The Impact of Polycomb Group Proteins on 3D Chromatin Structure and Environmental Stresses in Plants
Abstract
1. Introduction
2. Methods
3. The Relationship Between PRC1 and PRC2
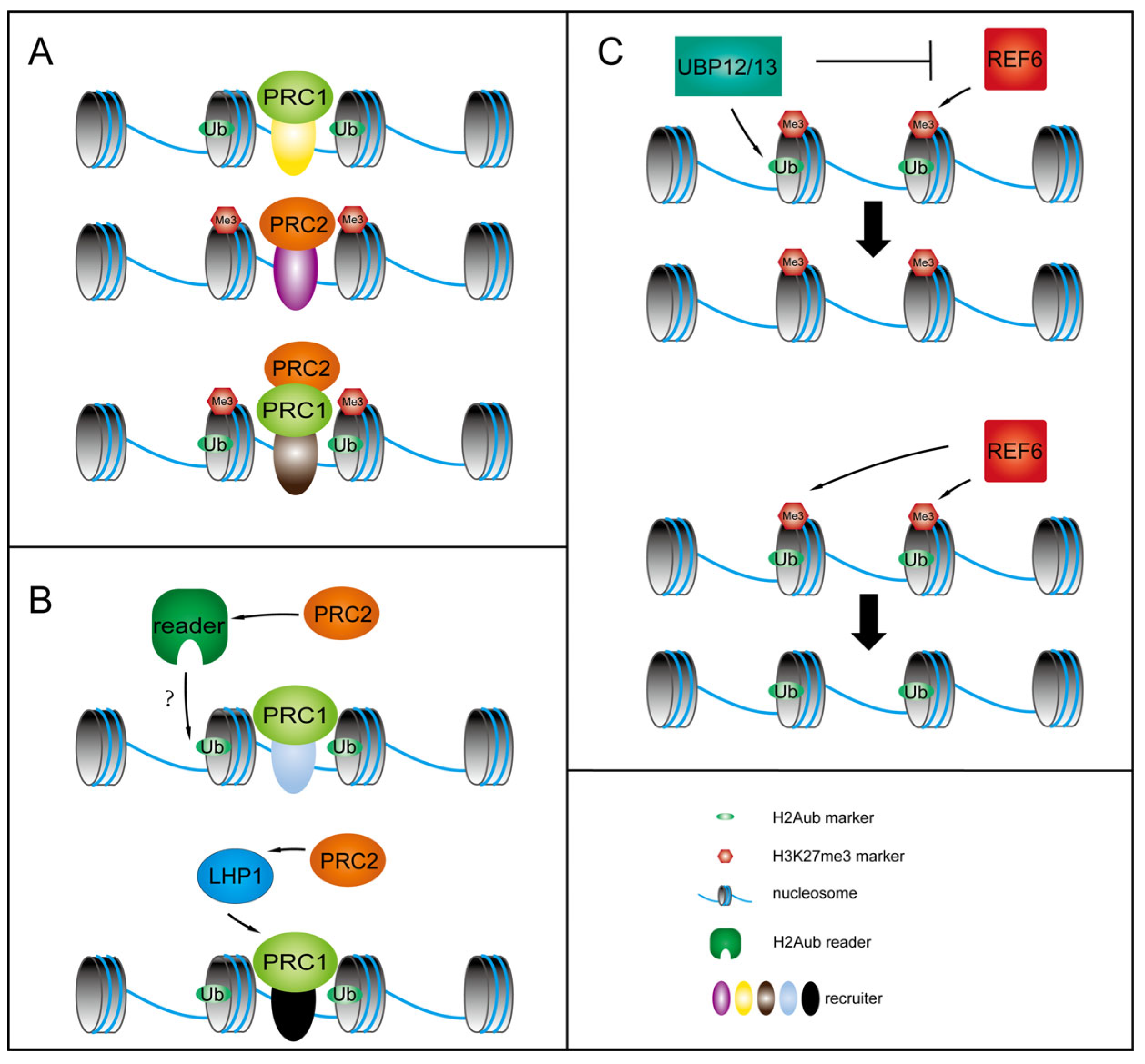
4. The Critical Role of H3K27me3 and H2Aub
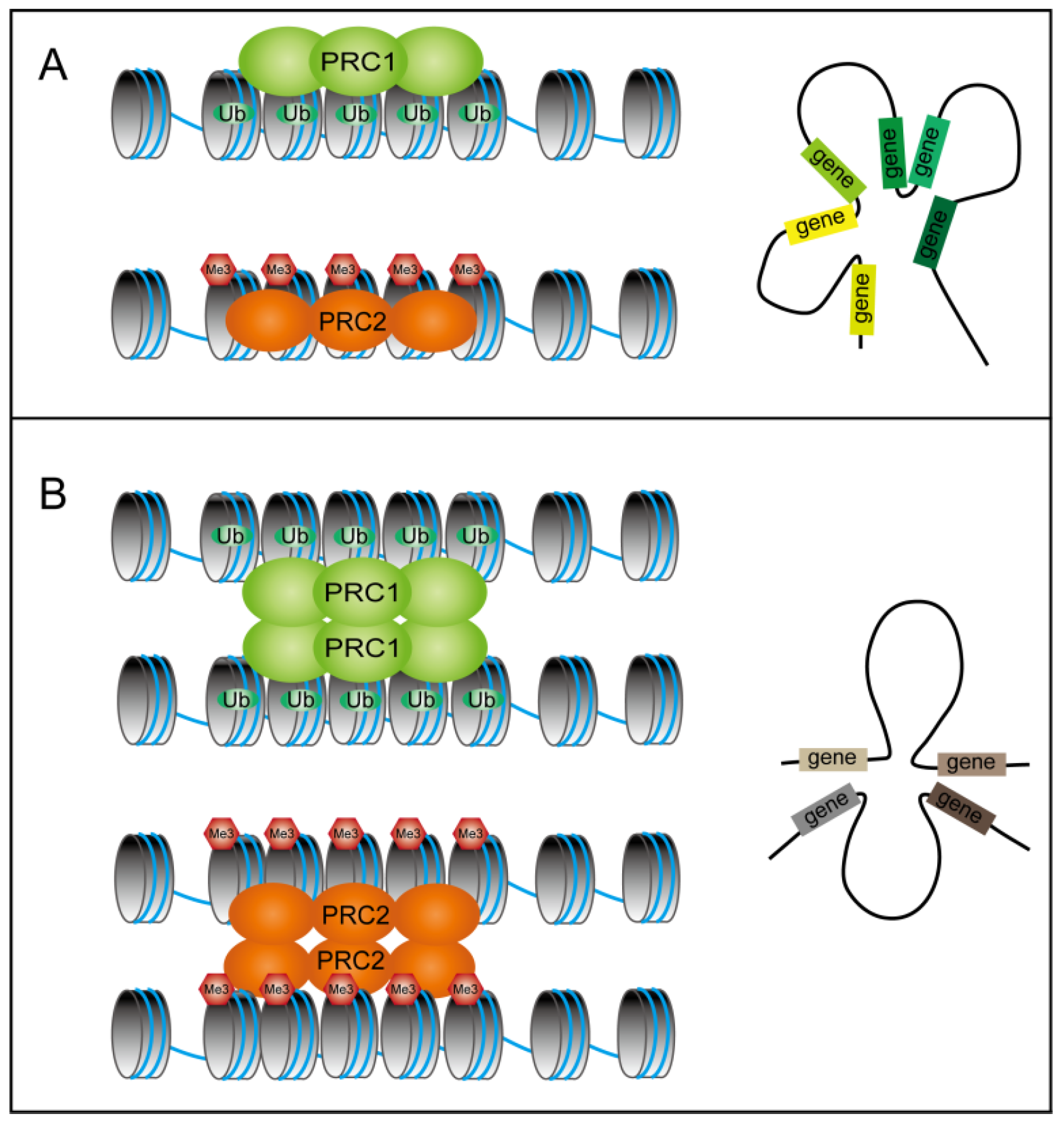
5. The Impact of PcG Proteins on the 3D Chromatin Structure
6. The Role of PcG Proteins in the Response to Environmental Stresses
7. Conclusions
8. Prospects
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Grossniklaus, U.; Paro, R. Transcriptional Silencing by Polycomb-Group Proteins. Cold Spring Harb. Perspect. Biol. 2014, 6, a019331. [Google Scholar] [CrossRef]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef]
- Gahan, J.M.; Rentzsch, F.; Schnitzler, C.E. The genetic basis for PRC1 complex diversity emerged early in animal evolution. Proc. Natl. Acad. Sci. USA 2020, 117, 22880–22889. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, D.-H.; Liu, B.-Y.; Shen, W.-H.; Ruan, Y. Conservation and diversification of polycomb repressive complex 2 (PRC2) proteins in the green lineage. Brief. Funct. Genom. 2017, 16, 106–119. [Google Scholar] [CrossRef]
- Huang, Y.; Jiang, L.; Liu, B.-Y.; Tan, C.-F.; Chen, D.-H.; Shen, W.-H.; Ruan, Y. Evolution and conservation of polycomb repressive complex 1 core components and putative associated factors in the green lineage. BMC Genom. 2019, 20, 533. [Google Scholar] [CrossRef]
- Vijayanathan, M.; Trejo-Arellano, M.G.; Mozgová, I. Polycomb Repressive Complex 2 in Eukaryotes—An Evolutionary Perspective. Epigenomes 2022, 6, 3. [Google Scholar] [CrossRef]
- Mozgova, I.; Hennig, L. The Polycomb Group Protein Regulatory Network. Annu. Rev. Plant Biol. 2015, 66, 269–296. [Google Scholar] [CrossRef]
- Baile, F.; Gómez-Zambrano, Á.; Calonje, M. Roles of Polycomb complexes in regulating gene expression and chromatin structure in plants. Plant Commun. 2022, 3, 100267. [Google Scholar] [CrossRef]
- Schuettengruber, B.; Bourbon, H.-M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef]
- Francis, N.J.; Kingston, R.E.; Woodcock, C.L. Chromatin compaction by a polycomb group protein complex. Science 2004, 306, 1574–1577. [Google Scholar] [CrossRef]
- Fischle, W.; Wang, Y.; Jacobs, S.A.; Kim, Y.; Allis, C.D.; Khorasanizadeh, S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003, 17, 1870–1881. [Google Scholar] [CrossRef]
- Min, J.; Zhang, Y.; Xu, R.-M. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003, 17, 1823–1828. [Google Scholar] [CrossRef]
- Buchwald, G.; van der Stoop, P.; Weichenrieder, O.; Perrakis, A.; van Lohuizen, M.; Sixma, T.K. Structure and E3-ligase activity of the Ring–Ring complex of Polycomb proteins Bmi1 and Ring1b. EMBO J. 2006, 25, 2465–2474. [Google Scholar] [CrossRef]
- Cao, R.; Tsukada, Y.-I.; Zhang, Y. Role of Bmi-1 and Ring1A in H2A Ubiquitylation and Hox Gene Silencing. Mol. Cell 2005, 20, 845–854. [Google Scholar] [CrossRef]
- Pengelly, A.R.; Kalb, R.; Finkl, K.; Müller, J. Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes Dev. 2015, 29, 1487–1492. [Google Scholar] [CrossRef]
- Vidal, M. Role of polycomb proteins Ring1A and Ring1B in the epigenetic regulation of gene expression. Int. J. Dev. Biol. 2009, 53, 355–370. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef]
- Wei, J.; Zhai, L.; Xu, J.; Wang, H. Role of Bmi1 in H2A Ubiquitylation and Hox Gene Silencing. J. Biol. Chem. 2006, 281, 22537–22544. [Google Scholar] [CrossRef]
- Bratzel, F.; López-Torrejón, G.; Koch, M.; Del Pozo, J.C.; Calonje, M. Keeping Cell Identity in Arabidopsis Requires PRC1 RING-Finger Homologs that Catalyze H2A Monoubiquitination. Curr. Biol. 2010, 20, 1853–1859. [Google Scholar] [CrossRef]
- Bratzel, F.; Yang, C.; Angelova, A.; López-Torrejón, G.; Koch, M.; del Pozo, J.C.; Calonje, M. Regulation of the New Arabidopsis Imprinted Gene AtBMI1C Requires the Interplay of Different Epigenetic Mechanisms. Mol. Plant 2012, 5, 260–269. [Google Scholar] [CrossRef]
- Li, J.; Wang, Z.; Hu, Y.; Cao, Y.; Ma, L. Polycomb Group Proteins RING1A and RING1B Regulate the Vegetative Phase Transition in Arabidopsis. Front. Plant Sci. 2017, 8, 867. [Google Scholar] [CrossRef]
- Baxter, I.; Li, W.; Wang, Z.; Li, J.; Yang, H.; Cui, S.; Wang, X.; Ma, L. Overexpression of AtBMI1C, a Polycomb Group Protein Gene, Accelerates Flowering in Arabidopsis. PLoS ONE 2011, 6, e21364. [Google Scholar] [CrossRef]
- Qin, F.; Sakuma, Y.; Tran, L.-S.P.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Fujita, M.; Umezawa, T.; Sawano, Y.; Miyazono, K.-i.; et al. Arabidopsis DREB2A-Interacting Proteins Function as RING E3 Ligases and Negatively Regulate Plant Drought Stress–Responsive Gene Expression. Plant Cell 2008, 20, 1693–1707. [Google Scholar] [CrossRef]
- El-Shemy, H.A.; Exner, V.; Aichinger, E.; Shu, H.; Wildhaber, T.; Alfarano, P.; Caflisch, A.; Gruissem, W.; Köhler, C.; Hennig, L. The Chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 Is Essential for H3K27me3 Binding and Function during Arabidopsis Development. PLoS ONE 2009, 4, e5335. [Google Scholar] [CrossRef]
- Gaudin, V.R.; Libault, M.; Pouteau, S.; Juul, T.; Zhao, G.; Lefebvre, D.; Grandjean, O. Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development 2001, 128, 4847–4858. [Google Scholar] [CrossRef]
- Smothers, J.F.; Henikoff, S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol. 2000, 10, 27–30. [Google Scholar] [CrossRef]
- Ferguson-Smith, A.C.; Turck, F.; Roudier, F.; Farrona, S.; Martin-Magniette, M.-L.; Guillaume, E.; Buisine, N.; Gagnot, S.; Martienssen, R.A.; Coupland, G.; et al. Arabidopsis TFL2/LHP1 Specifically Associates with Genes Marked by Trimethylation of Histone H3 Lysine 27. PLoS Genet. 2007, 3, e86. [Google Scholar] [CrossRef]
- Li, Z.; Fu, X.; Wang, Y.; Liu, R.; He, Y. Polycomb-mediated gene silencing by the BAH–EMF1 complex in plants. Nat. Genet. 2018, 50, 1254–1261. [Google Scholar] [CrossRef]
- Yin, X.; Romero-Campero, F.J.; de Los Reyes, P.; Yan, P.; Yang, J.; Tian, G.; Yang, X.; Mo, X.; Zhao, S.; Calonje, M.; et al. H2AK121ub in Arabidopsis associates with a less accessible chromatin state at transcriptional regulation hotspots. Nat. Commun. 2021, 12, 315. [Google Scholar] [CrossRef]
- Müller, J.; Hart, C.M.; Francis, N.J.; Vargas, M.L.; Sengupta, A.; Wild, B.; Miller, E.L.; O’Connor, M.B.; Kingston, R.E.; Simon, J.A. Histone Methyltransferase Activity of a Drosophila Polycomb Group Repressor Complex. Cell 2002, 111, 197–208. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Fursova, N.A.; Kelley, J.R.; Huseyin, M.K.; Feldmann, A.; Klose, R.J. PRC1 Catalytic Activity Is Central to Polycomb System Function. Mol. Cell 2020, 77, 857–874.e9. [Google Scholar] [CrossRef]
- Tamburri, S.; Lavarone, E.; Fernández-Pérez, D.; Conway, E.; Zanotti, M.; Manganaro, D.; Pasini, D. Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression. Mol. Cell 2020, 77, 840–856.e845. [Google Scholar] [CrossRef]
- Butenko, Y.; Ohad, N. Polycomb-group mediated epigenetic mechanisms through plant evolution. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2011, 1809, 395–406. [Google Scholar] [CrossRef]
- Brown, J.L.; Mucci, D.; Whiteley, M.; Dirksen, M.-L.; Kassis, J.A. The Drosophila Polycomb Group Gene pleiohomeotic Encodes a DNA Binding Protein with Homology to the Transcription Factor YY1. Mol. Cell 1998, 1, 1057–1064. [Google Scholar] [CrossRef]
- Erokhin, M.; Georgiev, P.; Chetverina, D. Drosophila DNA-Binding Proteins in Polycomb Repression. Epigenomes 2018, 2, 1. [Google Scholar] [CrossRef]
- Zhou, Y.; Tergemina, E.; Cui, H.; Förderer, A.; Hartwig, B.; Velikkakam James, G.; Schneeberger, K.; Turck, F. Ctf4-related protein recruits LHP1-PRC2 to maintain H3K27me3 levels in dividing cells in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 4833–4838. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Y.; Krause, K.; Yang, T.; Dongus, J.A.; Zhang, Y.; Turck, F. Telobox motifs recruit CLF/SWN–PRC2 for H3K27me3 deposition via TRB factors in Arabidopsis. Nat. Genet. 2018, 50, 638–644. [Google Scholar] [CrossRef]
- Yang, C.; Bratzel, F.; Hohmann, N.; Koch, M.; Turck, F.; Calonje, M. VAL- and AtBMI1-Mediated H2Aub Initiate the Switch from Embryonic to Postgerminative Growth in Arabidopsis. Curr. Biol. 2013, 23, 1324–1329. [Google Scholar] [CrossRef]
- Yuan, L.; Song, X.; Zhang, L.; Yu, Y.; Liang, Z.; Lei, Y.; Ruan, J.; Tan, B.; Liu, J.; Li, C. The transcriptional repressors VAL1 and VAL2 recruit PRC2 for genome-wide Polycomb silencing in Arabidopsis. Nucleic Acids Res. 2021, 49, 98–113. [Google Scholar] [CrossRef]
- de Bie, P.; Ciechanover, A. RING1B ubiquitination and stability are regulated by ARF. Biochem. Biophys. Res. Commun. 2012, 426, 49–53. [Google Scholar] [CrossRef]
- Fujisaki, S.; Ninomiya, Y.; Ishihara, H.; Miyazaki, M.; Kanno, R.; Asahara, T.; Kanno, M. Dimerization of the Polycomb-group protein Mel-18 is regulated by PKC phosphorylation. Biochem. Biophys. Res. Commun. 2003, 300, 135–140. [Google Scholar] [CrossRef]
- Gray, F.; Cho, H.J.; Shukla, S.; He, S.; Harris, A.; Boytsov, B.; Jaremko, Ł.; Jaremko, M.; Demeler, B.; Lawlor, E.R.; et al. BMI1 regulates PRC1 architecture and activity through homo- and hetero-oligomerization. Nat. Commun. 2016, 7, 13343. [Google Scholar] [CrossRef]
- Li, Z.; Cao, R.; Wang, M.; Myers, M.P.; Zhang, Y.; Xu, R.-M. Structure of a Bmi-1-Ring1B Polycomb Group Ubiquitin Ligase Complex. J. Biol. Chem. 2006, 281, 20643–20649. [Google Scholar] [CrossRef]
- Chen, D.; Molitor, A.; Liu, C.; Shen, W.-H. The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res. 2010, 20, 1332–1344. [Google Scholar] [CrossRef]
- Xu, L.; Shen, W.-H. Polycomb Silencing of KNOX Genes Confines Shoot Stem Cell Niches in Arabidopsis. Curr. Biol. 2008, 18, 1966–1971. [Google Scholar] [CrossRef]
- Arrigoni, R.; Alam, S.L.; Wamstad, J.A.; Bardwell, V.J.; Sundquist, W.I.; Schreiber-Agus, N. The Polycomb-associated protein Rybp is a ubiquitin binding protein. FEBS Lett. 2006, 580, 6233–6241. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Farcas, A.M.; Kondo, T.; King, H.W.; McGouran, J.F.; Hanssen, L.L.P.; Ito, S.; Cooper, S.; Kondo, K.; Koseki, Y.; et al. Variant PRC1 Complex-Dependent H2A Ubiquitylation Drives PRC2 Recruitment and Polycomb Domain Formation. Cell 2014, 157, 1445–1459. [Google Scholar] [CrossRef]
- Rose, N.R.; King, H.W.; Blackledge, N.P.; Fursova, N.A.; Ember, K.J.I.; Fischer, R.; Kessler, B.M.; Klose, R.J. RYBP stimulates PRC1 to shape chromatin-based communication between Polycomb repressive complexes. eLife 2016, 5, e18591. [Google Scholar] [CrossRef]
- Healy, E.; Mucha, M.; Glancy, E.; Fitzpatrick, D.J.; Conway, E.; Neikes, H.K.; Monger, C.; Van Mierlo, G.; Baltissen, M.P.; Koseki, Y.; et al. PRC2.1 and PRC2.2 Synergize to Coordinate H3K27 Trimethylation. Mol. Cell 2019, 76, 437–452.e436. [Google Scholar] [CrossRef]
- Zhou, Y.; Romero-Campero, F.J.; Gómez-Zambrano, Á.; Turck, F.; Calonje, M. H2A monoubiquitination in Arabidopsis thaliana is generally independent of LHP1 and PRC2 activity. Genome Biol. 2017, 18, 69. [Google Scholar] [CrossRef]
- Barbour, H.; Daou, S.; Hendzel, M.; Affar, E.B. Polycomb group-mediated histone H2A monoubiquitination in epigenome regulation and nuclear processes. Nat. Commun. 2020, 11, 5947. [Google Scholar] [CrossRef]
- Feng, J.; Chen, D.; Berr, A.; Shen, W.-H. ZRF1 Chromatin Regulators Have Polycomb Silencing and Independent Roles in Development. Plant Physiol. 2016, 172, 1746–1759. [Google Scholar] [CrossRef]
- Derkacheva, M.; Liu, S.; Figueiredo, D.D.; Gentry, M.; Mozgova, I.; Nanni, P.; Tang, M.; Mannervik, M.; Köhler, C.; Hennig, L. H2A deubiquitinases UBP12/13 are part of the Arabidopsis polycomb group protein system. Nat. Plants 2016, 2, 16126. [Google Scholar] [CrossRef]
- Kralemann, L.E.M.; Liu, S.; Trejo-Arellano, M.S.; Muñoz-Viana, R.; Köhler, C.; Hennig, L. Removal of H2Aub1 by ubiquitin-specific proteases 12 and 13 is required for stable Polycomb-mediated gene repression in Arabidopsis. Genome Biol. 2020, 21, 144. [Google Scholar] [CrossRef]
- Yan, W.; Chen, D.; Smaczniak, C.; Engelhorn, J.; Liu, H.; Yang, W.; Graf, A.; Carles, C.C.; Zhou, D.-X.; Kaufmann, K. Dynamic and spatial restriction of Polycomb activity by plant histone demethylases. Nat. Plants 2018, 4, 681–689. [Google Scholar] [CrossRef]
- Posfai, E.; Kunzmann, R.; Brochard, V.; Salvaing, J.; Cabuy, E.; Roloff, T.C.; Liu, Z.; Tardat, M.; van Lohuizen, M.; Vidal, M.; et al. Polycomb function during oogenesis is required for mouse embryonic development. Genes Dev. 2012, 26, 920–932. [Google Scholar] [CrossRef]
- Chen, D.; Molitor, A.M.; Xu, L.; Shen, W.-H. Arabidopsis PRC1 core component AtRING1 regulates stem cell-determining carpel development mainly through repression of class I KNOX genes. BMC Biol. 2016, 14, 112. [Google Scholar] [CrossRef]
- Lv, Y.; Li, J.; Wang, Z.; Liu, Y.; Jiang, Y.; Li, Y.; Lv, Z.; Huang, X.; Peng, X.; Cao, Y.; et al. Polycomb proteins RING1A/B promote H2A monoubiquitination to regulate female gametophyte development in Arabidopsis. J. Exp. Bot. 2024, 75, 4822–4836. [Google Scholar] [CrossRef]
- Lee, S.H.; Li, Y.; Kim, H.; Eum, S.; Park, K.; Lee, C.-H. The role of EZH1 and EZH2 in development and cancer. BMB Rep. 2022, 55, 595–601. [Google Scholar] [CrossRef]
- Shen, X.; Liu, Y.; Hsu, Y.-J.; Fujiwara, Y.; Kim, J.; Mao, X.; Yuan, G.-C.; Orkin, S.H. EZH1 Mediates Methylation on Histone H3 Lysine 27 and Complements EZH2 in Maintaining Stem Cell Identity and Executing Pluripotency. Mol. Cell 2008, 32, 491–502. [Google Scholar] [CrossRef]
- Jullien, P.E.; Katz, A.; Oliva, M.; Ohad, N.; Berger, F. Polycomb Group Complexes Self-Regulate Imprinting of the Polycomb Group Gene MEDEA in Arabidopsis. Curr. Biol. 2006, 16, 486–492. [Google Scholar] [CrossRef]
- Kinoshita, T.; Yadegari, R.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. Imprinting of the polycomb gene in the Arabidopsis endosperm. Plant Cell 1999, 11, 1945–1952. [Google Scholar] [CrossRef]
- Grossniklaus, U.; Vielle-Calzada, J.P.; Hoeppner, M.A.; Gagliano, W.B. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 1998, 280, 446–450. [Google Scholar] [CrossRef]
- Kiyosue, T.; Ohad, N.; Yadegari, R.; Hannon, M.; Dinneny, J.; Wells, D.; Katz, A.; Margossian, L.; Harada, J.J.; Goldberg, R.B.; et al. Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 1999, 96, 4186–4191. [Google Scholar] [CrossRef]
- Köhler, C.; Page, D.R.; Gagliardini, V.; Grossniklaus, U. The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat. Genet. 2004, 37, 28–30. [Google Scholar] [CrossRef]
- Arnaud, P.; Feil, R. MEDEA Takes Control of Its Own Imprinting. Cell 2006, 124, 468–470. [Google Scholar] [CrossRef][Green Version]
- Shu, J.; Chen, C.; Thapa, R.K.; Bian, S.; Nguyen, V.; Yu, K.; Yuan, Z.C.; Liu, J.; Kohalmi, S.E.; Li, C.; et al. Genome-wide occupancy of histone H3K27 methyltransferases CURLY LEAF and SWINGER in Arabidopsis seedlings. Plant Direct 2019, 3, e00100. [Google Scholar] [CrossRef]
- Shu, J.; Chen, C.; Li, C.; Cui, Y. The complexity of PRC2 catalysts CLF and SWN in plants. Biochem. Soc. Trans. 2020, 48, 2779–2789. [Google Scholar] [CrossRef]
- Tsuboi, M.; Kishi, Y.; Yokozeki, W.; Koseki, H.; Hirabayashi, Y.; Gotoh, Y. Ubiquitination-Independent Repression of PRC1 Targets during Neuronal Fate Restriction in the Developing Mouse Neocortex. Dev. Cell 2018, 47, 758–772.e5. [Google Scholar] [CrossRef]
- Chagraoui, J.; Hébert, J.; Girard, S.; Sauvageau, G. An anticlastogenic function for the Polycomb Group gene Bmi1. Proc. Natl. Acad. Sci. USA 2011, 108, 5284–5289. [Google Scholar] [CrossRef]
- Ginjala, V.; Nacerddine, K.; Kulkarni, A.; Oza, J.; Hill, S.J.; Yao, M.; Citterio, E.; van Lohuizen, M.; Ganesan, S. BMI1 Is Recruited to DNA Breaks and Contributes to DNA Damage-Induced H2A Ubiquitination and Repair. Mol. Cell. Biol. 2011, 31, 1972–1982. [Google Scholar] [CrossRef]
- Pan, M.-R.; Peng, G.; Hung, W.-C.; Lin, S.-Y. Monoubiquitination of H2AX Protein Regulates DNA Damage Response Signaling. J. Biol. Chem. 2011, 286, 28599–28607. [Google Scholar] [CrossRef]
- Arora, M.; Packard, C.Z.; Banerjee, T.; Parvin, J.D. RING1A and BMI1 bookmark active genes via ubiquitination of chromatin-associated proteins. Nucleic Acids Res. 2016, 44, 2136–2144. [Google Scholar] [CrossRef]
- Pemberton, H.; Anderton, E.; Patel, H.; Brookes, S.; Chandler, H.; Palermo, R.; Stock, J.; Rodriguez-Niedenführ, M.; Racek, T.; de Breed, L.; et al. Genome-wide co-localization of Polycomb orthologs and their effects on gene expression in human fibroblasts. Genome Biol. 2014, 15, R23. [Google Scholar] [CrossRef]
- Stock, J.K.; Giadrossi, S.; Casanova, M.; Brookes, E.; Vidal, M.; Koseki, H.; Brockdorff, N.; Fisher, A.G.; Pombo, A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 2007, 9, 1428–1435. [Google Scholar] [CrossRef]
- Liu, S.; Trejo-Arellano, M.S.; Qiu, Y.; Eklund, D.M.; Köhler, C.; Hennig, L. H2A ubiquitination is essential for Polycomb Repressive Complex 1-mediated gene regulation in Marchantia polymorpha. Genome Biol. 2021, 22, 253. [Google Scholar] [CrossRef]
- Gómez-Zambrano, Á.; Merini, W.; Calonje, M. The repressive role of Arabidopsis H2A.Z in transcriptional regulation depends on AtBMI1 activity. Nat. Commun. 2019, 10, 2828. [Google Scholar] [CrossRef]
- Lei, B.; Berger, F. H2A Variants in Arabidopsis: Versatile Regulators of Genome Activity. Plant Commun. 2020, 1, 100015. [Google Scholar] [CrossRef]
- Misteli, T. Beyond the Sequence: Cellular Organization of Genome Function. Cell 2007, 128, 787–800. [Google Scholar] [CrossRef]
- Domb, K.; Wang, N.; Hummel, G.; Liu, C. Spatial Features and Functional Implications of Plant 3D Genome Organization. Annu. Rev. Plant Biol. 2022, 73, 173–200. [Google Scholar] [CrossRef]
- Ea, V.; Baudement, M.-O.; Lesne, A.; Forné, T. Contribution of Topological Domains and Loop Formation to 3D Chromatin Organization. Genes 2015, 6, 734–750. [Google Scholar] [CrossRef]
- Jerković, I.; Cavalli, G. Understanding 3D genome organization by multidisciplinary methods. Nat. Rev. Mol. Cell Biol. 2021, 22, 511–528. [Google Scholar] [CrossRef]
- Sanyal, A.; Baù, D.; Martí-Renom, M.A.; Dekker, J. Chromatin globules: A common motif of higher order chromosome structure? Curr. Opin. Cell Biol. 2011, 23, 325–331. [Google Scholar] [CrossRef]
- Cheutin, T.; Cavalli, G. The multiscale effects of polycomb mechanisms on 3D chromatin folding. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 399–417. [Google Scholar] [CrossRef]
- Entrevan, M.; Schuettengruber, B.; Cavalli, G. Regulation of Genome Architecture and Function by Polycomb Proteins. Trends Cell Biol. 2016, 26, 511–525. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, G.G. Modulation of the high-order chromatin structure by Polycomb complexes. Front. Cell Dev. Biol. 2022, 10, 1021658. [Google Scholar] [CrossRef]
- Denholtz, M.; Bonora, G.; Chronis, C.; Splinter, E.; de Laat, W.; Ernst, J.; Pellegrini, M.; Plath, K. Long-Range Chromatin Contacts in Embryonic Stem Cells Reveal a Role for Pluripotency Factors and Polycomb Proteins in Genome Organization. Cell Stem Cell 2013, 13, 602–616. [Google Scholar] [CrossRef]
- Doyle, E.J.; Morey, L.; Conway, E. Know when to fold ‘em: Polycomb complexes in oncogenic 3D genome regulation. Front. Cell Dev. Biol. 2022, 10, 986319. [Google Scholar] [CrossRef]
- Illingworth, R.S. Chromatin folding and nuclear architecture: PRC1 function in 3D. Curr. Opin. Genet. Dev. 2019, 55, 82–90. [Google Scholar] [CrossRef]
- Kraft, K.; Yost, K.E.; Murphy, S.E.; Magg, A.; Long, Y.; Corces, M.R.; Granja, J.M.; Wittler, L.; Mundlos, S.; Cech, T.R.; et al. Polycomb-mediated genome architecture enables long-range spreading of H3K27 methylation. Proc. Natl. Acad. Sci. USA 2022, 119, e2201883119. [Google Scholar] [CrossRef]
- Krug, B.; Hu, B.; Chen, H.; Ptack, A.; Chen, X.; Gretarsson, K.H.; Deshmukh, S.; Kabir, N.; Andrade, A.F.; Jabbour, E.; et al. H3K27me3 spreading organizes canonical PRC1 chromatin architecture to regulate developmental programs. bioRxiv 2023. [Google Scholar] [CrossRef]
- Yu, L.; Shen, H.; Lyu, X. Roles of Polycomb Complexes in the Reconstruction of 3D Genome Architecture during Preimplantation Embryonic Development. Genes 2022, 13, 2382. [Google Scholar] [CrossRef]
- Huang, X.; Wei, C.; Li, F.; Jia, L.; Zeng, P.; Li, J.; Tan, J.; Sun, T.; Jiang, S.; Wang, J.; et al. PCGF6 regulates stem cell pluripotency as a transcription activator via super-enhancer dependent chromatin interactions. Protein Cell 2019, 10, 709–725. [Google Scholar] [CrossRef]
- Kondo, T.; Isono, K.; Kondo, K.; Endo, T.A.; Itohara, S.; Vidal, M.; Koseki, H. Polycomb Potentiates Meis2 Activation in Midbrain by Mediating Interaction of the Promoter with a Tissue-Specific Enhancer. Dev. Cell 2014, 28, 94–101. [Google Scholar] [CrossRef]
- Schoenfelder, S.; Sugar, R.; Dimond, A.; Javierre, B.-M.; Armstrong, H.; Mifsud, B.; Dimitrova, E.; Matheson, L.; Tavares-Cadete, F.; Furlan-Magaril, M.; et al. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat. Genet. 2015, 47, 1179–1186. [Google Scholar] [CrossRef]
- Vieux-Rochas, M.; Fabre, P.J.; Leleu, M.; Duboule, D.; Noordermeer, D. Clustering of mammalian Hox genes with other H3K27me3 targets within an active nuclear domain. Proc. Natl. Acad. Sci. USA 2015, 112, 4672–4677. [Google Scholar] [CrossRef]
- Del Prete, S.; Mikulski, P.; Schubert, D.; Gaudin, V. One, Two, Three: Polycomb Proteins Hit All Dimensions of Gene Regulation. Genes 2015, 6, 520–542. [Google Scholar] [CrossRef]
- Bendahmane, M.; Veluchamy, A.; Jégu, T.; Ariel, F.; Latrasse, D.; Mariappan, K.G.; Kim, S.-K.; Crespi, M.; Hirt, H.; Bergounioux, C.; et al. LHP1 Regulates H3K27me3 Spreading and Shapes the Three-Dimensional Conformation of the Arabidopsis Genome. PLoS ONE 2016, 11, e0158936. [Google Scholar] [CrossRef]
- Ariel, F.; Lucero, L.; Christ, A.; Mammarella, M.F.; Jegu, T.; Veluchamy, A.; Mariappan, K.; Latrasse, D.; Blein, T.; Liu, C.; et al. R-Loop Mediated trans Action of the APOLO Long Noncoding RNA. Mol. Cell 2020, 77, 1055–1065.e1054. [Google Scholar] [CrossRef]
- Huang, Y.; Sicar, S.; Ramirez-Prado, J.S.; Manza-Mianza, D.; Antunez-Sanchez, J.; Brik-Chaouche, R.; Rodriguez-Granados, N.Y.; An, J.; Bergounioux, C.; Mahfouz, M.M.; et al. Polycomb-dependent differential chromatin compartmentalization determines gene coregulation in Arabidopsis. Genome Res. 2021, 31, 1230–1244. [Google Scholar] [CrossRef]
- Sun, L.; Cao, Y.; Li, Z.; Liu, Y.; Yin, X.; Deng, X.W.; He, H.; Qian, W. Conserved H3K27me3-associated chromatin looping mediates physical interactions of gene clusters in plants. J. Integr. Plant Biol. 2023, 65, 1966–1982. [Google Scholar] [CrossRef]
- Dong, P.; Tu, X.; Chu, P.-Y.; Lü, P.; Zhu, N.; Grierson, D.; Du, B.; Li, P.; Zhong, S. 3D Chromatin Architecture of Large Plant Genomes Determined by Local A/B Compartments. Mol. Plant 2017, 10, 1497–1509. [Google Scholar] [CrossRef]
- Feng, S.; Cokus, S.J.; Schubert, V.; Zhai, J.; Pellegrini, M.; Jacobsen, S.E. Genome-wide Hi-C Analyses in Wild-Type and Mutants Reveal High-Resolution Chromatin Interactions in Arabidopsis. Mol. Cell 2014, 55, 694–707. [Google Scholar] [CrossRef]
- Yin, X.; Romero-Campero, F.J.; Yang, M.; Baile, F.; Cao, Y.; Shu, J.; Luo, L.; Wang, D.; Sun, S.; Yan, P.; et al. Binding by the Polycomb complex component BMI1 and H2A monoubiquitination shape local and long-range interactions in the Arabidopsis genome. Plant Cell 2023, 35, 2484–2503. [Google Scholar] [CrossRef]
- Shu, J.; Sun, L.; Wang, D.; Yin, X.; Yang, M.; Yang, Z.; Gao, Z.; He, Y.; Calonje, M.; Lai, J.; et al. EMF1 functions as a 3D chromatin modulator in Arabidopsis. Mol. Cell 2024, 84, 4729–4739.e4726. [Google Scholar] [CrossRef]
- Bsteh, D.; Moussa, H.F.; Michlits, G.; Yelagandula, R.; Wang, J.; Elling, U.; Bell, O. Loss of cohesin regulator PDS5A reveals repressive role of Polycomb loops. Nat. Commun. 2023, 14, 8160. [Google Scholar] [CrossRef]
- Tatavosian, R.; Kent, S.; Brown, K.; Yao, T.; Duc, H.N.; Huynh, T.N.; Zhen, C.Y.; Ma, B.; Wang, H.; Ren, X. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 2019, 294, 1451–1463. [Google Scholar] [CrossRef]
- Isono, K.; Endo, T.A.; Ku, M.; Yamada, D.; Suzuki, R.; Sharif, J.; Ishikura, T.; Toyoda, T.; Bernstein, B.E.; Koseki, H. SAM Domain Polymerization Links Subnuclear Clustering of PRC1 to Gene Silencing. Dev. Cell 2013, 26, 565–577. [Google Scholar] [CrossRef]
- Yuan, L.; Liu, X.; Luo, M.; Yang, S.; Wu, K. Involvement of Histone Modifications in Plant Abiotic Stress Responses. J. Integr. Plant Biol. 2013, 55, 892–901. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic stress responses in plants. Nat. Rev. Genet. 2021, 23, 104–119. [Google Scholar] [CrossRef]
- Kleinmanns, J.A.; Schubert, D. Polycomb and Trithorax group protein-mediated control of stress responses in plants. Biol. Chem. 2014, 395, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Molitor, A.M.; Latrasse, D.; Zytnicki, M.; Andrey, P.; Houba-Hérin, N.; Hachet, M.; Battail, C.; Del Prete, S.; Alberti, A.; Quesneville, H.; et al. The Arabidopsis hnRNP-Q Protein LIF2 and the PRC1 Subunit LHP1 Function in Concert to Regulate the Transcription of Stress-Responsive Genes. Plant Cell 2016, 28, 2197–2211. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Prado, J.S.; Latrasse, D.; Rodriguez-Granados, N.Y.; Huang, Y.; Manza-Mianza, D.; Brik-Chaouche, R.; Jaouannet, M.; Citerne, S.; Bendahmane, A.; Hirt, H.; et al. The Polycomb protein LHP1 regulates Arabidopsis thaliana stress responses through the repression of the MYC2-dependent branch of immunity. Plant J. 2019, 100, 1118–1131. [Google Scholar] [CrossRef]
- Hayashi, K.; Fernie, A.R. A neat wheat trick to hide genes from selection. Trends Plant Sci. 2024, 29, 837–838. [Google Scholar] [CrossRef]
- Wei, W.; Tao, J.-J.; Chen, H.-W.; Li, Q.-T.; Zhang, W.-K.; Ma, B.; Lin, Q.; Zhang, J.-S.; Chen, S.-Y. A Histone Code Reader and a Transcriptional Activator Interact to Regulate Genes for Salt Tolerance. Plant Physiol. 2017, 175, 1304–1320. [Google Scholar] [CrossRef]
- Pu, L.; Liu, M.-S.; Kim, S.Y.; Chen, L.-F.O.; Fletcher, J.C.; Sung, Z.R. EMBRYONIC FLOWER1 and ULTRAPETALA1 Act Antagonistically on Arabidopsis Development and Stress Response. Plant Physiol. 2013, 162, 812–830. [Google Scholar] [CrossRef]
- Wu, J.; Mei, X.; Zhang, J.; Ye, L.; Hu, Y.; Chen, T.; Wang, Y.; Liu, M.; Zhang, Y.; Xin, X.-F. CURLY LEAF modulates apoplast liquid water status in Arabidopsis leaves. Plant Physiol. 2023, 193, 792–808. [Google Scholar] [CrossRef] [PubMed]
- Kleinmanns, J.A.; Schatlowski, N.; Heckmann, D.; Schubert, D. BLISTER Regulates Polycomb-Target Genes, Represses Stress-Regulated Genes and Promotes Stress Responses in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1530. [Google Scholar] [CrossRef]
- Alexandre, C.; Möller-Steinbach, Y.; Schönrock, N.; Gruissem, W.; Hennig, L. Arabidopsis MSI1 Is Required for Negative Regulation of the Response to Drought Stress. Mol. Plant 2009, 2, 675–687. [Google Scholar] [CrossRef]
- Roy, S.; Gupta, P.; Rajabhoj, M.P.; Maruthachalam, R.; Nandi, A.K. The Polycomb-Group Repressor MEDEA Attenuates Pathogen Defense. Plant Physiol. 2018, 177, 1728–1742. [Google Scholar] [CrossRef]
- Gupta, P.; Roy, S.; Nandi, A.K. MEDEA-interacting protein LONG-CHAIN BASE KINASE 1 promotes pattern-triggered immunity in Arabidopsis thaliana. Plant Mol. Biol. 2020, 103, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Nugroho, A.B.D.; Kim, S.; Lee, S.W.; Kim, D.-H. Transcriptomic and epigenomic analyses revealed that polycomb repressive complex 2 regulates not only developmental but also stress responsive metabolism in Brassica rapa. Front. Plant Sci. 2023, 14, 1079218. [Google Scholar] [CrossRef]
- Folsom, J.J.; Begcy, K.; Hao, X.; Wang, D.; Walia, H. Rice Fertilization-Independent Endosperm1 Regulates Seed Size under Heat Stress by Controlling Early Endosperm Development. Plant Physiol. 2014, 165, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Kwon, C.S.; Lee, D.; Choi, G.; Chung, W.I. Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J. 2009, 60, 112–121. [Google Scholar] [CrossRef]
- Shen, Q.; Lin, Y.; Li, Y.; Wang, G. Dynamics of H3K27me3 Modification on Plant Adaptation to Environmental Cues. Plants 2021, 10, 1165. [Google Scholar] [CrossRef]
- Wang, H.; Yin, C.; Zhang, G.; Yang, M.; Zhu, B.; Jiang, J.; Zeng, Z. Cold-induced deposition of bivalent H3K4me3-H3K27me3 modification and nucleosome depletion in Arabidopsis. Plant J. 2024, 118, 549–564. [Google Scholar] [CrossRef]
- Zeng, Z.; Zhang, W.; Marand, A.P.; Zhu, B.; Buell, C.R.; Jiang, J. Cold stress induces enhanced chromatin accessibility and bivalent histone modifications H3K4me3 and H3K27me3 of active genes in potato. Genome Biol. 2019, 20, 123. [Google Scholar] [CrossRef]

| PcG | Components | Plants | Mammals | Flies |
|---|---|---|---|---|
| PRC1 | core subunits | RING1A/B | RING1A/B | dRing |
| BMI1A/B/C | PCGF2/4(cPRC1) PCGF1/3/5/6(vPRC1) | Psc Su(z)2 | ||
| cPRC1 specific | / | CBX2/4/6/7/8 | Pc | |
| PH1/2/3 | Ph | |||
| SCM | Scm | |||
| vPRC1 specific | VAL1/2 AL1/2/3/4/5/6/7 NDX VRN1 HDAC SAP18 | RYBP/YAF2 KDM2 | dRybp dKdm2 | |
| PRC2 | core subunits | CLF SWN MEA | EZH1/2 | E(z) |
| EMF2 VRN2 FIS2 | SUZ12 | Su(z)12 | ||
| FIE | EED | Esc | ||
| MSI1 | RBBP4/7 | Nurf55 | ||
| Plant specific PcG | subunits | LHP1 EMF1 | / | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Xiao, S.; Yang, M.; Guo, G.; Zhou, Y. The Impact of Polycomb Group Proteins on 3D Chromatin Structure and Environmental Stresses in Plants. Plants 2025, 14, 1038. https://doi.org/10.3390/plants14071038
Liu Y, Xiao S, Yang M, Guo G, Zhou Y. The Impact of Polycomb Group Proteins on 3D Chromatin Structure and Environmental Stresses in Plants. Plants. 2025; 14(7):1038. https://doi.org/10.3390/plants14071038
Chicago/Turabian StyleLiu, Yali, Suxin Xiao, Minqi Yang, Guangqin Guo, and Yue Zhou. 2025. "The Impact of Polycomb Group Proteins on 3D Chromatin Structure and Environmental Stresses in Plants" Plants 14, no. 7: 1038. https://doi.org/10.3390/plants14071038
APA StyleLiu, Y., Xiao, S., Yang, M., Guo, G., & Zhou, Y. (2025). The Impact of Polycomb Group Proteins on 3D Chromatin Structure and Environmental Stresses in Plants. Plants, 14(7), 1038. https://doi.org/10.3390/plants14071038






