Abstract
Calmodulin-like proteins (CMLs) are essential for calcium signal transduction in plants, influencing growth, development, stress responses, and the regulation of medicinal secondary metabolites. Despite their importance, the roles of CML genes in B. striata have not been characterized. This study aimed to elucidate the composition and function of the BsCML gene family in B. striata, identifying 38 genes across eight subfamilies. Evolutionary analysis showed that BsCML genes are stable and conserved, while functional predictions indicated involvement in environmental stress response, hormone regulation, and circadian rhythms. Expression profiling revealed that BsCML27 and BsCML16 were highly expressed during callus culture, suggesting their involvement in growth and development. Notably, BsCML32 and BsCML37 exhibited bidirectional regulation of militarine synthesis under sodium acetate (NaAc) and salicylic acid (SA) treatments, with tissue-specific expression strongly correlated (p < 0.01) with metabolite accumulation. These findings highlight the significant roles of BsCML genes in stress response and secondary metabolite synthesis, providing a foundation for enhancing the medicinal quality of B. striata.
1. Introduction
The Ca2+ signaling system plays a crucial role in regulating plant growth and development, ensuring stability under biotic and abiotic stress conditions [1]. Calcium signals are characterized by the frequency, amplitude, and distribution of calcium concentration fluctuations, which exhibit both spatial and temporal specificity. In response to various environmental stressors or internal variations, plants typically modulate their biological activities by integrating the spatiotemporal dynamics of calcium ion changes [2,3,4]. For instance, the opening and closing of plant stomata, as well as the formation of root hair symbiosis under high-temperature conditions, are regulated by calcium signals [5]. Calcium signaling plays a central role in various aspects of biological regulation. The precise transmission of these extensive signals relies on the involvement of multiple calcium-sensing proteins, including calmodulin (CaMs), calmodulin-like proteins (CMLs), calcineurin B-like proteins (CBLs), and calcium-dependent protein kinases (CDPKs) [6]. The fluctuations of calcium signals are decoded by these calcium sensors, which subsequently interact with downstream transcription factors (TFs), kinases, ion channels, and other components to complete the process of biological signal transduction [7]. The EF-hand motif—a helix-loop-helix structure critical for Ca2+ binding—is the defining feature of calcium-sensing proteins [8]. Upon binding to Ca2+, this structure undergoes a conformational change, enabling its signal-mediated functions [9,10,11]. Based on these characteristic structural motifs, researchers have identified CML family members in various plant species, thus establishing a foundation for exploring the functional roles of CMLs.
Calmodulin-like proteins (CMLs) play a crucial role in plant growth and development, stress resistance, and the regulation of substance metabolism. For instance, CML38 interacts with PEP1 receptor 2 to negatively regulate root growth in Arabidopsis thaliana, thereby maintaining nitrogen resource balance [12]. Additionally, CML38 has been identified as a key regulator of root response to hypoxia stress [13]. In addition, AtCML9 has been shown to promote innate immunity in A. thaliana [14]. MsCML10 interacts with glutathione S-transferase (MsGSTU8) and fructose 1,6-bisphosphate aldolase (MsFBA6) to positively regulate cold tolerance in Medicago truncatula [15]. The role of CMLs in enhancing plant stress resistance has been widely documented. Overexpression of MdCML3 increases the resistance of Malus domestica callus to high salinity and abscisic acid [16]. Similarly, overexpression of SlCML37 enhances cold tolerance in Solanum lycopersicum [17]. Furthermore, AhCML69 in Arachis hypogaea positively regulates plant disease resistance [18]. To adapt to stress factors or changes in the growth environment, plants often activate specific adaptation mechanisms. As a key medium for plant–environment interactions, the generation and metabolic regulation of secondary metabolites play a critical role in enabling plants to cope with stress. In A. thaliana, under oxidative stress, the synthesis of AsA is positively regulated by AtCML10 to mitigate such stress [19]. For instance, in Gossypium spp., GbCML45 and GbCML50 positively regulate lignin accumulation to enhance resistance to Verticillium wilt [20]. These studies suggest that the CML family may represent one of the key hub genes involved in the synthesis of secondary metabolites. Furthermore, in studies addressing abiotic stress, it was observed that hormones such as salicylic acid, jasmonic acid (JA), methyl jasmonic acid (MeJA), and abscisic acid (ABA) play significant regulatory roles in the expression of CMLs. This finding indicates that CMLs may regulate the synthesis of secondary metabolites by responding to these hormonal signals.
To investigate the synthesis mechanism of militarine in B. striata, salicylic acid (SA) and sodium acetate (NaAc) were used to treat the callus of B. striata in previous studies. Transcriptome and metabolome analyses revealed that the expression of CML was significantly altered at certain time points. A comprehensive analysis indicated that CML is one of the key genes involved in regulating the synthesis of militarine [21]. Militarine is a glycosidic active compound found in B. striata, known for its anti-tumor, anti-inflammatory, hemostatic, whitening, and antioxidant effects [22]. It is listed as the sole medicinal component of B. striata in the 2020 edition of the ‘Chinese Pharmacopoeia’ [23], underscoring its significant research value. Despite the pharmacological importance of militarine and the established roles of CMLs in other plants, the CML gene family in B. striata (BsCML) remains uncharacterized, and its regulatory role in militarine biosynthesis is unknown. Additionally, the regulatory effects of SA and NaAc on BsCML remain poorly understood. In this study, we identified 38 CML members from B. striata and systematically analyzed their encoded proteins, including physicochemical properties, gene structures, evolutionary relationships, and simple sequence repeat (SSR) loci. Also investigated were the temporal expression patterns of BsCML members under SA and NaAc treatments and their regulatory functions in militarine biosynthesis. These results provide valuable insights for further functional characterization of the BsCML family and establish an experimental foundation for elucidating the genetic mechanisms underlying militarine biosynthesis.
2. Results
2.1. Functional Annotation of BsCML
A total of 9996/18,309 differentially expressed genes (DEGs) and 1561/2107 differential metabolites (DEs) were identified from the suspension cells of B. striata, including 42 BsCML. KEGG and GO annotation analyses indicated that BsCML plays a significant role in calcium ion binding, the MAPK signaling pathway (mitogen-activated protein kinase), and plant–pathogen interactions. These findings suggest that BsCML is involved in plant signal transduction, immune defense, and stress response, thereby maintaining normal physiological functions. Considering the effects of NaAc and SA on growth stress and induction in B. striata callus, BsCML was selected as a candidate gene for further investigation into the mechanism of secondary metabolite accumulation in B. striata.
2.2. Identification and Proteins Physicochemical Property Analysis of BsCML
A total of 42 CML gene sequences were initially obtained from the transcriptome data of B. striata. The CML sequences from three species and the HMM profile were used for alignment and screening. Ultimately, 38 unigenes related to CML were identified and designated as BsCML1 to BsCML38. In terms of the physicochemical properties of the BsCML family proteins (Table 1), the ORF length ranged from 71 to 271 amino acids (aa), and the molecular weight varied from 7983.08 to 29,322.21 Daltons (Da). Among these, BsCML8 had the smallest ORF, while BsCML17 had the largest. The theoretical isoelectric point (pI) of the BsCML family ranged from 3.71 to 6.31, with BsCML20 having the lowest and BsCML17 having the highest pI, suggesting that BsCML family proteins are generally acidic. The instability index of most BsCML proteins was greater than 40, while the aliphatic index was less than 100, and the Grand Average of Hydropathy (GRAVY) value was negative, indicating that the BsCML family proteins are generally unstable and hydrophilic. Cellular localization predictions indicated that the family members were primarily distributed in the nucleus and cytoplasm (Table 1). These findings suggest that BsCML proteins may play roles in regulating physiological processes, such as energy metabolism, growth and development, and light responses. Furthermore, signal peptide prediction identified that only BsCML31 contained a signal peptide. In the transmembrane structure prediction, BsCML10 and BsCML31 were found to possess transmembrane regions, suggesting that BsCML31 may be involved in the regulation of light response processes.

Table 1.
Physicochemical properties and cellular localization of BsCML family proteins.
2.3. Amino Acid Conserved Motifs and Gene Structure Analysis
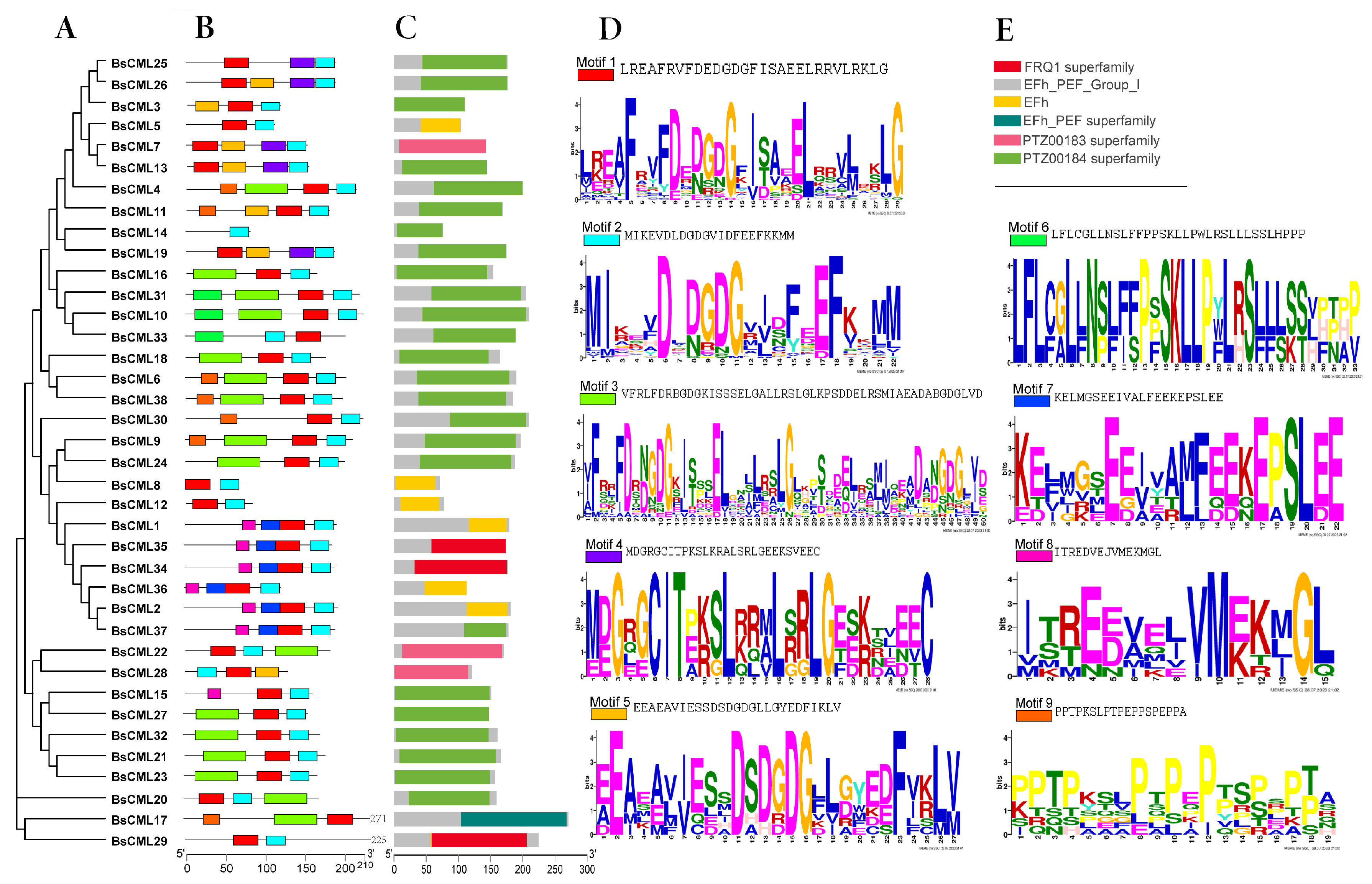
Analysis of the protein structure of BsCML revealed that the conserved motif structures were similar among members of closely related gene families in the evolutionary tree (Figure 1A). It was found that each BsCML member contained between two and four conserved motifs (Figure 1B). A total of nine motifs were identified through the analysis of conserved motif sequences using MEME (Figure 1D). Among these, the conserved EF-hand domain characteristic sequence (D-x-D-x-D) was found in motifs 1, 2, 3, and 5, and was present in all family members (BsCML1 to BsCML38), further indicating that the BsCML family shared characteristics typical of calcium-sensing proteins. In subsequent Conserved Domain (CD) predictions, three EF-hand conserved domains were identified (Figure 1C), which confirmed these findings.
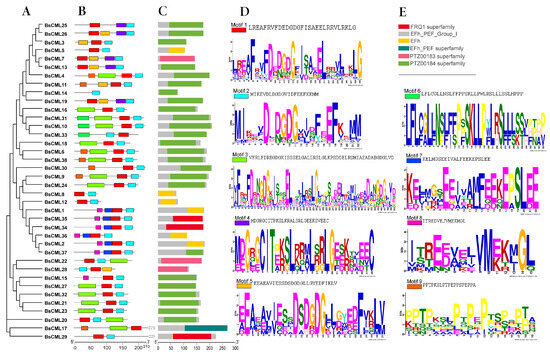
Figure 1.
Phylogenetic and structural analysis of BsCML proteins. (A) Phylogenetic tree of the BsCML family. (B) Motif analysis of BsCML proteins. (C) Prediction of conserved domains (CDs) in BsCML proteins. (D) Sequence logos corresponding to panel (B), where the height of each letter represents the bit score of the respective amino acid, with higher bit scores indicating greater conservation. (E) Six predicted structural motifs identified in panel (C).
2.4. Phylogenetic Analysis of the BsCML Family
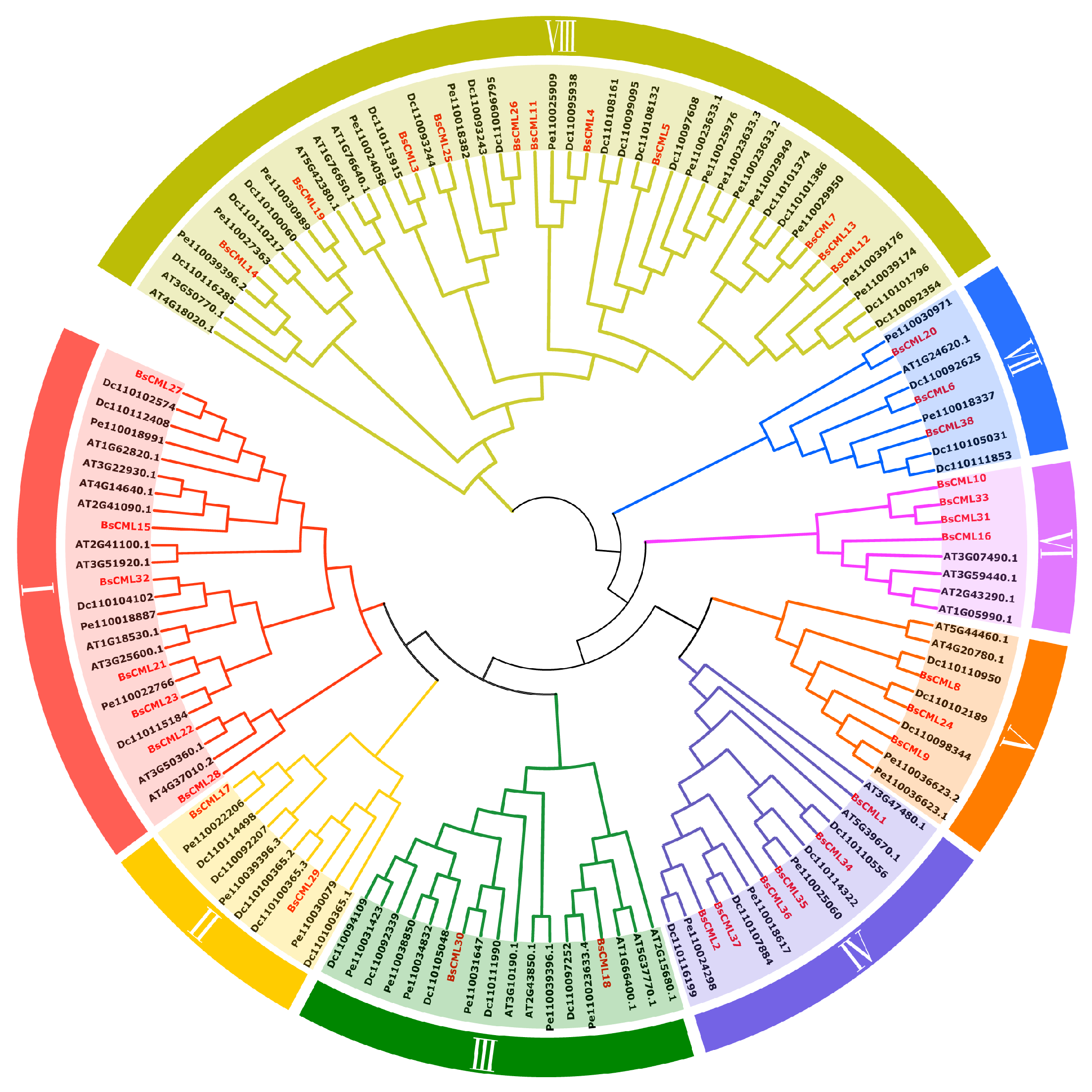
In the phylogenetic analysis of BsCML, the BsCML family was divided into eight subgroups (I-VIII) by comparing the conserved domain characteristics of CML sequences across four plant species using TBtools v1.132 (Figure 2). Within subgroup VI, BsCML10, BsCML33, BsCML16, and BsCML31 displayed a close genetic relationship, which is reflected in their structural similarity, as shown in Figure 1B. Additionally, BsCML members are dispersed among various subfamilies, suggesting that BsCML has maintained relatively complete genetic information throughout evolution.
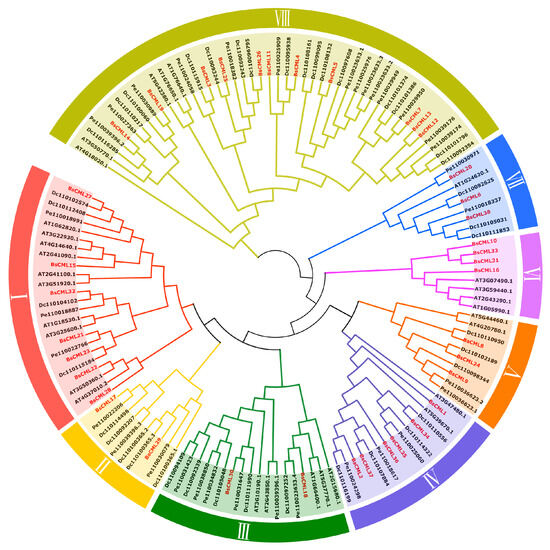
Figure 2.
Phylogenetic relationships of BsCML with CML families from A. thaliana (At), D. catenatum (Dc), and P. equestris (Pe). Members of the BsCML family are highlighted in red and distributed across subfamilies I–VIII, with each subfamily denoted by a distinct color.
2.5. Detection of EST-SSR
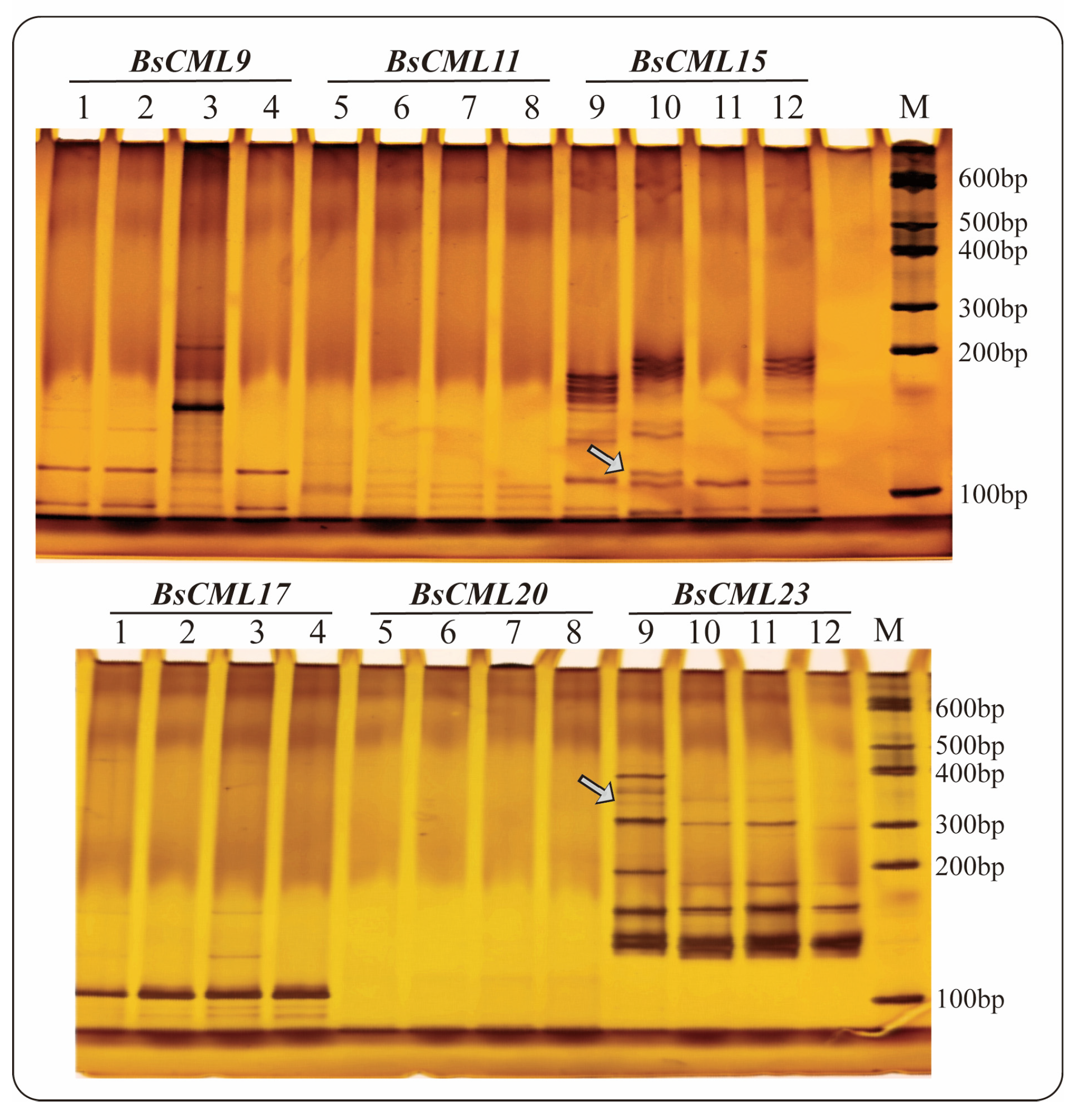
Seven sequences containing SSR loci were identified in 38 BsCML members, of which three sequences were dinucleotide repeats and four sequences were trinucleotide repeats, with repeat numbers ranging from a minimum of 3 to a maximum of 11 (Table 2). The SSR frequency (i.e., the ratio of the number of unigenes containing SSRs to the total number of unigenes) of BsCML in B. striata was 18.42%. The results of PAGE detection of SSR molecular markers (Figure 3) showed that BsCML could stably amplify bands, and it was observed that BsCML11, BsCML15, and BsCML23 exhibited different banding patterns among different strains of B. striata. Additionally, multiple allele bands were isolated for BsCML15 and BsCML23. These results indicate that BsCML in B. striata exhibits polymorphism at SSR loci, potentially playing an important role in the identification of different B. striata germplasm resources.

Table 2.
EST-SSR Fragment Information.
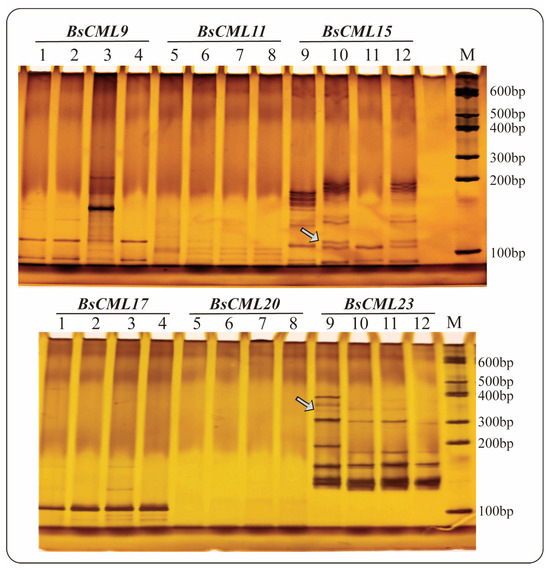
Figure 3.
PAGE detection of SSR molecular marker amplification results for BsCML genes in four B. striata varieties. Lane numbering in the figure corresponds to the following PCR templates: Lanes 1, 5, 9: Genomic DNA from ZMU-Bs001. Lanes 2, 6, 10: Genomic DNA from ZMU-Bs002. Lanes 3, 7, 11: Genomic DNA from ZMU-Bs003. Lanes 4, 8, 12: Genomic DNA from ZMU-Bs004. BsCML15 and BsCML23 show polymorphic bands (arrows), indicating SSR locus diversity.
2.6. Cis-Acting Elements of the BsCML Gene
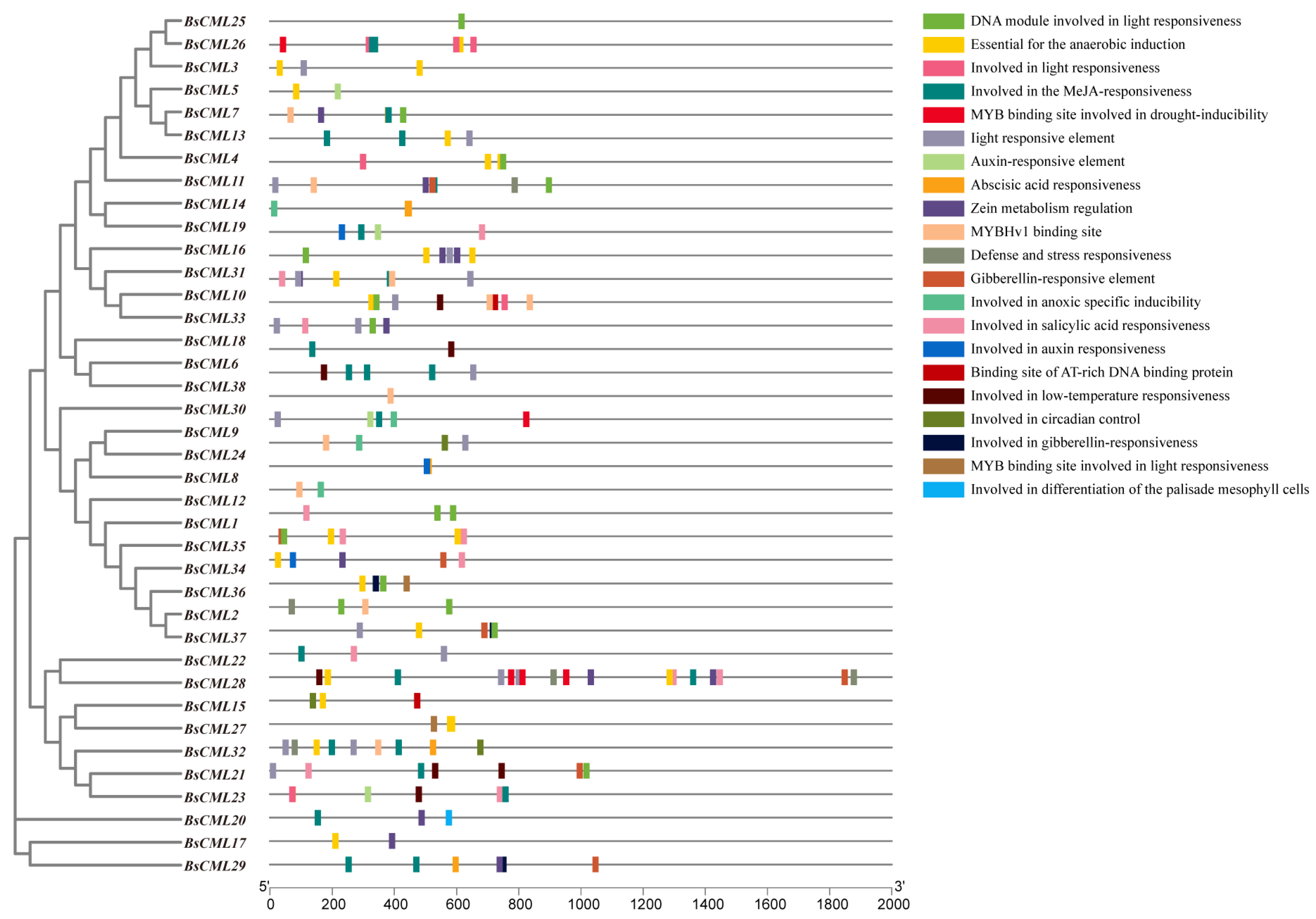
The CML family is a key signaling protein mediated by Ca2+, playing an essential role in plant responses and defense against various stress factors. In this study, the upstream 2000 bp promoter region of 38 BsCML family members was analyzed, revealing a variety of cis-regulatory elements, including elements responsive to physical factors such as low temperature, drought, hypoxia, and light, as well as hormone-related regulatory elements responsive to SA, auxin, and gibberellin. Additionally, elements involved in the regulation of circadian rhythm were identified (Figure 4). The distribution of these elements suggests that the BsCML family may have complex and diverse roles in plant responses to environmental stress, hormone regulation, and circadian clock regulation. Through the presence of these regulatory elements in their promoter regions, BsCML genes likely play a significant role in influencing plant physiological responses, metabolic regulation, and enhancing adaptability to environmental changes.
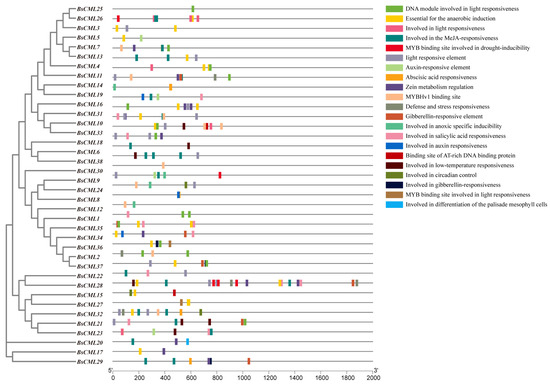
Figure 4.
Potential cis-acting elements in the BsCML gene.
2.7. Interactions Among BsCML Proteins
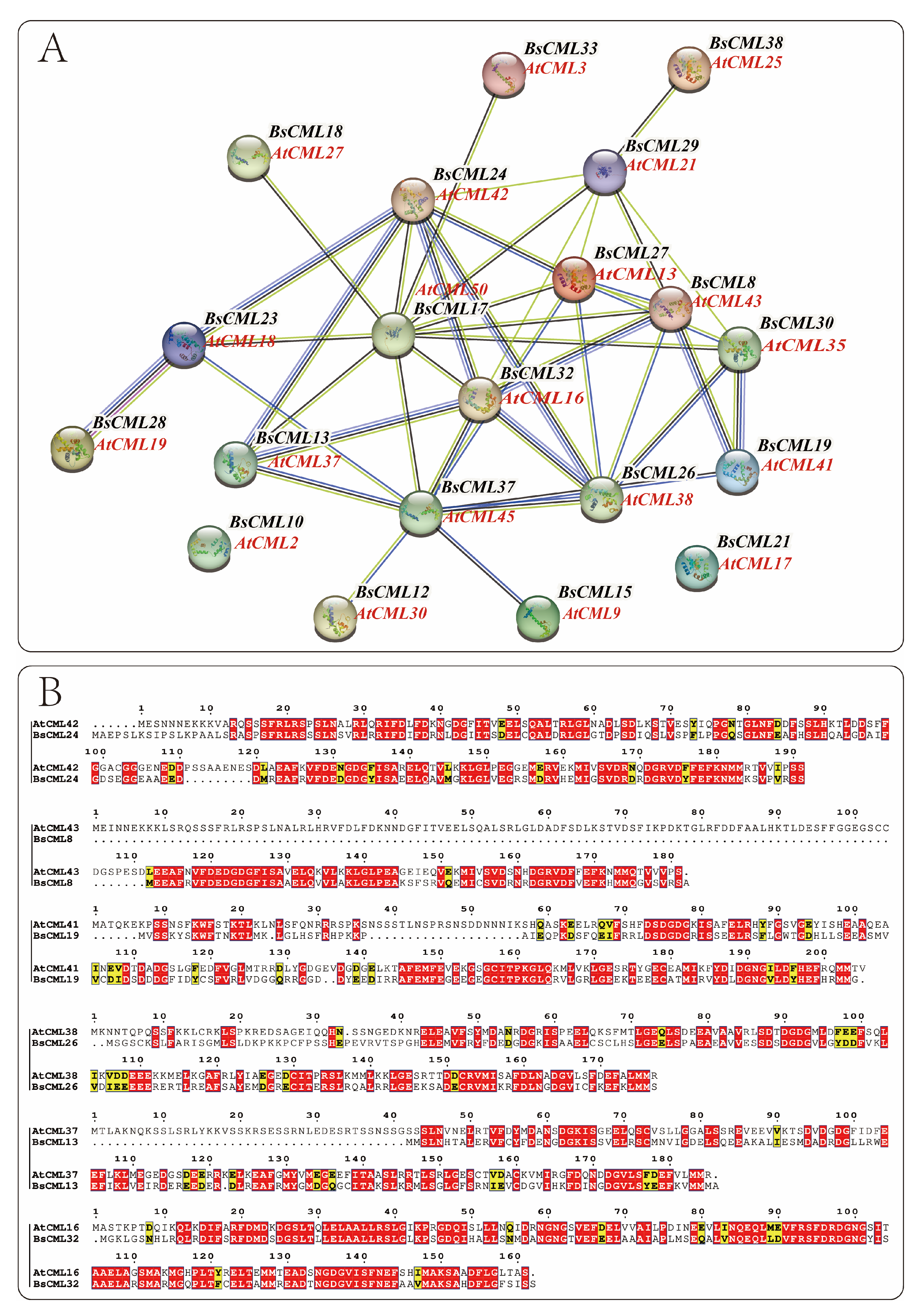
During plant growth and metabolism, regulation often involves multiple genes. The regulatory proteins or biocatalytic enzymes encoded by these genes play synergistic or antagonistic roles to ensure normal organismal development. The function and interactions of BsCML proteins were preliminarily examined by comparison with the CML genome proteins of the model plant A. thaliana. From the BsCML protein–protein interaction (PPI) network (Figure 5A), it was observed that BsCML32, BsCML24, BsCML23, BsCML28, BsCML13, BsCML37, BsCML26, BsCML19, BsCML18, and BsCML30 exhibit close relationships, with BsCML32 at the core. These results suggest a potential close interaction among BsCML genes.
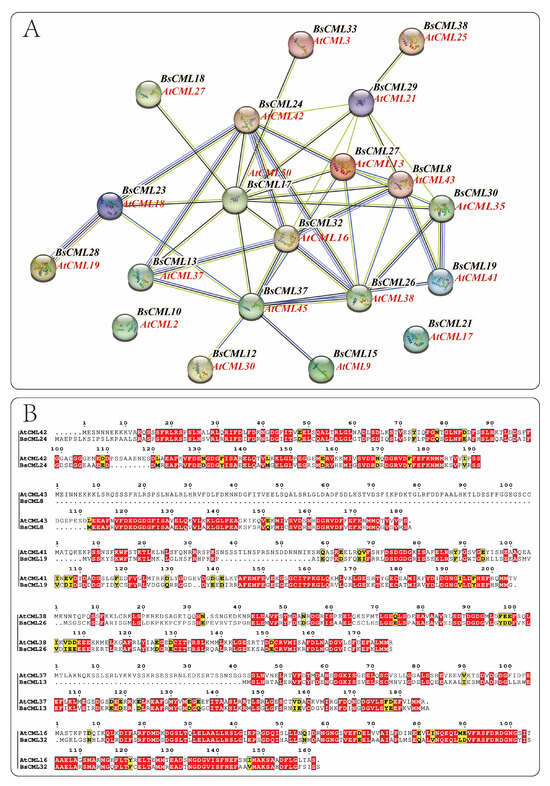
Figure 5.
Protein interaction network of BsCML and comparative analysis of sequence similarity with Arabidopsis CML. (A) Protein–protein interaction network of BsCML: Using AtCML as the reference genome, spheres (nodes) represent BsCML and AtCML proteins from the model plant Arabidopsis. The thickness of the connecting lines indicates the strength of interactions between two proteins. (B) Comparative analysis of protein sequence similarity between B. striata and Arabidopsis: Based on the protein interaction relationships shown in panel A, six pairs of protein sequences were analyzed. Red regions indicate highly conserved domains or critical functional sites, while yellow regions represent areas with moderate conservation or weak similarity.
The structure serves as the foundation for functional expression. By analyzing the similarity between the protein sequences of A. thaliana and B. striata (Figure 5B), insights can be gained to further explore the functions encoded by BsCML genes. The results indicated that among these interacting proteins, the corresponding protein structures of A. thaliana and B. striata displayed high similarity and conservation, suggesting that their functions are also similar.
2.8. Expression Patterns of BsCML Under NaAc and SA Treatment and Their Effects on Growth and Secondary Metabolite Accumulation in B. striat
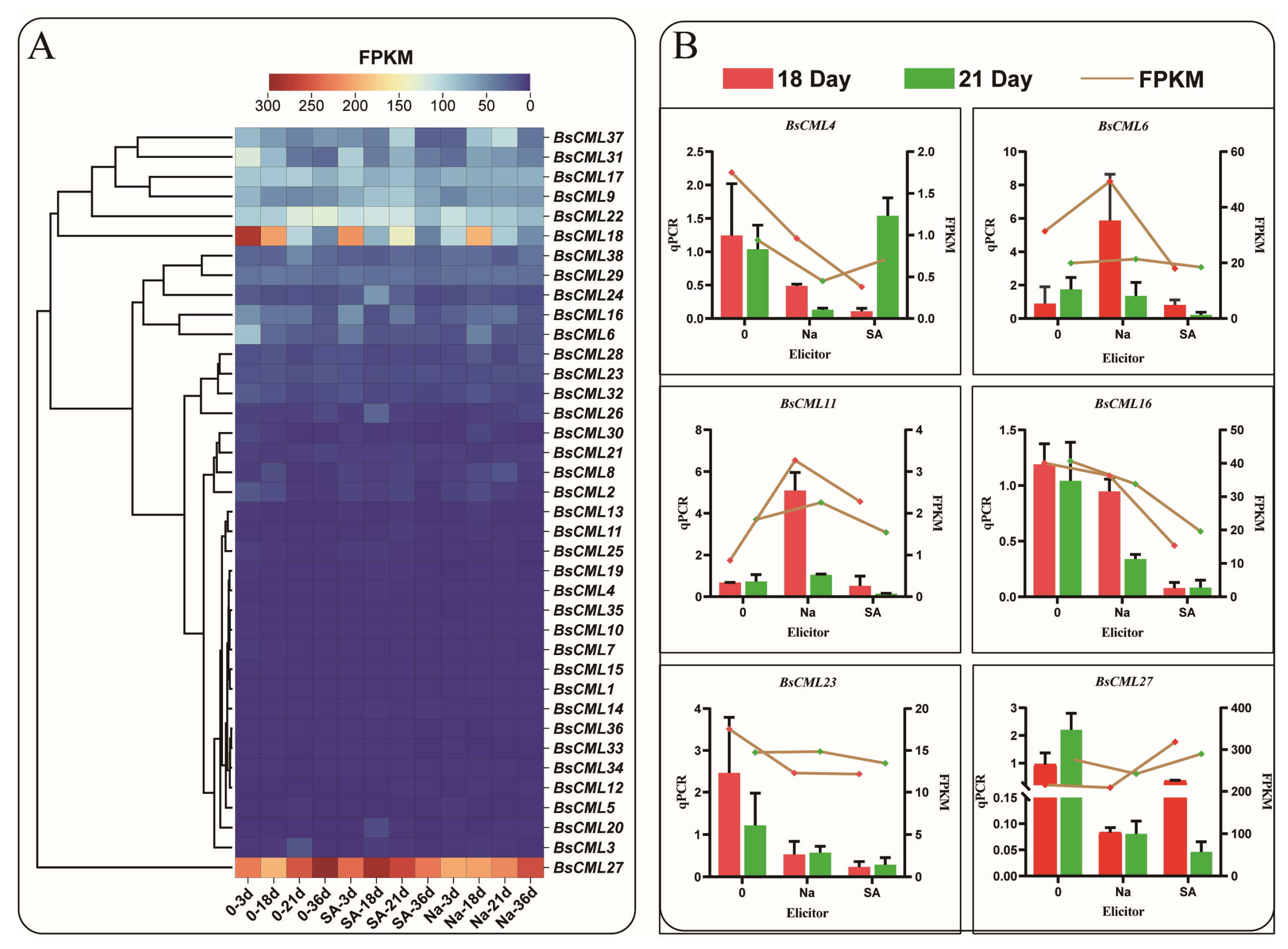
To explore the expression differences in BsCML family genes at different growth stages and under elicitor treatments, the expression levels of BsCML family genes in B. striata were analyzed using transcriptomic FPKM values to generate a heatmap. The results indicated that the expression of the BsCML gene family was generally silenced or exhibited a low response to SA and NaAc treatments (Figure 6A). Among these genes, the expression levels of BsCML1-5, BsCML7-8, BsCML10-15, BsCML19-21, BsCML25-26, BsCML30, and BsCML32-36 were relatively low (FPKM < 150), whereas BsCML27 showed high expression at various developmental stages. It is speculated that BsCML27 is involved in the growth and development of B. striata callus. These findings suggest that the BsCML family is in a low-response state during B. striata callus culture and that specific BsCML members may play roles in the growth and development of B. striata cell suspensions. To verify the reliability of the transcriptome data, six genes were randomly selected for validation, with the β-actin gene used as a reference control (Figure 6B). The results showed that, among the six randomly selected genes, the expression trends of five genes were largely consistent between FPKM and qPCR data. However, an inconsistency was observed in the expression trend of BsCML27. For example, at 18 days, the FPKM expression pattern of BsCML27 was SA > control > NaAc, whereas the qPCR pattern was control > SA > NaAc. Overall, the qPCR validation results confirm the reliability of the transcriptomic FPKM data.
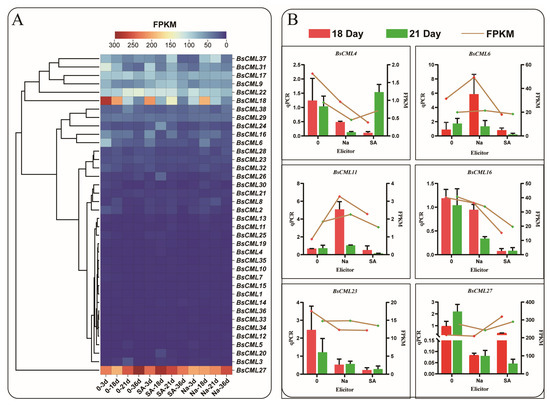
Figure 6.
Expression profiles of BsCML during suspension culture of callus and qPCR validation. (A) Expression patterns of BsCML family members at 3 dpi, 18 dpi, 21 dpi, and 36 dpi: ‘Na’ represents treatment with 150 μmol/LNaAc, ‘SA’ denotes treatment with 15 μmol/L SA, and ’0’ indicates the control group without any inducer. (B) qPCR validation of the expression levels of six selected BsCML genes.
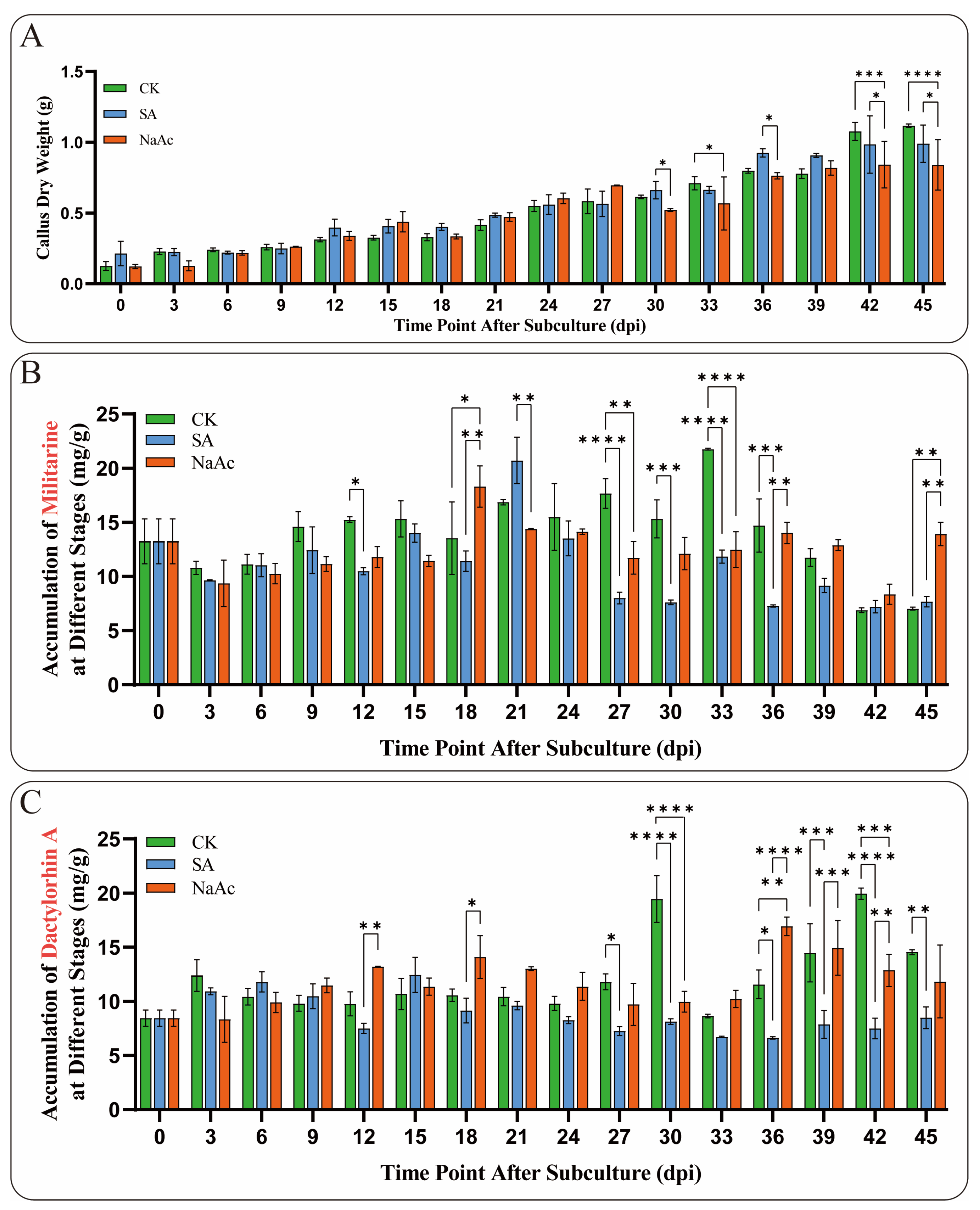
In this study, NaAc and SA were used as inducers during the suspension culture of B. striata callus. The results indicated that NaAc and SA, as exogenous stress factors, could significantly slow the growth of B. striata callus to varying extents. Notably, the inhibitory effect of NaAc was highly significant at 42 and 45 dpi (p < 0.005) (Figure 7A). Moreover, the accumulation of metabolites in the callus treated with NaAc and SA was notably altered. It is worth mentioning that the elicitors significantly inhibited (p < 0.01) militarine accumulation at 27 and 33 dpi, whereas SA treatment significantly (p < 0.01) promoted militarine accumulation at 45 dpi (Figure 7B). For the accumulation of Dactylorhin A, the inhibitory effects of NaAc and SA were also significant (p < 0.01), particularly at 30 and 42 dpi (Figure 7C). This alteration in secondary metabolite levels reflects the regulation of callus response to stress factors. Notably, the significant effects of the elicitors on the accumulation of militarine and Dactylorhin A were observed after 12 dpi, indicating the time required for the callus to respond to stress factors. These results demonstrate that NaAc and SA can significantly influence the growth and metabolism of B. striata, exhibiting varying degrees of promotion and inhibition on the synthesis of secondary metabolites.
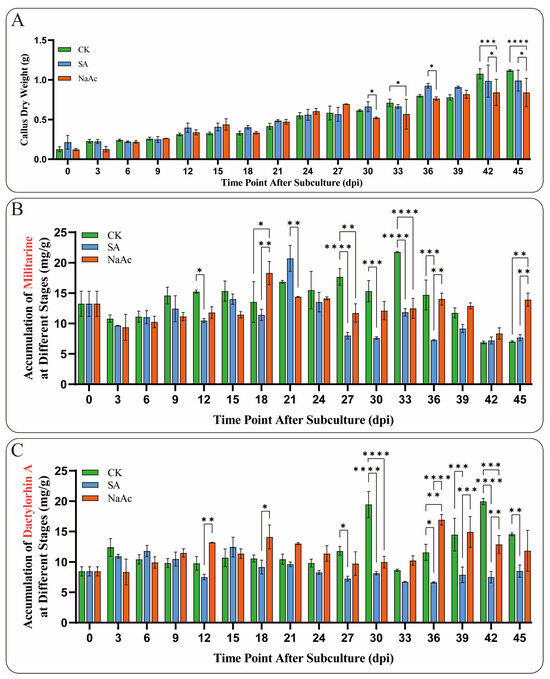
Figure 7.
Growth and metabolite accumulation of B. striata callus treated with 15 μmol/L SA and 150 μmol/L NaAc. (A) Dry weight growth trend of B. striata callus cells in the elicitor and control (CK) groups from 0 to 45 dpi. (B) Changes in the accumulation of militarine in the induced callus. (C) Changes in the accumulation of Dactylorhin A in the callus treated with induction. Asterisks denote significance levels: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 (one-way ANOVA).
2.9. Analysis of BsCML Regulation on Militarine and Dactylorhin A Synthesis Under Elicitor Treatment
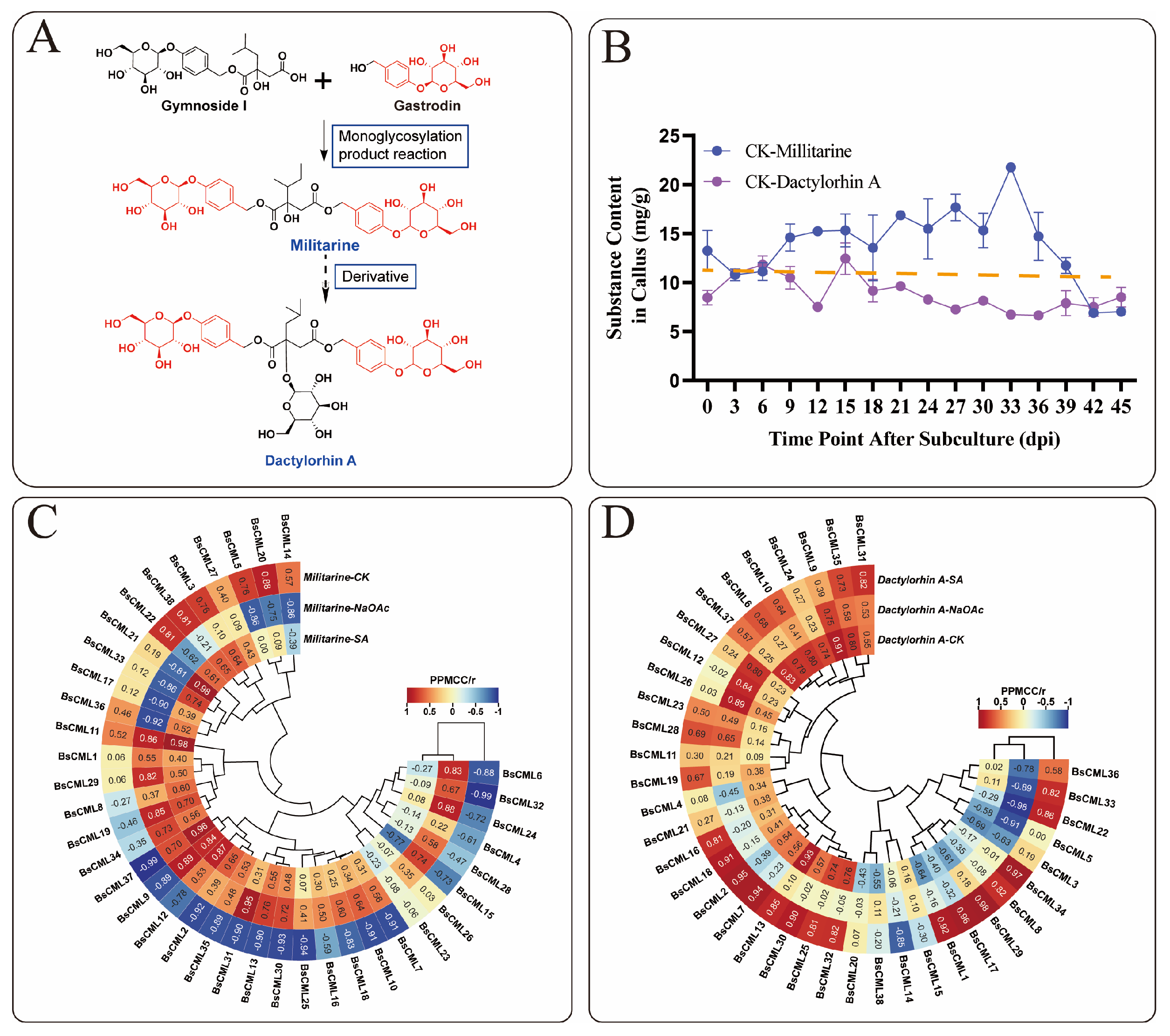
Based on the metabolic relationships of militarine [22] (Figure 8A), this study analyzed militarine and its derivative, dactylorhin A. A significant inverse accumulation trend between the two compounds was observed during the 0–45 dpi cultivation period (Figure 8B). Correlation heatmap analysis of BsCML expression and militarine accumulation in B. striata callus at four time points (Figure 8C) revealed distinct expression patterns under different treatments. In the CK group, 21 BsCML genes negatively regulated militarine biosynthesis, with BsCML32 (r = −0.992) and BsCML37 (r = −0.993) showing significant negative correlations (p < 0.01). In the NaAc treatment group, 28 genes were positively correlated with militarine biosynthesis, with BsCML31 (r = 0.955) showing a significant correlation (p < 0.05). In the SA treatment group, 29 genes positively regulated militarine biosynthesis, with BsCML11 (r = 0.984), BsCML21 (r = 0.985), and BsCML37 (r = 0.958) demonstrating significant effects at p < 0.05. In the dactylorhin A biosynthesis correlation analysis (Figure 8D), BsCML genes in the CK, NaAc, and SA groups mainly exhibited positive regulatory effects. In the SA group, BsCML17 (r = 0.985), BsCML29 (r = 0.976), and BsCML34 (r = 0.971) significantly promoted dactylorhin A biosynthesis (p < 0.05). Notably, BsCML11 strongly regulated both militarine and dactylorhin A biosynthesis.
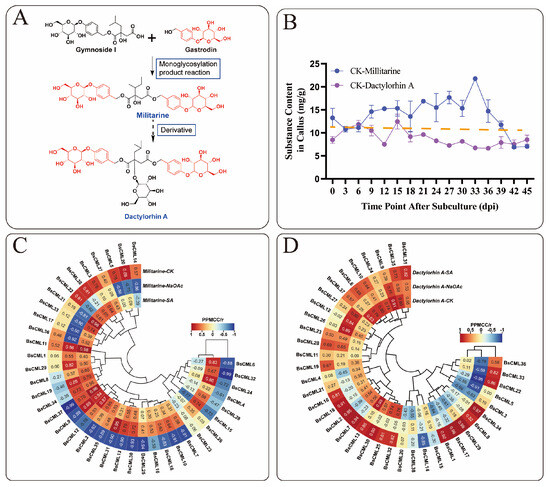
Figure 8.
Correlation of BsCML expression with accumulations of militarine and dactylorhin A. (A) Relationship between militarine, gastrodin, and dactylorhin A. (B) The accumulation trend of militarine and the derivative dactylorhin A in the 0–45 dpi control group, with the dotted line representing the virtual symmetry axis. (C) Correlation heatmap of BsCML expression in B. striata callus and militarine synthesis. (D) Correlation Heatmap of BsCML expression in B.striata callus and dactylorhin A synthesis.
These results identify BsCML11, BsCML32, and BsCML37 as key regulators of militarine synthesis, with bidirectional responses to SA and NaAc treatments. These genes represent prime candidates for metabolic engineering to enhance B. striata’s medicinal value. Overall, the regulatory patterns of BsCML on militarine and its derivatives lacked consistency, likely due to differences in accumulation patterns and indirect metabolic relationships. These findings suggest that BsCML family members play a key role in regulating militarine biosynthesis, which can be either inhibited or promoted under SA and NaAc treatments. This phenomenon may be linked to the activation or suppression of specific transcription factors. Moreover, cis-acting element prediction (Figure 4) identified SA-responsive elements, suggesting that the shift from negative regulation (CK) to positive regulation (SA and NaAc) in most genes could be mediated by these elements.
2.10. Analysis of BsCML Expression and Militarine Accumulation in Different B. striata Tissues
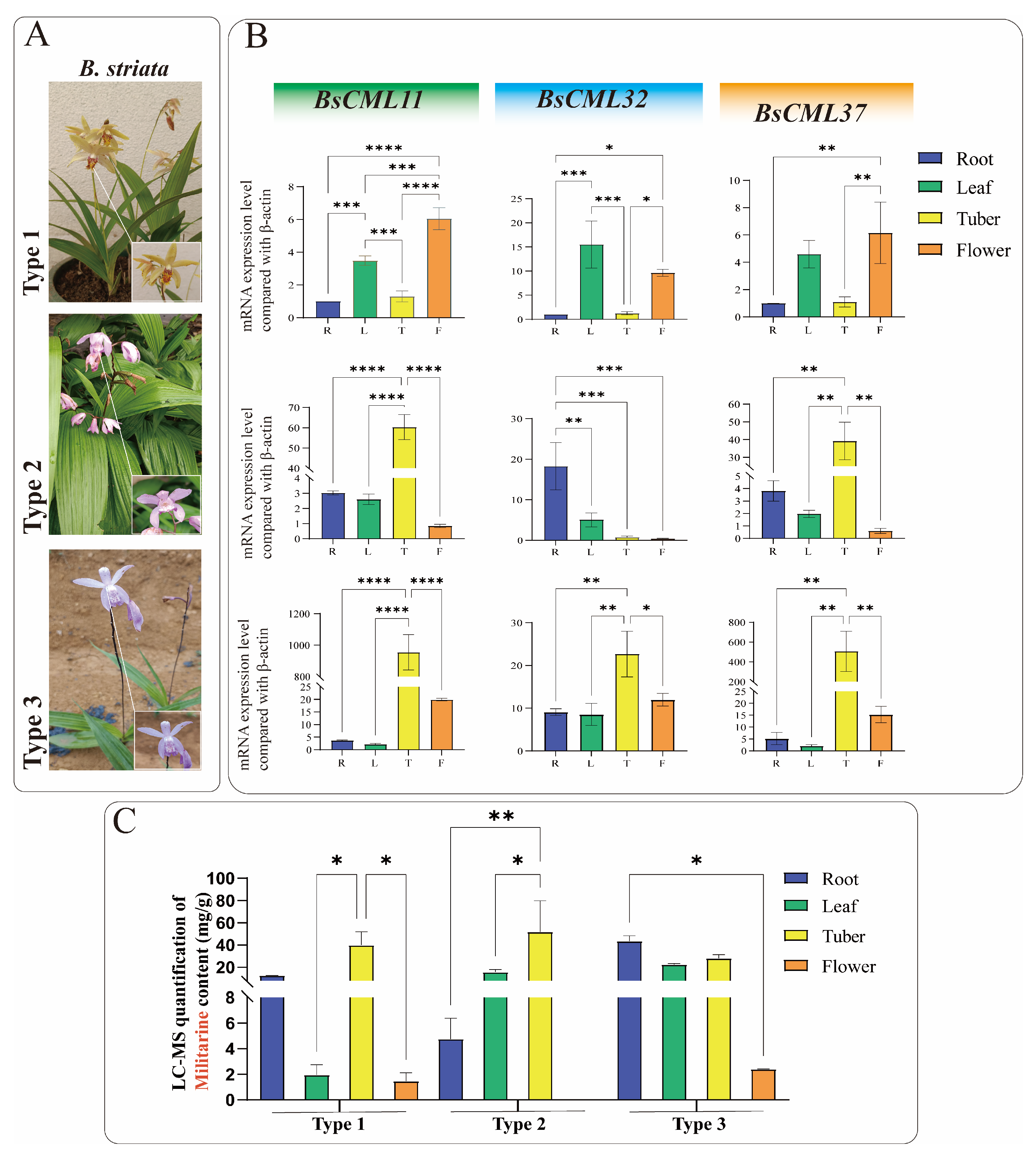
To investigate the regulatory roles of BsCML11, BsCML32, and BsCML37 in militarine biosynthesis, their expression patterns were analyzed in the roots, tubers, flowers, and leaves of three B. striata varieties with distinct phenotypes (Figure 9A). The results revealed significant tissue-specific expression patterns for all three genes (Figure 9B). Notably, the expression levels of BsCML11 and BsCML37 in tubers followed the order type3 > type2 > type1, with significantly higher expression in the tubers of purple-flowered varieties (type2 and type3) compared to other tissues (p < 0.01 or p < 0.0001). Since tubers are the primary medicinal part of B. striata, the high expression of BsCML11 and BsCML37 may contribute to the superior medicinal value of purple-flowered varieties over yellow-flowered ones. Additionally, BsCML11 and BsCML37 exhibited similar expression trends and levels across the three B. striata varieties, suggesting that their functions may be conserved or involved in essential plant metabolic processes. For example, BsCML11 and BsCML37 showed extremely low expression in type2 flowers but significantly higher expression in the tubers of type2 and type3 compared to type1, which may correlate with differences in flower color and medicinal efficacy. In contrast, the expression of BsCML32 in the tubers and roots of type1 was significantly lower than in type3, implying its potential role in the differential metabolite accumulation between these varieties. These findings underscore the diversity of BsCML expression patterns and their tissue-specific variations, suggesting their regulatory roles in the accumulation and metabolic processes of B. striata.
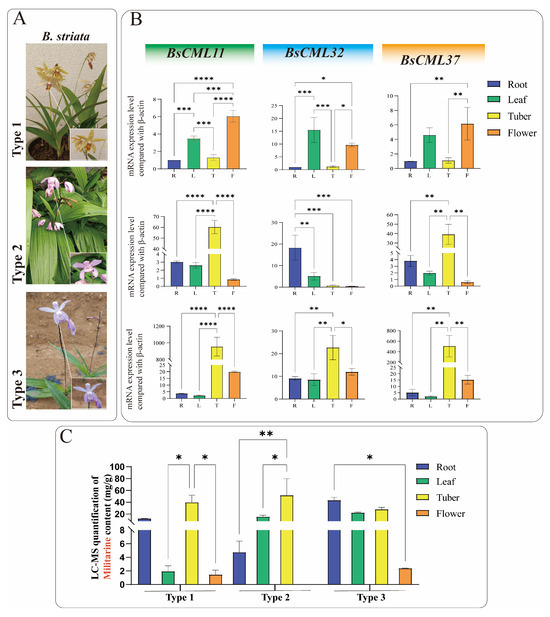
Figure 9.
Expression levels of three BsCML genes and militarine content in three B. striata varieties. (A) Three B. striata varieties with distinct agronomic traits. (B) Expression profiles of three BsCML genes in different tissues of the three B. striata varieties. (C) LC-MS quantification of militarine in different tissues of the three B. striata varieties. Asterisks denote significance levels: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 (one-way ANOVA).
Militarine (tuber content > 2.0%) is the primary marker for assessing the medicinal value of B. striata. LC-MS analysis showed that militarine accumulates in roots, tubers, flowers, and leaves, with the highest concentrations found in rootlets and tubers (Figure 9C). The militarine content in the tubers of all three B. striata varieties exceeded 20 mg/g, suggesting that, in addition to purple-flowered varieties, certain yellow-flowered varieties also have medicinal value.
Integrated analysis of gene expression (Figure 9B) and militarine content (Figure 9C) provides evidence for the regulatory role of BsCML in militarine biosynthesis. For instance, the militarine content in the flowers of all three B. striata varieties was extremely low, with nearly undetectable levels in type2 flowers, consistent with the expression patterns of BsCML11 and BsCML37. Additionally, the higher expression of BsCML32 in Type3 correlates with its greater militarine accumulation compared to type1, further supporting its regulatory function. In type1 and type2 B. striata, the expression of BsCML32 exhibited an inverse relationship with militarine accumulation. Pearson correlation analysis revealed a negative correlation between the two (r = −0.418, n = 12), further confirming that BsCML32 negatively regulates militarine biosynthesis at the p < 0.01 level. Furthermore, NaAc treatment induced BsCML expression in callus tissues (Figure 8B, CK group). These findings indicate that BsCML is involved in the regulation of militarine biosynthesis, affecting its tissue-specific accumulation, and that this process is influenced by external factors.
3. Discussion
3.1. Evolutionary Analysis of the BsCML Family
In this study, 38 BsCML family members with EF-hand motifs were identified in B. striata (Table 1). Changes in the number of gene family members are often considered important indicators of gene evolution. Among monocotyledonous plants, 32 CML genes were identified in Oryza sativa [24], 39 in Saccharum officinarum [25], 54 in P. equestris and D. officinale [26], and 80 in Hordeum vulgare [27]. For dicotyledonous plants, 44 CML genes were identified in Cucumis sativus [28], 52 in S. lycopersicum [29], and 58 in M. domestica [16]. These studies indicate that the number of BsCML members has not undergone a significant reduction or expansion. Furthermore, the structural characteristics of the proteins encoded by BsCML are consistent with the evolutionary patterns observed in various plant lineages. For instance, the ring structure represented by the D-x-D-G-D-G sequence in motif 1 (Figure 1D) is consistent with the EF-hand domains found in CMLs of monocots, bryophytes, and early-diverging eudicots. Similarly, the D-x-D-G-D structure in motif 2 (Figure 1D) aligns with the EF-hand domains present in early-diverging angiosperms and superasterids [30]. During the evolution of B. striata, the evolutionary trends of the eight BsCML subfamilies (Figure 2) show similarities to the phylogenetic tree constructed for 41 plant species [31]. These results suggest that the BsCML gene family has remained relatively stable throughout evolution.
3.2. Expression Pattern and Functional Analysis of the BsCML Gene Family
As a widely cultivated perennial medicinal orchid, the growth and development of B. striata, as well as the accumulation of its medicinal components, are influenced by various factors, including temperature, light quality [32], endophytic fungi [33], and soil microorganisms [34]. The diversity of these external environmental factors is a key contributor to the differential expression of CML genes in different tissues and organs of plants. In this study, most members of the BsCML gene family in B. striata callus under suspension induction exhibited low expression levels (Figure 6A). It is speculated that, under conditions characterized by a short cultivation period and mild environmental factors, the activation signals for the BsCML gene family were limited, resulting in their overall low expression. It is noteworthy that BsCML27 (Figure 6A) is highly expressed at all developmental stages, suggesting its involvement in the growth and development of B. striata callus. Several studies have indicated that CMLs play significant roles in regulating plant growth and development. For instance, Arabidopsis CML39 is involved in seed development, germination, and fruit development [35]. Additionally, CpCML15 interacts with PP2C46 and PP2C65 to promote the fruit ripening of Pseudocydonia sinensis [36]. Moreover, MtCML42 promotes the flowering of M. truncatula by regulating the expression of ABI5 (Abscisic Acid Insensitive 5) and FT (Flowering Locus T) genes [37]. These findings underscore the crucial role of CMLs in plant growth and suggest that their function is modulated through interactions with multiple genes. In plants, the expression of CMLs exhibits tissue- and organ-specific patterns, as demonstrated by the differential expression observed in Passiflora edulis [38] and M. truncatula [39]. This evidence further suggests that the spatial expression patterns of CML genes are closely linked to the synthesis of secondary metabolites. In this study, the expression levels of BsCML11 and BsCML37 in the tubers of two B. striata specimens were significantly higher than those in other tissues (Figure 9B). This suggests that BsCML11 and BsCML37 may be involved in the accumulation of secondary metabolites in tubers. Previous studies have shown that overexpression of VaCML65 in wild grape (V. amurensis) cell cultures can increase the content of stilbenes [40], while overexpression of RrCML13 in the roots of R. roxburghii enhances the accumulation of AsA [41]. These studies highlight the significance of CML expression in the biosynthesis of secondary metabolites.
It is well established that gene families of the same type in different plant species often share similar structures, which typically results in functional similarities. This characteristic provides a useful approach for studying the functions of newly identified gene families. In this study, the BsCML protein interaction network analysis revealed close interactions among family members (Figure 5A), and structural similarities between BsCML proteins and the corresponding Arabidopsis CML members were also observed (Figure 5B). Previous studies have demonstrated that AtCML13 and AtCML14 interact with proteins containing isoleucine–glutamine domains in A. thaliana [42]. These interactions are essential for regulatory roles in growth and development, contributing to cytoskeletal remodeling and intracellular transport [43]. Through structural similarity matching, AtCML13 was found to correspond to BsCML27 in their interaction networks, providing strong evidence that BsCML27 may play a role in regulating the growth and development of B. striata. Additionally, AtCML37 and AtCML42 exhibit antagonistic interactions in response to drought and salt stress, modulating the plant’s stress response via the regulation of gibberellin and auxin signaling pathways [44]. By examining the relationships where AtCML37 corresponds to BsCML13 and AtCML42 corresponds to BsCML24 (Figure 5A), these findings further illustrate the potential interactions among BsCML members and support their involvement in plant stress responses.
3.3. Prospective Applications of Inducer-Mediated CML Expression in Regulating Secondary Metabolite Biosynthesis
Secondary metabolites are a class of small molecules produced by plants to adapt to environmental stresses, typically serving as mediators of plant–environment interactions [45,46,47]. The involvement of CML genes in responses to various environmental stress factors suggests their critical role in the biosynthesis of secondary metabolites. For example, overexpression of VaCML92 in Vitis amurensis improves cold stress tolerance and enhances stilbene synthesis by 7.8- to 8.7-fold [48]. Similarly, exogenous calcium treatment induces the expression of CmCML11 and CmCAMTA5, resulting in elevated accumulation of γ-aminobutyric acid in melon [49]. These studies highlight the importance of exploring plant CML functions through the application of inducers. In the current study, treatment with NaAc and SA altered the accumulation of militarine and dactylorhin A in callus cultures at specific time points (Figure 7B,C), reflecting the responsiveness of BsCML to hormones and salt stress, and suggesting its potential regulatory role in militarine biosynthesis under stress conditions. Several studies have demonstrated the role of CMLs in regulating glycoside biosynthesis. For instance, overexpression of CML42 in A.thaliana enhances the accumulation of glucosinolates [50], whereas CML42 also exerts a negative regulatory effect on the synthesis of specific glycosides.
SA is a crucial chemical signal in plants, playing an important regulatory role. Studies have shown that SA can enhance plant tolerance to salt stress, drought, and heavy metal stress by activating the antioxidant enzyme system, regulating stomatal movement, stabilizing cell membranes, and improving photosynthetic efficiency [51,52]. SA plays a crucial role in plant stress regulation and has been widely utilized to induce the synthesis of secondary metabolites. For example, SA induces the synthesis of total phenols and anthocyanins in V. vinifera fruits [53], significantly promotes the accumulation of glucosinolates in Brassica oleracea [54], and enhances the accumulation of alkaloids in Hemerocallis citrina [55] and the marine microalga Arthrospira platensis [56]. In this study, it was found that the BsCML gene family can respond to SA and potentially influence the synthesis of secondary metabolites. For instance, after the CK group was treated with SA and NaAc (Figure 8B), the role of BsCML2, BsCML9, BsCML12, and BsCML37 in regulating militarine synthesis shifted from negative to positive regulation. This finding suggests that SA induction plays a crucial role in the regulation of militarine synthesis by BsCML genes and highlights the diverse expression patterns of BsCML members in the regulation of secondary metabolite synthesis. This result aligns with the presumed presence of SA response elements (Figure 4). Furthermore, environmental factors such as light conditions, plant hormones, and various ions also affect the expression of CML genes [11]. In the present study, the sensitivity of CMLs to inducer responses was also observed under NaAc salt treatment. These observations indicate that CML genes are highly sensitive to external environmental cues and can play a significant role in regulating plant growth and metabolism when subjected to appropriate biotic or abiotic stimuli.
4. Materials and Methods
4.1. Experimental Materials
The capsules of B. striata were collected from the nursery at Zunyi Medical University (27°42′ N, 107°01′ E), located in the Xinpu District of Zunyi City, Guizhou Province, China. The powdered seeds were cultured in suspension for 30 days using Murashige and Skoog (MS) medium supplemented with 1 mg/L 6-benzylaminopurine (6-BA), 2 mg/L 2,4-dichlorophenoxyacetic acid (2,4-D), 0.5 mg/L naphthaleneacetic acid (NAA), and 30 g/L sucrose. The cultures were maintained in sterile bottles (35 mL medium per 200 mL bottle) at a temperature of 25 ± 1 °C under dark conditions, shaking at 120 rpm. After the initial culture period, well-grown calli were selected for suspension subculture at 1 g per bottle. The subculture medium consisted of half-strength MS (1/2MS) supplemented with 1 mg/L 6-BA, 3 mg/L 2,4-D, 0.5 mg/L NAA, and 30 g/L sucrose. Subcultures were maintained under the same conditions (25 ± 1 °C, dark culture, 120 rpm shaking speed), and the medium was refreshed every 15 days [57]. From the second subculture (0 days post-inoculation, 0 dpi) to 45 dpi, samples were collected at random every 3 days, and growth was monitored throughout the period. After the second subculture, 150 μmol/L NaAc and 15 μmol/L SA were added separately, and three samples were randomly selected at 3 dpi, 18 dpi, 21 dpi, and 36 dpi [21]. Total RNA was extracted from each sample for transcriptome sequencing, and the CML gene set of B. striata was screened from the sequencing results for subsequent analysis.
4.2. Detection of Secondary Metabolite Accumulation in Callus by HPLC
Metabolites from suspension-cultured cells were collected at 3, 18, 21, and 36 dpi, with three biological replicates at each time point, and quantified using the Waters e2695 High-Performance Liquid Chromatography (HPLC) system (Waters, Milford, MA, USA). After precise weighing, the standards (militarine and dactylorhin A) were dissolved in methanol to prepare individual high-concentration standard solutions. The concentration of both standard solutions was 0.5 mg/mL, and they were stored according to the manufacturer’s instructions. Separation of the two components was performed on an ACQUITY UPLC BEH C18 column (2.1 × 100 mm, 1.7 µm). The column temperature was maintained at 30 °C, and the injection volume was set to 5.00 µL. The flow rate of the mobile phase was 0.3 mL/min. Mobile phase A consisted of 0.1% formic acid aqueous solution, while mobile phase B was acetonitrile. The gradient elution program was as follows: 0–10 min, 20% A and 80% B; 10–25 min, 50% A and 50% B; 25–27 min, 95% A and 5% B; 27–30 min, 95% A and 5% B; 30–32 min, 20% A and 80% B.
4.3. Characteristic Analysis Based on Nucleotide and Amino Acid Sequences
4.3.1. Identification of BsCML Family Members in B. striata
In order to identify the CML gene family members of B. striata, the CML gene family sequences of A. thaliana were obtained from the TAIR database (https://www.arabidopsis.org/), and the CML gene family sequences of Dendrobium catenatum and Phalaenopsis equestris were downloaded from NCBI. The CML members of B. striata were screened using TBtools v1.132 for conserved domain alignment. To further verify the screening results, the hidden Markov model (PF13499) of CML was downloaded from InterPro95.0 (http://www.ebi.ac.uk/interpro/entry/pfam/#table (accessed on 2 August 2023)), and TBtools [58] was used to search all CML sequences of B. striata.
4.3.2. Analysis of Conserved Motifs and Domains of BsCML
To explore the sequence characteristics of BsCML and the evolutionary relationships among its family members, a phylogenetic tree of the BsCML gene family was constructed and visualized using TBtools. The MEMESuite5.5.3 tool (https://meme-suite.org/meme/index.html (accessed on 3 August 2023)) was employed to analyze the conserved motifs of CML sequences in B. striata, with the number of motifs set to 20. Subsequently, the EF-hand domain of BsCML was identified using the CD-Search function of NCBI and visualized by TBtools.
4.3.3. Analysis of Physical and Chemical Properties of Proteins
To understand the properties of the proteins encoded by B. striata CML, ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/ (accessed on 2 August 2023)) was used to analyze the open reading frames (ORFs) of BsCML. ExPASy (https://web.expasy.org/protparam/ (accessed on 2 August 2023)) was employed to predict molecular weight (MW/Da), amino acid size (AA), isoelectric point (pI), instability index, alpha-helix content, and other physical and chemical properties [59]. The secondary structure of the proteins was predicted using SOPMA (https://npsa.lyon.inserm.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html (accessed on 2 August 2023)) [60].
4.3.4. Detection and Validation of EST-SSR
To explore the conservation of CML in different strains of B. striata, the genes containing SSR loci were analyzed. Genomic DNA was extracted using the CTAB method and diluted to a concentration of 50 ng/μL for EST-SSR detection. The SSR loci of 38 CML sequences were detected using the NWISRL online server (https://ssr.nwisrl.ars.usda.gov/ (accessed on 4 August 2023)), with parameters set to default values. The online tool Primer3 Plus (https://www.primer3plus.com/index.html (accessed on 4 August 2023)) was used to design specific primers for SSR loci (Table 2), followed by PCR amplification and PAGE detection of the results. After electrophoresis, the bands were visualized using silver staining and photographed.
4.4. Functional Prediction Based on BsCML Sequence Features
4.4.1. Subcellular Localization, Signal Peptide Identification, Transmembrane Structure, and Cis-Element Analysis of the BsCML Gene
The subcellular localization of the CML gene was predicted using WoLF PSORT (https://wolfpsort.hgc.jp/ (accessed on 22 January 2024)) [61]. The signal peptide of the CML was identified using SignaIP-4.1 (https://services.healthtech.dtu.dk/services/SignalP-4.1/ (accessed on 22 January 2024)). The transmembrane structure was predicted using Deep TMHMM (https://dtu.biolib.com/DeepTMHMM (accessed on 22 January 2024)). To analyze the homeostatic elements within the 2000 bp promoter region upstream of the BsCMLs gene, the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 21 January 2024)) was used, and the results were visualized in Tbtools, with each element represented by a 20 bp window for localization purposes.
4.4.2. Analysis of Evolutionary Characteristics of the BsCML Gene Family
To analyze the evolutionary relationships within the 38 CML gene families of B. striata, 102 CML amino acid sequences from A. thaliana, Dendrobium nobile, and Phalaenopsis were used as references. Specifically, 29 A. thaliana, 40 D. catenatum, and 33 P. equestris CML sequences were downloaded, along with the identified BsCML protein sequences, to construct an interspecies phylogenetic tree. MAGE11 was used to process the data (parameter settings: Neighbor-Joining method, Bootstrap replications set to 1000, with other parameters set to default), followed by visualization using iTOL (https://itol.embl.de/ (accessed on 31 July 2023)). Finally, subfamily classification was performed based on the conserved domain characteristics of these genes.
4.4.3. Interaction Network Analysis of BsCML Family Proteins
To analyze the protein–protein interaction (PPIs) and gene ontology (GO) of the BsCML family proteins, the online tool STRING 12.0 (http://string-db.org/ (accessed on 22 January 2024)) [62] was used with default parameters, using A. thaliana as a reference plant. Subsequently, CLUSTALW (https://www.genome.jp/tools-bin/clustalw (accessed on 28 September 2024)) and ESPript 3.0 (https://espript.ibcp.fr/ESPript/ESPript/index.php (accessed on 28 September 2024)) [63] were employed for sequence alignment analysis of potentially homologous proteins.
4.5. Functional Verification and Analysis of the BsCML Gene Family
4.5.1. Expression Pattern Analysis and Quantitative Fluorescence PCR Validation of BsCML
In a previous study, the content of militarine was measured across multiple growth stages, revealing that four specific time points (3 dpi, 18 dpi, 21 dpi, 36 dpi) served as critical inflection points for militarine synthesis [57]. At these time points, the expression pattern of the BsCML gene family was visualized using ChiPlot (https://www.chiplot.online/ (accessed on 28 April 2024)), based on the FPKM values of CML transcripts in B. striata callus.
To verify the expression of CML genes represented by the FPKM values from the transcriptome, the first strand of cDNA was synthesized by reverse transcription using the TIANGEN kit, with 1 µg of total RNA from qualified B. striata suspension cells at 18 dpi and 21 dpi as the template. The qPCR reaction was performed on the AGS4800 real-time quantitative PCR instrument. The total volume of the qPCR reaction mixture was 10 µL, consisting of 1 µL cDNA template (with a concentration of 55–60 ng/µL), 5 µL SYBR Green qPCR Master Mix, 0.2 µL of each primer (10 µmol/L), and 3.6 µL ddH2O. Primer information is shown in Table 3. Thermal cycling conditions were as follows: initial denaturation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 10 s, 55–60 °C for 30 s, and 72 °C for 30 s (fluorescence acquisition). The final step was at 95 °C for 15 s. Gene expression was normalized to the β-actin reference gene (GenBank: XM_045392100).

Table 3.
RT-qPCR primer information for the B. striata BsCML gene family.
To explore the regulatory role in the synthesis of militarine and dactylorhin A, Pearson correlation analysis (PPMCC) was performed using SPSS 29.0 between the measured metabolite concentrations (3 biological replicates and 3 technical replicates) and the expression of BsCML transcripts. A correlation heatmap was then generated to visualize the data. Through comprehensive analysis, genes significantly associated with the accumulation of secondary metabolites were identified within the BsCML family.
4.5.2. Expression Profile Analysis of BsCML in Different B. striata Tissues
To investigate the regulation of BsCML family members in the synthesis of secondary metabolites and their expression patterns in perennial B. striata plants, three BsCML family members—BsCML11 *, BsCML32 **, and BsCML37 ** (* p < 0.05, ** p < 0.01 level correlation)—which were significantly associated with militarine synthesis, were selected for further study. RT-PCR was conducted on four tissues: flowers, leaves, tubers, and roots, from three types of B. striata plants that had been cultivated for over three years. The 2−ΔΔCt method was used to calculate gene expression relative to β-actin.
4.5.3. LC-MS Detection of Secondary Metabolite Accumulation in Different B. striata Tissues
The roots, tubers, leaves, and flowers from three different B. striata plants were collected, and the metabolites were quantitatively analyzed using an Agilent 6545 Q-TOF liquid chromatography-mass spectrometry (LC-MS) system. The analytes were quantified in both positive and negative ion detection modes using multiple reaction monitoring (MRM). The three standards were accurately weighed and dissolved in methanol to prepare individual high-concentration standard solutions (militarine at 0.25 mg/mL and dactylorhin A at 0.5 mg/mL). The following conditions were used: Agilent Eclipse Plus C18 column (2.1 × 100 mm, 1.8 µm); flow rate: 0.5 mL/min; mobile phase: water with 0.1% formic acid (A) and 100% acetonitrile (B); column temperature: 30 °C; detection wavelengths: 220 nm and 254 nm; injection volume: 5.00 µL. The gradient elution was as follows: 0–0.5 min, 90% A and 10% B; 0.5–6 min, 100% B; 6–8 min, 90% A and 10% B.
By comprehensively analyzing the relationship between gene expression and metabolite accumulation in BsCML family members, we identified BsCML genes closely involved in militarine synthesis, providing an experimental basis for future studies on militarine biosynthesis.
5. Conclusions
In this study, 38 BsCML family members containing EF-hand motifs were identified from B. striata based on genomic sequencing and were classified into eight subfamilies (I–VIII). Analysis of BsCML gene structures, conserved protein domains, and phylogenetic relationships indicated that the inheritance of BsCML genes during evolution is highly conserved. Interaction analysis revealed a close interaction network within the BsCML family. Although most BsCML members exhibit low or negligible expression, they show significant tissue-specific differences and can be activated to varying degrees by elicitors. Upon treatment with elicitors such as NaAc and SA, the regulatory roles of BsCML in the accumulation of secondary metabolites changed significantly, suggesting that the metabolite accumulation in B. striata is regulated through the interaction between BsCML genes and the external environment. Furthermore, the expression patterns of BsCML11, BsCML32, and BsCML37 exhibited a high degree of similarity to the trends observed in militarine accumulation, suggesting that the differential expression of BsCML genes is closely related to the accumulation of metabolites in B. striata. These genes may serve as candidate genes for further investigation into the biosynthetic mechanisms of militarine. The results of this study have identified the BsCML family in B. striata and systematically explored their potential functions, thereby laying a foundation for the future utilization of BsCML molecular resources.
Author Contributions
K.L.: Writing—original draft, Formal analysis, Software, Validation, Visualization, Methodology; M.X.: Methodology, Software; Q.L.: Methodology, Data curation, Investigation; H.L.: Methodology; Y.X.: Methodology; D.X.: Writing—review editing, Funding acquisition, Supervision, Project administration, Conceptualization. All authors have read and agreed to the published version of the manuscript.
Funding
This research received financial support from various sources, including the National Natural Science Foundation of China (32260089); the Science and Technology Department Foundation of Guizhou Province, China (QKHJC-MS[2025]371, QKPTRC2019-027); the “12345 Future Talent Training Plan” of Zunyi Medical University (XJ2023-JX-01-06); the Undergraduate Education and Teaching Reform Project of Zunyi Medical University (XJKCSZ2023-9, XJJG2022-22, XJJG2024-09); and the first batch of Class Advisor Studios at Zunyi Medical University (2024BZR-01).
Data Availability Statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA1009214.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Ranty, B.; Aldon, D.; Cotelle, V.; Galaud, J.P.; Thuleau, P.; Mazars, C. Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 2016, 7, 327. [Google Scholar] [CrossRef]
- Jiang, Y.; Ding, P. Calcium signaling in plant immunity: A spatiotemporally controlled symphony. Trends Plant Sci. 2023, 28, 74–89. [Google Scholar] [CrossRef]
- Wang, C.; Luan, S. Calcium homeostasis and signaling in plant immunity. Curr. Opin. Plant Biol. 2024, 77, 102485. [Google Scholar] [CrossRef]
- Zheng, Z.; Shen, J.; Pan, W.; Pan, J. Calcium sensors and their stress signaling pathways in plants. Hereditas 2013, 35, 875–884. [Google Scholar] [CrossRef]
- Kudla, J.; Batistič, O.; Hashimoto, K. Calcium signals: The lead currency of plant information processing. Plant Cell 2010, 22, 541–563. [Google Scholar] [CrossRef]
- Luan, S.; Kudla, J.; Rodriguez-Concepcion, M.; Yalovsky, S.; Gruissem, W. Calmodulins and calcineurin B-like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 2002, 14, S389–S400. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Z.; Han, S.; Liu, P.; Sadeghnezhad, E.; Liu, M. Growth or survival: What is the role of calmodulin-like proteins in plant? Int. J. Biol. Macromol. 2023, 242, 124733. [Google Scholar] [CrossRef] [PubMed]
- Mohanta, T.K.; Yadav, D.; Khan, A.L.; Hashem, A.; Abd Allah, E.F.; Al-Harrasi, A. Molecular players of EF-hand containing calcium signaling event in plants. Int. J. Mol. Sci. 2019, 20, 1476. [Google Scholar] [CrossRef] [PubMed]
- Young, B.D.; Cook, M.E.; Costabile, B.K.; Samanta, R.; Zhuang, X.; Sevdalis, S.E.; Varney, K.M.; Mancia, F.; Matysiak, S.; Lattman, E.; et al. Binding and functional folding (BFF): A physiological framework for studying bio-molecular interactions and allostery. J. Mol. Biol. 2022, 434, 167872. [Google Scholar] [CrossRef]
- Delk, N.A.; Johnson, K.A.; Chowdhury, N.I.; Braam, J. CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol. 2005, 139, 240–253. [Google Scholar] [CrossRef]
- Köster, P.; DeFalco, T.A.; Zipfel, C. Ca signals in plant immunity. EMBO J. 2022, 41, e110741. [Google Scholar] [CrossRef]
- Song, X.; Li, J.; Lyu, M.; Kong, X.; Hu, S.; Song, Q.; Zuo, K. CALMODULIN-LIKE-38 and PEP1 RECEPTOR 2 integrate nitrate and brassinosteroid signals to regulate root growth. Plant Physiol. 2021, 187, 1779–1794. [Google Scholar] [CrossRef] [PubMed]
- Lokdarshi, A.; Conner, W.C.; McClintock, C.; Li, T.; Roberts, D.M. Arabidopsis CML38, a calcium sensor that localizes to ribonucleoprotein complexes under hypoxia stress. Plant Physiol. 2016, 170, 1046–1059. [Google Scholar] [CrossRef]
- Leba, L.J.; Cheval, C.; Ortiz-Martín, I.; Ranty, B.; Beuzón, C.R.; Galaud, J.P.; Aldon, D. CML9, an Arabidopsis calmodulin-like protein, contributes to plant innate immunity through a flagellin-dependent signalling pathway. Plant J. 2012, 71, 976–989. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Wu, J.; Sun, Y.; Zhu, H.; Sun, Q.; Zhao, P.; Huang, R.; Guo, Z. A calmodulin-like protein (CML10) interacts with cytosolic enzymes GSTU8 and FBA6 to regulate cold tolerance. Plant Physiol. 2022, 190, 1321–1333. [Google Scholar] [CrossRef]
- Li, C.; Meng, D.; Zhang, J.; Cheng, L. Genome-wide identification and expression analysis of calmodulin and calmodulin-like genes in apple (Malus × domestica). Plant Physiol. Biochem. 2019, 139, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Xu, C.; Cao, H.; Shi, Y.; Chen, J.; Chai, Y.; Li, Z. Tomato calmodulin-like protein SlCML37 is a calcium (Ca2+) sensor that interacts with proteasome maturation factor SlUMP1 and plays a role in tomato fruit chilling stress tolerance. J. Plant Physiol. 2021, 258–259, 153373. [Google Scholar] [CrossRef]
- Yang, D.; Chen, T.; Wu, Y.; Tang, H.; Yu, J.; Dai, X.; Zheng, Y.; Wan, X.; Yang, Y.; Tan, X. Genome-wide analysis of the peanut CaM/CML gene family reveals that the AhCML69 gene is associated with resistance to Ralstonia solanacearum. BMC Genom. 2024, 25, 200. [Google Scholar] [CrossRef]
- Cho, K.M.; Nguyen, H.T.; Kim, S.Y.; Shin, J.S.; Cho, D.H.; Hong, S.B.; Shin, J.S.; Ok, S.H. CML10, a variant of calmodulin, modulates ascorbic acid synthesis. New Phytol. 2016, 209, 664–678. [Google Scholar] [CrossRef]
- Yi, F.; Li, Y.; Song, A.; Shi, X.; Hu, S.; Wu, S.; Shao, L.; Chu, Z.; Xu, K.; Li, L.; et al. Positive roles of the Ca2+ sensors GbCML45 and GbCML50 in improving cotton Verticillium wilt resistance. Mol. Plant Pathol. 2024, 25, e13483. [Google Scholar] [CrossRef]
- Li, Q.; Xu, M.; Wu, F.; Guo, Z.; Yang, N.; Li, L.; Wen, W.; Xu, D. Integrated transcriptomics and metabolomics provide insights into the biosynthesis of militarine in the cell suspension culture system of Bletilla striata. Adv. Biotechnol. 2024, 2, 25. [Google Scholar] [CrossRef]
- Jia, X.; Li, Q.; Xu, M.; Zhang, J.; Xu, D. Advances in militarine: Pharmacology, synthesis, molecular regulation and regulatory mechanisms. Heliyon 2024, 10, e24341. [Google Scholar] [CrossRef] [PubMed]
- Commission, C.P. Pharmacopoeia of the People’s Republic of China, 2020th ed.; China Medical Science Press: Beijing, China, 2020. [Google Scholar]
- Boonburapong, B.; Buaboocha, T. Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biol. 2007, 7, 4. [Google Scholar] [CrossRef]
- Qian, Z.; Gu, S.; Zhao, X.; Rao, X.; Zeng, D.; Shen, Q.; Zhang, R.; Chen, S.; He, L.; Li, F. Identification and cold stress expression analysis of CML gene family in Erianthus fulvus based on transcriptome. J. Agric. Biotechnol. 2022, 30, 885–895. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, T.; Liang, L.; Li, L.; Liu, L. Genome-wide analysis of CaM/CML gene family in two Orchidaceae species. Forest Res. 2018, 31, 15–25. [Google Scholar] [CrossRef]
- Cai, K.; Kuang, L.; Yue, W.; Xie, S.; Xia, X.; Zhang, G.; Wang, J. Calmodulin and calmodulin-like gene family in barley: Identification, characterization and expression analyses. Front. Plant Sci. 2022, 13, 964888. [Google Scholar] [CrossRef]
- Liu, Y.; Yin, F.; Liao, L.; Shuai, L. Genome-wide identification and expression analysis of calmodulin-like proteins in cucumber. PeerJ 2023, 11, e14637. [Google Scholar] [CrossRef]
- Munir, S.; Khan, M.R.; Song, J.; Munir, S.; Zhang, Y.; Ye, Z.; Wang, T. Genome-wide identification, characterization and expression analysis of calmodulin-like (CML) proteins in tomato (Solanum lycopersicum). Plant Physiol. Biochem. 2016, 102, 167–179. [Google Scholar] [CrossRef]
- Li, Q.; Gao, L.; Yu, F.; Lü, S.; Yang, P. Evolution and diversification of CaM/CML gene family in green plants. Plant Physiol. Biochem. 2023, 202, 107922. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Kumar, P.; Bae, H. Genomics and evolutionary aspect of calcium signaling event in calmodulin and calmodulin-like proteins in plants. BMC Plant Biol. 2017, 17, 38. [Google Scholar] [CrossRef]
- Zhang, M.; Luo, D.; Fang, H.; Zhao, W.; Zheng, Y. Effect of light quality on the growth and main chemical composition of Bletilla striata. J. Plant Physiol. 2022, 272, 153690. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Guo, J.; Cui, L.J.; Yan, P.D.; Yan, Y.P. Effects of rhizosphere soil physical, chemical factors and fungal community on yield and quality of Bletilla striata. Shaanxi Agric. Sci. 2022, 68, 64–70. [Google Scholar] [CrossRef]
- Xiao, C.; Xu, C.; Zhang, J.; Jiang, W.; Zhang, X.; Yang, C.; Xu, J.; Zhang, Y.; Zhou, T. Soil microbial communities affect the growth and secondary metabolite accumulation in Bletilla striata (Thunb.) Rchb. f. Front. Microbiol. 2022, 13, 916418. [Google Scholar] [CrossRef]
- Midhat, U.; Ting, M.K.Y.; Teresinski, H.J.; Snedden, W.A. The calmodulin-like protein, CML39, is involved in regulating seed development, germination, and fruit development in Arabidopsis. Plant Mol. Biol. 2018, 96, 375–392. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Tan, Q.; Gao, Q.; Zheng, S.; Chen, W.; Galaud, J.P.; Li, X.; Zhu, X. Calmodulin-like protein CML15 interacts with PP2C46/65 to regulate papaya fruit ripening via integrating calcium, ABA and ethylene signals. Plant Biotechnol. J. 2024, 22, 1703–1723. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Huang, R.; Zhu, H.; Sun, Y.; Guo, Z. A novel Medicago truncatula calmodulin-like protein (MtCML42) regulates cold tolerance and flowering time. Plant J. 2021, 108, 1069–1082. [Google Scholar] [CrossRef]
- Zhang, D.; Du, L.; Lin, J.; Wang, L.; Zheng, P.; Deng, B.; Zhang, W.; Su, W.; Liu, Y.; Lu, Y.; et al. Genome-wide identification and expression analysis of calmodulin and calmodulin-like genes in passion fruit (Passiflora edulis) and their involvement in flower and fruit development. BMC Plant Biol. 2024, 24, 626. [Google Scholar] [CrossRef]
- Sun, Q.; Yu, S.; Guo, Z. Calmodulin-like (CML) gene family in Medicago truncatula: Genome-wide identification, characterization and expression analysis. Int. J. Mol. Sci. 2020, 21, 7142. [Google Scholar] [CrossRef]
- Aleynova, O.A.; Suprun, A.R.; Ananev, A.A.; Nityagovsky, N.N.; Ogneva, Z.V.; Dubrovina, A.S.; Kiselev, K.V. Effect of calmodulin-like gene (CML) overexpression on stilbene biosynthesis in cell cultures of Vitis amurensis Rupr. Plants 2022, 11, 171. [Google Scholar] [CrossRef]
- Yin, F.; Zhao, M.; Gong, L.; Nan, H.; Ma, W.; Lu, M.; An, H. Genome-wide identification of Rosa roxburghii CML family genes identifies an RrCML13-RrGGP2 interaction involved in calcium-mediated regulation of ascorbate biosynthesis. Plant Physiol. Biochem. 2024, 214, 108874. [Google Scholar] [CrossRef]
- Teresinski, H.J.; Hau, B.; Symonds, K.; Kilburn, R.; Munro, K.A.; Doner, N.M.; Mullen, R.; Li, V.H.; Snedden, W.A. Arabidopsis calmodulin-like proteins CML13 and CML14 interact with proteins that have IQ domains. Plant Cell Environ. 2023, 46, 2470–2491. [Google Scholar] [CrossRef] [PubMed]
- Symonds, K.; Teresinski, H.J.; Hau, B.; Dwivedi, V.; Belausov, E.; Bar-Sinai, S.; Tominaga, M.; Haraguchi, T.; Sadot, E.; Ito, K.; et al. Functional characterization of calmodulin-like proteins, CML13 and CML14, as novel light chains of Arabidopsis class VIII myosins. J. Exp. Bot. 2024, 75, 2313–2329. [Google Scholar] [CrossRef]
- Heyer, M.; Scholz, S.S.; Reichelt, M.; Kunert, G.; Oelmüller, R.; Mithöfer, A. The Ca2+ sensor proteins CML37 and CML42 antagonistically regulate plant stress responses by altering phytohormone signals. Plant Mol. Biol. 2022, 109, 611–625. [Google Scholar] [CrossRef]
- Erb, M.; Kliebenstein, D.J. Plant secondary metabolites as defenses, regulators, and primary metabolites: The blurred functional trichotomy. Plant Physiol. 2020, 184, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xiong, K.; Wen, W.; Li, L.; Xu, D. Functional endophytes regulating plant secondary metabolism: Current status, prospects and applications. Int. J. Mol. Sci. 2023, 24, 1153. [Google Scholar] [CrossRef]
- Koprivova, A.; Kopriva, S. Plant secondary metabolites altering root microbiome composition and function. Curr. Opin. Plant Biol. 2022, 67, 102227. [Google Scholar] [CrossRef]
- Aleynova, O.A.; Kiselev, K.V.; Suprun, A.R.; Ananev, A.A.; Dubrovina, A.S. Involvement of the calmodulin-like protein gene VaCML92 in grapevine abiotic stress response and stilbene production. Int. J. Mol. Sci. 2023, 24, 15827. [Google Scholar] [CrossRef] [PubMed]
- You, W.; Zhang, J.; Ru, X.; Xu, F.; Wu, Z.; Jin, P.; Zheng, Y.; Cao, S. CmCML11 interacts with CmCAMTA5 to enhance γ-aminobutyric acid (GABA) accumulation by regulating GABA shunt in fresh-cut cantaloupe. Plant Physiol. Biochem. 2024, 206, 108217. [Google Scholar] [CrossRef]
- Vadassery, J.; Reichelt, M.; Hause, B.; Gershenzon, J.; Boland, W.; Mithöfer, A. CML42-mediated calcium signaling co-ordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol. 2012, 159, 1159–1175. [Google Scholar] [CrossRef]
- Tiwari, S.; Lata, C.; Chauhan, P.S. Salicylic acid: Metabolism, regulation, and functions in crop abiotic stress tolerance. In Augmenting Crop Productivity in Stress Environment; Ansari, S.A., Ansari, M.I., Husen, A., Eds.; Springer Nature: Singapore, 2022; pp. 257–274. [Google Scholar]
- Liu, J.; Qiu, G.; Liu, C.; Li, H.; Chen, X.; Fu, Q.; Lin, Y.; Guo, B. Salicylic acid, a multifaceted hormone, combats abiotic stresses in plants. Life 2022, 12, 886. [Google Scholar] [CrossRef]
- Oraei, M.; Panahirad, S.; Zaare-Nahandi, F.; Gohari, G. Pre-véraison treatment of salicylic acid to enhance anthocyanin content of grape (Vitis vinifera L.) berries. J. Sci. Food Agric. 2019, 99, 5946–5952. [Google Scholar] [CrossRef]
- Yi, G.E.; Robin, A.H.; Yang, K.; Park, J.I.; Hwang, B.H.; Nou, I.S. Exogenous methyl jasmonate and salicylic acid induce subspecies-specific patterns of glucosinolate accumulation and gene expression in Brassica oleracea L. Molecules 2016, 21, 1417. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Li, H.; Pan, J.; Zhou, B.; He, T.; Wu, Y.; Zhou, D.; He, W.; Chen, L. Salicylic acid modulates secondary metabolism and enhanced colchicine accumulation in long yellow daylily (Hemerocallis citrina). AoB Plants 2024, 16, plae029. [Google Scholar] [CrossRef] [PubMed]
- Hadizadeh, M.; Ofoghi, H.; Kianirad, M.; Amidi, Z. Elicitation of pharmaceutical alkaloids biosynthesis by salicylic acid in marine microalgae Arthrospira platensis. Algal Res. 2019, 42, 101597. [Google Scholar] [CrossRef]
- Liu, H.; Huang, C.; Li, Q.; Wang, M.; Xiao, S.; Shi, J.; He, Y.; Wen, W.; Li, L.; Xu, D. Genome-wide identification of genes related to biosynthesis of phenolic acid derivatives in Bletilla striata at different suspension culture stages. Front. Plant Sci. 2022, 13, 875404. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Saeed, F.; Hashmi, M.H.; Aksoy, E.; Demirel, U.; Bakhsh, A. Identification and characterization of RNA polymerase II (RNAP) C-terminal domain phosphatase-like 3 (SlCPL3) in tomato under biotic stress. Mol. Biol. Rep. 2023, 50, 6783–6793. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).