-5205-1. [Google Scholar]
- Srivasta, A.; Mehta, S.; Lindlof, A.; Bhargava, S. Over-Represented Promoter Motifs in Abiotic Stress-Induced DREB Genes of Rice and Sorghum and Their Probable Role in Regulation of Gene Expression. Plant Signal Behav. 2010, 5, 775–784. [Google Scholar] [CrossRef]
- Phillips, T.; Hoopes, L. Transcription Factors and Transcriptional Control in Eukaryotic Cells. Nat. Educ. 2008, 1, 119. [Google Scholar]
- Vera Hernández, P.F.; Mendoza Onofre, L.E.; Rosas Cárdenas, F. de F. Responses of Sorghum to Cold Stress: A Review Focused on Molecular Breeding. Front. Plant Sci. 2023, 14, 1124335. [Google Scholar] [CrossRef]
- Yang, T.; Wang, Y.; Li, Y.; Liang, S.; Yang, Y.; Huang, Z.; Li, Y.; Gao, J.; Ma, N.; Zhou, X. The Transcription Factor RhMYB17 Regulates the Homeotic Transformation of Floral Organs in Rose (Rosa hybrida) under Cold Stress. J. Exp. Bot. 2024, 75, 2965–2981. [Google Scholar] [CrossRef]
- Lei, P.; Jiang, Y.; Zhao, Y.; Jiang, M.; Ji, X.; Ma, L.; Jin, G.; Li, J.; Zhang, S.; Kong, D.; et al. Functions of Basic Helix-Loop-Helix (bHLH) Proteins in the Regulation of Plant Responses to Cold, Drought, Salt, and Iron Deficiency: A Comprehensive Review. J. Agric. Food Chem. 2024, 72, 10692–10709. [Google Scholar] [CrossRef]
- Zhang, L.; Xing, L.; Dai, J.; Li, Z.; Zhang, A.; Wang, T.; Liu, W.; Li, X.; Han, D. Overexpression of a Grape WRKY Transcription Factor VhWRKY44 Improves the Resistance to Cold and Salt of Arabidopsis thaliana. Int. J. Mol. Sci. 2024, 25, 7437. [Google Scholar] [CrossRef]
- Liu, Y.; Khan, A.R.; Gan, Y. C2H2 Zinc Finger Proteins Response to Abiotic Stress in Plants. Int. J. Mol. Sci. 2022, 23, 2730. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, P.; Kummari, D.; Palakolanu, S.R.; Nagasai Tejaswi, U.; Nagaraju, M.; Rajasheker, G.; Jawahar, G.; Jalaja, N.; Rathnagiri, P.; Kavi Kishor, P.B. Genome-Wide Identification and Expression Profile Analysis of Nuclear Factor Y Family Genes in Sorghum bicolor L. (Moench). PLoS ONE 2019, 14, e0222203. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R.J.; Srivastava, A.; Tsimilli-Michael, M. The Fluorescence Transient as a Tool to Characterize and Screen Photosynthetic Samples. Probing Photosynth. Mech. Regul. Adapt. 2000, 25, 445–483. [Google Scholar]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient Expression Vectors for Functional Genomics, Quantification of Promoter Activity and RNA Silencing in Plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef]
- Kidokoro, S.; Hayashi, K.; Haraguchi, H.; Ishikawa, T.; Soma, F.; Konoura, I.; Toda, S.; Mizoi, J.; Suzuki, T.; Shinozaki, K.; et al. Posttranslational Regulation of Multiple Clock-Related Transcription Factors Triggers Cold-Inducible Gene Expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2021048118. [Google Scholar] [CrossRef]
- Rawat, N.; Singla-Pareek, S.L.; Pareek, A. Membrane Dynamics during Individual and Combined Abiotic Stresses in Plants and Tools to Study the Same. Physiol. Plant 2021, 171, 653–676. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Dong, F.; Zou, J.; Gao, C.; Zhu, Z.; Liu, Y. Multiple Roles of Wheat Calmodulin Genes during Stress Treatment and TaCAM2-D as a Positive Regulator in Response to Drought and Salt Tolerance. Int. J. Biol. Macromol. 2022, 220, 985–997. [Google Scholar] [CrossRef]
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Network of Plant Cold-Stress Responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef]
- Brar, G.S.; Steiner, J.L.; Unger, P.W.; Prihar, S.S. Modeling Sorghum Seedling Establishment from Soil Wetness and Temperature of Drying Seed Zones. Agron. J. 1992, 84, 905–910. [Google Scholar] [CrossRef]
- Lee, S.H.; Singh, A.P.; Chung, G.C.; Kim, Y.S.; Kong, I.B. Chilling Root Temperature Causes Rapid Ultrastructural Changes in Cortical Cells of Cucumber (Cucumis sativus L.) Root Tips. J. Exp. Bot. 2002, 53, 2225–2237. [Google Scholar] [CrossRef]
- Payne, W.A.; Balota, M.; Rosenow, D.T. Sorghum Diversity for Germination and Coleoptile Elongation under Cool Conditions. Int. Sorghum Millets Newsl. 2003, 44, 76–79. [Google Scholar]
- Hund, A.; Fracheboud, Y.; Soldati, A.; Frascaroli, E.; Salvi, S.; Stamp, P. QTL Controlling Root and Shoot Traits of Maize Seedlings under Cold Stress. Theor. Appl. Genet. 2004, 109, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Zhi-Hong, Z.; Li, S.; Wei, L.; Wei, C.; Ying-Guo, Z. A Major QTL Conferring Cold Tolerance at the Early Seedling Stage Using Recombinant Inbred Lines of Rice (Oryza sativa L.). Plant Sci. 2005, 168, 527–534. [Google Scholar] [CrossRef]
- Patanè, C.; Cavallaro, V.; Avola, G.; D’Agosta, G. Seed Respiration of Sorghum [Sorghum bicolor (L.) Moench] during Germination as Affected by Temperature and Osmoconditioning. Seed Sci. Res. 2006, 16, 251–260. [Google Scholar] [CrossRef]
- Fiedler, K.; Bekele, W.A.; Friedt, W.; Snowdon, R.; Stützel, H.; Zacharias, A.; Uptmoor, R. Genetic Dissection of the Temperature Dependent Emergence Processes in Sorghum Using a Cumulative Emergence Model and Stability Parameters. Theor. Appl. Genet. 2012, 125, 1647–1661. [Google Scholar] [CrossRef]
- Goswami, A.K.; Maurya, N.K.; Goswami, S.; Bardhan, K.; Singh, S.K.; Prakash, J.; Pradhan, S.; Kumar, A.; Chinnusamy, V.; Kumar, P.; et al. Physio-Biochemical and Molecular Stress Regulators and Their Crosstalk for Low-Temperature Stress Responses in Fruit Crops: A Review. Front. Plant Sci. 2022, 13, 1022167. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, Y.; Qiu, X.; Lu, H.; Hwang, I.; Wang, T. Tolerant Mechanism of Model Legume Plant Medicago truncatula to Drought, Salt, and Cold Stresses. Front. Plant Sci. 2022, 13, 847166. [Google Scholar] [CrossRef]
- Xu, G.-Y.; Rocha, P.S.C.F.; Wang, M.-L.; Xu, M.-L.; Cui, Y.-C.; Li, L.-Y.; Zhu, Y.-X.; Xia, X. A Novel Rice Calmodulin-like Gene, OsMSR2, Enhances Drought and Salt Tolerance and Increases ABA Sensitivity in Arabidopsis. Planta 2011, 234, 47–59. [Google Scholar] [CrossRef]
- Xu, G.; Cui, Y.; Li, M.; Wang, M.; Yu, Y.; Zhang, B.; Huang, L.; Xia, X. OsMSR2, a Novel Rice Calmodulin-like Gene, Confers Enhanced Salt Tolerance in Rice (Oryza sativa L.). Aust. J. Crop Sci. 2013, 7, 368–373. [Google Scholar]
- Zhao, H.; Gao, Y.; Du, Y.; Du, J.; Han, Y. Genome-Wide Analysis of the CML Gene Family and Its Response to Melatonin in Common Bean (Phaseolus vulgaris L.). Sci. Rep. 2023, 13, 1196. [Google Scholar] [CrossRef]
- Aleynova, O.A.; Kiselev, K.V.; Suprun, A.R.; Ananev, A.A.; Dubrovina, A.S. Involvement of the Calmodulin-like Protein Gene VaCML92 in Grapevine Abiotic Stress Response and Stilbene Production. Int. J. Mol. Sci. 2023, 24, 15827. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.-C.; Nose, A.; Wasano, K. Effects of Chilling Temperature on Photosynthetic Rates, Photosynthetic Enzyme Activities and Metabolite Levels in Leaves of Three Sugarcane Species. Plant Cell Environ. 1999, 22, 317–324. [Google Scholar] [CrossRef]
- Wang, D.; Portis, A.R.; Moose, S.P.; Long, S.P. Cool C4 Photosynthesis: Pyruvate Pi Dikinase Expression and Activity Corresponds to the Exceptional Cold Tolerance of Carbon Assimilation in Miscanthus x giganteus. Plant Physiol. 2008, 148, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Wang, Y.; Li, Y.; Zhu, R.; Gu, Y.; Li, J.; Guo, H.; Ye, W.; Nabi, H.G.; Yang, T.; et al. The OsNAC41-RoLe1-OsAGAP Module Promotes Root Development and Drought Tolerance in Upland Rice. Mol. Plant 2024, 17, 1573–1593. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. Regulons Involved in Osmotic Stress-Responsive and Cold Stress-Responsive Gene Expression in Plants. Physiol. Plant. 2006, 126, 62–71. [Google Scholar] [CrossRef]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.-H.; Fujii, H.; Zheng, X.; Zhu, J.-K. A R2R3 Type MYB Transcription Factor Is Involved in the Cold Regulation of CBF Genes and in Acquired Freezing Tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef]
- Achard, P.; Gong, F.; Cheminant, S.; Alioua, M.; Hedden, P.; Genschik, P. The Cold-Inducible CBF1 Factor-Dependent Signaling Pathway Modulates the Accumulation of the Growth-Repressing DELLA Proteins via Its Effect on Gibberellin Metabolism. Plant Cell 2008, 20, 2117–2129. [Google Scholar] [CrossRef]
- Marla, S.R.; Burow, G.; Chopra, R.; Hayes, C.; Olatoye, M.O.; Felderhoff, T.; Hu, Z.; Raymundo, R.; Perumal, R.; Morris, G.P. Genetic Architecture of Chilling Tolerance in Sorghum Dissected with a Nested Association Mapping Population. G3 Genes Genomes Genet. 2019, 9, 4045–4057. [Google Scholar] [CrossRef]
- Ye, K.; Li, H.; Ding, Y.; Shi, Y.; Song, C.; Gong, Z.; Yang, S. BRASSINOSTEROID-INSENSITIVE2 Negatively Regulates the Stability of Transcription Factor ICE1 in Response to Cold Stress in Arabidopsis. Plant Cell 2019, 31, 2682–2696. [Google Scholar] [CrossRef]
- Barrero-Gil, J.; Salinas, J. CBFs at the Crossroads of Plant Hormone Signaling in Cold Stress Response. Mol. Plant 2017, 10, 542–544. [Google Scholar] [CrossRef]
- Zhou, M.; Chen, H.; Wei, D.; Ma, H.; Lin, J. Arabidopsis CBF3 and DELLAs Positively Regulate Each Other in Response to Low Temperature. Sci. Rep. 2017, 7, 39819. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate Regulates the Inducer of CBF Expression–c-Repeat Binding Factor/DRE Binding Factor1 Cascade and Freezing Tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.J.; Seo, Y.W. Identification of Novel C-Repeat Binding Factor (CBF) Genes in Rye (Secale cereale L.) and Expression Studies. Gene 2019, 684, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Zhang, L.; Zhang, T.; Zhang, H.; Xu, S.; An, L. Transcriptional Regulation Network of Cold-Responsive Genes in Higher Plants. Plant Sci. 2005, 169, 987–995. [Google Scholar] [CrossRef]
- Zhou, L.; Li, J.; He, Y.; Liu, Y.; Chen, H. Functional Characterization of SmCBF Genes Involved in Abiotic Stress Response in Eggplant (Solanum melongena). Sci. Hortic. 2018, 233, 14–21. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Hiei, Y.; Ishida, Y.; Komari, T. Progress of Cereal Transformation Technology Mediated by Agrobacterium tumefaciens. Front. Plant Sci. 2014, 5, 628. [Google Scholar] [CrossRef]
- Fu, X.-Z.; Chen, C.-W.; Wang, Y.; Liu, J.-H.; Moriguchi, T. Ectopic Expression of MdSPDS1 in Sweet Orange (Citrus sinensis Osbeck) Reduces Canker Susceptibility: Involvement of H2O2 Production and Transcriptional Alteration. BMC Plant Biol. 2011, 11, 55. [Google Scholar] [CrossRef]
- Zhong, Z.; Zhang, Y. Protoplast Isolation and Transfection in Rice. In Protoplast Technology: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2022; pp. 83–90. [Google Scholar]
- Shin, J.; Park, E.; Choi, G. PIF3 Regulates Anthocyanin Biosynthesis in an HY5-Dependent Manner with Both Factors Directly Binding Anthocyanin Biosynthetic Gene Promoters in Arabidopsis. Plant J. 2007, 49, 981–994. [Google Scholar] [CrossRef]
- Dahro, B.; Wang, F.; Peng, T.; Liu, J.-H. PtrA/NINV, an Alkaline/Neutral Invertase Gene of Poncirus trifoliata, Confers Enhanced Tolerance to Multiple Abiotic Stresses by Modulating ROS Levels and Maintaining Photosynthetic Efficiency. BMC Plant Biol. 2016, 16, 76. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lou, Q.; Wang, P.; Yu, M.; Xie, Z.; Xu, C.; Chen, S.; Yu, H.; Zhang, R.; Tian, G.; Hao, D.; et al. Transcriptome Analysis Reveals the Pivotal Genes and Regulation Pathways Under Cold Stress and Identifies SbERF027, an AP2/ERF Gene That Confers Cold Tolerance in Sorghum. Plants 2025, 14, 879. https://doi.org/10.3390/plants14060879
Lou Q, Wang P, Yu M, Xie Z, Xu C, Chen S, Yu H, Zhang R, Tian G, Hao D, et al. Transcriptome Analysis Reveals the Pivotal Genes and Regulation Pathways Under Cold Stress and Identifies SbERF027, an AP2/ERF Gene That Confers Cold Tolerance in Sorghum. Plants. 2025; 14(6):879. https://doi.org/10.3390/plants14060879
Chicago/Turabian StyleLou, Qijin, Peifeng Wang, Miao Yu, Zhigan Xie, Chen Xu, Shengyu Chen, Hao Yu, Rui Zhang, Guangling Tian, Di Hao, and et al. 2025. "Transcriptome Analysis Reveals the Pivotal Genes and Regulation Pathways Under Cold Stress and Identifies SbERF027, an AP2/ERF Gene That Confers Cold Tolerance in Sorghum" Plants 14, no. 6: 879. https://doi.org/10.3390/plants14060879
APA StyleLou, Q., Wang, P., Yu, M., Xie, Z., Xu, C., Chen, S., Yu, H., Zhang, R., Tian, G., Hao, D., Ke, X., Yu, S., Zhou, J., Zhao, Y., Ye, C., Guo, J., Zhang, H., Chen, M., & Liu, X. (2025). Transcriptome Analysis Reveals the Pivotal Genes and Regulation Pathways Under Cold Stress and Identifies SbERF027, an AP2/ERF Gene That Confers Cold Tolerance in Sorghum. Plants, 14(6), 879. https://doi.org/10.3390/plants14060879




