Abstract
Early flowering refers to the phenomenon in which the first flower appears in fewer days than normal, regardless of the sex of the flower. It is a significant feature impacting the early maturity and economic yield of cucumbers. The early flowering trait of cucumber is influenced by several factors. Considering its heritability, technologies such as whole-genome sequencing, genetic modification, bioinformatics analysis, quantitative trait locus (QTL) mapping, molecular marker-assisted selection, and gene editing are widely used to explore the regulatory genes and molecular mechanisms of the early flowering trait in cucumbers. This review aimed to summarize the factors, QTL mapping, molecular regulation mechanisms, and omics analysis related to early flowering traits in cucumbers. This review contributes theoretical insights to support both cucumber breeding for early flowering and fundamental research on early flowering traits.
1. Introduction
Cucumber (Cucumis sativus L., 2n = 14) belongs to the Cucurbitaceae family and Cucumis genus; it is an important vegetable crop with the largest planting area and yield among vegetables and plays a vital role in the annual production and supply of vegetables [1,2]. The wild type of cucumber, C. sativus var. hardwickei, was originated in India [3]. Cucumbers have adapted to various climatic conditions due to natural and artificial selection. The bitter taste of cucumber fruit disappears, the morphological diversity and adaptability change, and both yield and quality are improved during domestication. Additionally, the sensitivity of cucumber plants to extended photoperiods has changed due to domestication. Different types of cucumbers can bloom rapidly only under suitable photoperiod conditions. However, the molecular processes of cucumber domestication remain unclear [4]. The phylogenetic analysis shows that cucumbers are classified into four geographic groups: the wild Indochina group, the semi-wild Xishuangbanna group, and two cultivated cucumber groups (Eurasia and East Asia) [5].
Yunnan, China, is considered the secondary center of the origin of cucumbers. Cucumbers have been planted on a large scale for many years; however, their planting areas and yields continue to increase steadily worldwide. The total area of cucumber cultivation worldwide was 2.25 million hectares in 2020, yielding 9.035 million tons. China accounted for 56.4% (1.27 million hectares) of the global cucumber production area and 81.2% (73.36 million tons) of the global cucumber production [6]. As a fruit and a vegetable, early maturing cucumber offers high economic benefits; therefore, early maturity has become a crucial focus for efficient and high-yield breeding [7]. Cucumber breeders have also shifted their focus to developing early maturing and high-yielding varieties [8,9]. Previous studies have extensively investigated the agronomic traits related to the early maturity of cucumbers. These traits related to the early maturity of cucumbers mainly include the position of the first flowering node, the position of the first female flower, and the number of days from sowing to the opening of the first female flower during the initial flowering period [10]. Cucumber domestication has significantly improved yield, quality, and photoperiod adaptation. However, the underlying molecular mechanisms need complete elucidation. The increasing global demand for cucumbers, particularly in China as the dominant producer, has led to breeding efforts focused on early maturing varieties with enhanced economic value.
Early flowering refers to the phenomenon in which the first flower appears in fewer days than normal, regardless of the sex of the flower; it is a significant feature impacting the early maturity and economic yield of cucumbers [7,11,12]. The flowering transformation is triggered by multiple factors combining endogenous genetic pathways with environmental stimuli [13,14,15]. Early flowering can improve the economic benefits of crops; hence, the mechanisms of flowering in many crops have been explored, and early flowering is a crucial breeding goal [16]. Various pathways influencing flowering have been reported in Arabidopsis [14]. These include endogenous (autonomous, gibberellin, circadian rhythm, and age) and environmental (vernalization, environmental temperature, and photoperiod) factors [17,18,19,20]. In addition, factors that can cause early flowering have been detected in crops such as tomatoes [16], rice [21], wheat [22], Chinese cabbage [23], and cassava [24]. These findings indicate that early flowering is governed by conserved genetic pathways across diverse plant species while exhibiting species-specific regulatory adaptations. Understanding these shared and unique mechanisms in cucumbers may be crucial for developing targeted breeding strategies to optimize flowering time and enhance crop productivity.
The data on germplasm resources obtained through physiological and molecular investigations on cucumbers are limited. This review summarizes the environmental factors, genetic QTL mapping, molecular mechanisms, and omics research related to the early flowering of cucumbers. It has significant application value for shortening the growth period of cucumbers, balancing and optimizing the industrial demand, and increasing cucumber yields.
2. Factors Influencing Cucumber Early Flowering Traits
2.1. Light
Light is the primary driver of plant photosynthesis and one of the most important environmental cues to regulate plant growth and development [25,26]. Both the length of the photoperiod and the intensity of light can impact the flowering time of plants. Long-day (LD) plants flower when the sunshine duration exceeds the critical length, whereas short-day (SD) plants flower when the sunshine duration is shorter than the critical length. The flowering of day-neutral plants is independent of the length of daylight hours [13]. Arabidopsis thaliana is an LD plant, with LD accelerating flowering and SD delaying flowering [27]. Xishuangbanna (XIS) cucumber (C. sativus L. var. xishuangbannesis) is a typical SD plant requiring a certain number of SD to induce flowering [28,29]. Weak light leads to delayed flowering time, delayed peak flowering period, prolonged flowering period, and decreased flowering quality in cucumbers [12]. Plants sense light mainly via photoreceptors [30,31,32,33,34,35,36,37,38]. Among these, photosensitive pigments significantly influencing the flowering time of plants have been identified [39,40,41,42,43]. For example, Phytochrome B (PHYB) can directly or indirectly interact with the Highly Expressed Osmotically Responsive Genes 1 (HOS1), Phytochrome and Flowering Time 1 (PFT1), and Vascular Plant One-zinc Finger 1/2 (VOZ1/2) to promote or inhibit FLOWERING LOCUS T (FT) and CONSTANS (CO) expression, ultimately mediating flowering [44,45,46,47]. CsPHYB regulates cucumber flowering time in a photoperiod-dependent manner [48]. The changes in the genes involved in the photoperiod pathway can promote flowering in photoperiod-sensitive crops [27,29]. Previous studies found that PHYB-deficient mutants exhibited early flowering [49,50]. Light is essential for the normal development of plants, significantly influencing crop growth, development, and quality. The flowering of cucumbers is impacted by the length of light exposure; therefore, the early flowering process of cucumbers can be regulated through reasonable control of light exposure during production.
2.2. Hormones
Hormones play vital roles in plant growth and development. Flowering is regulated by several hormones, such as gibberellins (GAs), jasmonic acid (JA), abscisic acid (ABA), brassinosteroids (BRs), and salicylic acid (SA) [51,52,53,54].
2.2.1. GAs
GAs have multiple physiological functions, including promoting stem elongation, breaking dormancy, promoting fruit development, affecting gender expression, delaying aging, and improving stress resistance. It has also been reported that GAs promote flowering [55,56,57,58,59,60]. A previous study showed that treating cucumbers with 100 μmol GA promoted flowering whereas the treatment of cucumbers with 100 μmol paclobutrazol (PAC) delayed flowering [48]. CsPHYB can delay cucumber flowering time by inhibiting the biosynthesis of GA.
2.2.2. JA
JA is vital in plant growth, development, and stress resistance. It can induce the expression of early JA response genes and flower development-related proteins, thereby influencing plant flowering time [61,62,63,64,65,66]. The JA Zinc-finger protein expressed in Inflorescence Meristem (ZIM) domain protein, CsJAZ1, is linked to the Homeodomain-Leucine Zipper IV (HD-ZIP IV) protein to regulate the flowering time of male cucumber flowers [67].
2.2.3. ABA
ABA mainly participates in plant stress response and growth and development regulation. It alleviates drought, salt, and low-temperature stresses; promotes seed development and dormancy; enhances root growth; and regulates stomatal closure. It also plays an essential role in plant flowering [68]. Further, it inhibits flowering [69]. It not only promotes plant dormancy but also affects flower bud differentiation [70,71]. ABA can induce the expression of the C. sativus SHATTERPROOF (CsSHP) gene, thereby influencing the formation and development of cucumber floral organs [72].
2.2.4. BR and SA
BR is widely involved in plant growth, development, and stress response. It promotes cell elongation and division; regulates the development of plant stems, leaves, and roots; enhances plant drought, salt, and disease resistance; and regulates plant photosynthesis. SA mainly participates in the defense response of plants. It improves their drought resistance, stress resistance, and antioxidant effects and promotes seed germination and root development. Both BR and SA can also influence the flowering time in cucumbers [73]. Plant hormones have diverse effects on the early flowering of cucumbers. Therefore, the early flowering process of cucumbers can be modulated by regulating the concentration and synthetic pathways of hormones during production, thereby providing strong support for cucumber production.
2.3. Other Factors
The early flowering of cucumbers is influenced by various factors, including temperature and nutritional conditions, which can be reasonably controlled in production. The reproductive stage has the most stringent temperature requirements during plant growth and development [74]. Temperature impacts the flowering time of cucumbers; high temperatures can promote early flowering [75,76]. Moreover, treatments such as nutrient composition, inorganic fertilizers (phosphate solubilizing bacteria, PSB), and organic measures (integrated nutrient management, INM) can lead to cucumber flowering [77,78]. A previous study showed that cucumber plants treated with organic and biological fertilizers exhibited a better early flowering shape; early flowering was better than that in the control group, especially in plots treated with farmyard manure, FYM, at a concentration of 20 t ha−1 [79].
In conclusion, regulating cucumber flowering time involves a complex interplay of environmental factors (light and temperature), hormonal pathways (GA, JA, ABA, BR, and SA), and nutritional management. These findings indicate that the targeted manipulation of photoperiod conditions, hormone balance, and organic fertilization can effectively promote early flowering in cucumbers. Future studies should further explore the molecular mechanisms underlying the aforementioned interactions to develop more precise cultivation strategies for optimizing flowering time and yield.
3. Current Status of Breeding Early Flowering Cucumbers
The breeding of early flowering cucumbers is an essential direction in cucumber breeding. The growth cycle of cucumbers can be shortened and the yield and economic benefits can be improved by selecting early flowering varieties. A total of 1215 cucumber varieties were registered as non-major crop varieties in China from 2017 to 2020 [80]. Multiple breeding methods have been adopted in cucumber breeding, including hybrid, mutagenesis, molecular marker-assisted selection, and genetic engineering breeding [80]. The breeding of early flowering cucumbers also achieved significant results through the continuous efforts of breeders. For example, “Jinchun No.4” takes about 34 days from emergence to flowering [81]. “Zhongnong No.5” is a new female cucumber variety, with the first flowering occurring at two to three nodes [82]. “Kaohsiung No.3” was obtained through the hybridization of KSL009 and KSL017 inbred lines; it manifested as an early flowering variety tolerant to heat and moisture [83]. In summary, the breeding of early flowering cucumbers breeding has demonstrated significant potential in enhancing productivity and economic returns. The successful development and application of these varieties highlight the practical achievements in this field.
4. QTL Mapping of Cucumber Early Flowering Traits
QTL mapping is used for determining the position of quantitative trait genes on chromosomes. It is based on Morgan’s linkage inheritance law. It uses molecular markers to classify the genotype of each individual, analyzes the relationship between the genotype of each individual in the mapping population and the phenotype of the target trait, and predicts the genetic linkage and distance between QTL loci and molecular markers of the target trait [84]. Screening molecular markers is an effective means of successfully determining the relative positions of related genes. Biotechnological advances have led to the successful development of various molecular markers, such as restriction fragment length polymorphisms (RFLP), sequence-characterized amplified region (SCAR), sequence tagged sites (STS), simple sequence repeats (SSR), single primer amplified region (SPAR), single strand conformation polymorphism (SSCP), random amplified polymorphic DNA (RAPD), inter-simple sequence repeat (ISSR), sequence-related amplified polymorphism (SRAP), cleaved amplified polymorphic sequence (CAPS), and amplified fragment length polymorphism (AFLP) [85]. Employing these molecular markers is of great significance for QTL mapping and crop breeding.
Early flowering is essential for improving the early maturity and economic yield of cucumbers [7]. The early flowering of cucumbers is a typical quantitative trait [12,86]. Many studies have successfully mapped QTLs for the early flowering of cucumbers (Table 1). The gynoecious C. sativus var. sativus line “GY14” and wild C. sativus var. hardwickii (R) Alef. “PI183967” plants were used to backcross BC, S3, and F2 populations for the QTL mapping analysis of early flowering [87]. Two QTLs controlling flowering time were identified: one on linkage group 5 near the RFLP marker CsC029 and the other on linkage group 2 near the F-locus. The recombinant inbred line (RIL) and F2 population constructed through “G421” and “H-19” were used to conduct the genetic mapping and QTL analysis of the agronomic traits of cucumbers [88]. Four QTLs for flowering time (ant1.1, ant2.1, ant5.1, and ant6.1) were mapped to linkage groups 1, 2, 5, and 6. An F2 population was developed by crossing cucumber inbred lines S06 and S52 [89]. The use of 64 sequences SRAP markers indicated that the first flower node trait control gene ffn was located on the linkage group 9, with distances of 10.3 and 12.1 cM from the two side markers DC1EM5 and ME7EM2A, respectively. The F9 generation RIL populations were generated using the North China protected area-type cucumber “9930” and the European greenhouse-type cucumber “9110Gt”, and a QTL Da1.1 located on Chr1, which is related to flowering time, was identified [10].
The phenotypic investigation and genetic analysis were conducted on the F2 population constructed using “9930” (late flowering) and “Muromskij” (early flowering) [86]. The results showed that the early flowering traits were quantitative traits controlled by multiple genes, and the main flowering gene in “Muromskij” was dominant. The QTL Ef1.1, which controls the early flowering trait, was located in the 890 kb interval of Chr1. It was speculated that Csa1G651710, homologous to FT, was a candidate gene for the cucumber early flowering QTL Ef1.1. A total of 124 RILs derived from the cross of the XIS cucumber with cultivated inbred lines of cucumbers (“CC3” and “SWCC8”) were used to create a linkage map. Further, fft1.1 major-effect QTL (R2 = 25.8%) which was repeatedly identified in all four environments and had a highly consistent peak position on the genetic map, and fft6.1 minor QTL (R2 ≈ 6%) was detected in two seasons on Chr1 and Chr6, respectively [90]. Both QTLs led to an early flowering phenotype in cucumber plants.
The F2 and F2:3 populations were constructed using the all-female cucumber line “S1000” and the strong male cucumber line “S1002” as parents, and a high-density genetic linkage map was drawn for QTL mapping [85]. A total of six QTLs were detected on Chr3, Chr5, Chr6, and Chr7: EF3.1-1, EF3.1-2, EF5.1, EF6.1, EF6.2, and EF7.1. Csa6G382930, Csa7G430750, and Csa7G431330 were preliminarily identified as candidate genes for EF6.1 and EF7.1. Another set of F2 and F2:3 populations was constructed using early flowering inbred line “WI7200” and late flowering inbred line “WI7167” of cucumber, and three QTLs controlling flowering time were identified: FT1.1, FT5.1, and FT6.2 [91]. Among these, FT6.2 (R2 = 71.9–83.3%), which was located on Chr6, played the most important role in early flowering with low photoperiod sensitivity during domestication. FT1.1 (R2 = 9.1%), which was located on Chr1, was more involved in regulating the flowering time of cultivated cucumbers, with FT5.1 (R2 = 4.8–16.1%) moderation influencing flowering time. The cultivated cucumber line “Gy14” and the wild cucumber line “WI7221” were used to construct RIL, F2, and F2:3 populations [29]. Two QTLs related to flowering time were identified through QTL analysis. Among these, the main QTL, FT1.1 (R2 = 42.8%), played an essential role in regulating flowering, whereas the minor QTL, FT6.3 (R2 = 8%), contributed to photoperiod-sensitive flowering time during domestication. One major QTL (FT1.1, R2 = 13.9–44.9%) and one minor QTL (FT6.4, R2 = 14.8%) related to flowering time were identified using segregation F2 and RIL populations from the cross between “WI2757” and “TL” [92].
FT1.1 is located in the overlapping region on Chr1 [10,29,90,91,92], indicating that they may belong to the same locus. The flowering time was assessed in cucumber lines “CG5479” and “9930” with different day lengths to construct NILs; an early flowering gene Ef1.1 was finely located [93]. RILs and F9 populations were constructed using “CC3” and “SWCC8” as parents, and the main QTL DFF1.1 regulating cucumber flowering time was located on Chr1 through QTL sequence analysis [27]. Many studies have identified QTL related to early flowering in cucumbers, laying a solid foundation for subsequent mapping of early flowering genes and revealing gene functions.
Extensive QTL mapping studies have successfully identified multiple genomic regions (particularly on Chr1 and Chr6) governing early flowering traits in cucumbers. FT1.1 has emerged as a consistently detected major-effect locus across diverse populations. Integrating high-density genetic maps and candidate gene analysis (e.g., Csa1G651710 homologous to FT) has significantly advanced the understanding of the genetic architecture underlying flowering time regulation. These findings provide valuable tools for molecular marker-assisted breeding and pave the way for the functional characterization of key genes to enhance precision in improving the early flowering traits in cucumbers.


Table 1.
QTL mapping of early flowering traits in cucumbers.
Table 1.
QTL mapping of early flowering traits in cucumbers.
| Parents | Populations | QTLs | References |
|---|---|---|---|
| GY14 * PI183967 | BC, S3 | Two chromosome regions | [87] |
| G421 * H-19 | RIL | ant1.1, ant2.1, ant5.1, ant6.1 | [88] |
| S06 * S52 | F2 | ffn | [89] |
| 9930 * 9110Gt | F9 | Da1.1 | [10] |
| 9930 * Muromskij | F2 | Ef1.1 | [85] |
| CC3 * SWCC8 | RILs | fft1.1, fft6.1 | [90] |
| S1000 * S1002 | F2 and F2:3 | EF3.1-1, EF3.1-2, EF5.1, EF6.1, EF6.2, EF7.1 | [85] |
| WI7200 * WI7167 | F2 and F2:3 | FT1.1, FT5.1, FT6.2 | [91] |
| Gy14 * WI7221 | RIL, F2, and F2:3 | FT1.1, FT6.3 | [29] |
| WI2757 * TL | F2 | FT1.1, FT6.4 | [92] |
| CG5479 * 9930 | NILs | Ef1.1 | [93] |
| CC3 * SWCC8 | RILs and F9 | DFF1.1 | [27] |
The asterisk * represents hybridization.
5. Molecular Regulatory Mechanism of Cucumber Early Flowering Traits
Flowering is an essential indicator of the transition from vegetative to reproductive growth in higher plants [94], ensuring the production of seeds necessary for species survival [95]. It is regulated by a complex genetic pathway responding to both endogenous and environmental stimuli [13,14]. Research on Arabidopsis has revealed six regulatory pathways related to flowering time: vernalization, photoperiod, gibberellin, autonomy, age, and environmental temperature pathways [96,97,98], which are interconnected to form a complex regulatory network influencing flowering. Several genes related to flowering time have been identified in A. thaliana [96], all of which are specific flowering-related genes. These genes are named flowering integration genes [99]. They include FT [100], SUPER PRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) [101], and LEAFY (LFY) [102].
Many early flowering genes in cucumbers homologous to those in Arabidopsis have been identified (Table 2). For example, the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) gene was first discovered in Antirrhinum majus [103] and plays a critical role in various stages of plant transition from vegetative to reproductive growth [104,105]. SPL genes directly or indirectly regulate the flowering time in A. thaliana [106,107,108,109,110,111]. However, whether cucumber CsSPL mediates flowering remains largely unclear. Therefore, the SQUAMOSA PROMOTER-BINDING-LIKE PROTEIN 13A (CsSPL13A) gene was cloned, and CsSPL13A-OE plants were constructed. The results showed that the CsSPL13A-OE plants exhibited early flowering compared with the wild-type plants. The yeast one-hybrid and dual-luciferase reporter assays indicate that CsSPL13A protein directly binds to the promoters of CsFT and β-AMYLASE (CsBAM), up-regulates their expression, and mediates cucumber flowering [6].
FT belongs to the phosphatidylethanolamine-binding protein (PEBP) family and regulates the flowering time in A. thaliana [112,113,114,115,116]. It is an activator of flowering transition [117,118]. An early flowering QTL Ef1.1 was mapped using the F2 population with “Muromskij” and “9930” hybridization [86]. The results of fine mapping and cloning showed that large genetic structural variations from CsFT upstream led to early flowering in cucumbers [93]. The pan-genomic analysis showed a consistent correlation between the variance of sequences located upstream of FT and the flowering time of the cucumber germplasm [119]. FT regulates SD flowering in XIS cucumbers, and the cis-regulation of FT is likely due to TE insertion [120]. The hybrid of “PI183967” and “9930” was used to cultivate and identify the photosensitive site sp-1 under different light lengths (LD and SD) [4]. The mapping results showed that CsFT, which was the only member of the FT/TSF-like clade in the PEBP family, was a candidate gene. The overexpression of CsFT significantly accelerated flowering in Arabidopsis [121]. Therefore, CsFT is a crucial gene involved in cucumber photoperiod domestication.
MADS-box genes are key transcription factors involved in plant development, particularly flower development [122,123]. MADS-box genes, such as Cucumber MADS box gene 1 (CUM1), Cucumber MADS box gene 10 (CUM10), Cucumber MADS box gene 26 (CUM26), CsAPETALA3 (CsAP3), and CsEPALLATA2 (CsSEP2), can affect the development of cucumber flowers; however, their functions still need confirmation [124,125,126,127]. A comprehensive analysis was conducted on 43 MADS-box genes in cucumber, which were compared with the genes in Arabidopsis, grape, and poplar. The expression analysis showed that 42 of 43 MADS-box members in cucumbers were expressed in various plant tissues, suggesting their varying roles in plants [128]. The MADS-box gene CsSHP was specifically enriched in stamens and carpels, and its overexpression resulted in early flowering in Arabidopsis. A yeast two-hybrid (Y2H) assay result showed the interaction of CsSHP with CsSEPs [72]. Moreover, the ectopic overexpression of CsMADS02 and CsMADS08 caused earlier flowering in Arabidopsis [129,130,131].
Branched-chain amino acids and transferases (BCATs) play crucial roles in the metabolism of branched-chain amino acids (BCAAs). The overexpression of BCATs promotes flowering in Arabidopsis by regulating the expression of genes regulating flowering time. BCAT can affect flowering in cucumbers [132]. Also, the overexpression of CsBCATs led to early flowering in Arabidopsis. CsBCATs can up-regulate the expression levels of FT and down-regulated the expression levels of SOC1 in the GIGANTEA/CONSTANS (GI/CO) and SHORT VEGETATIVE PHASE (SVP)/FLC modules, thereby promoting early flowering in Arabidopsis [133].
LFY is a key gene influencing the development and flowering of plant floral organs [134]. It can also interact with genes such as AGAMOUS (AG) and TERMINAL FLOWER 1 (TFL1) to regulate the onset of flowering [135]. Cucumber-FLO-LFY (CFL) in cucumber, which is homologous to Arabidopsis LFY, may participate in flowering [136]. The results of mRNA in situ hybridization indicate that the expression of CFL is related to the development of flower organs in cucumbers [137]. Transferring the CFL into gloxinia (Sinningia speciosa) for ectopic expression revealed that CFL could promote early flowering in gloxinia [138].
PHYB plays an essential role in regulating flowering in a photoperiod-dependent manner in Arabidopsis [139]; it degrades the factor promoting flowering, CO, to delay flowering [139,140,141]. A novel early flowering mutant was screened in cucumber. The mapping-based cloning revealed that CsPHYB (homolog of Arabidopsis PHYB) had a 5.5 kb long-terminal-repeat (LTR) retrotransposon insertion responsible for the mutation phenotype [48]. The ectopic expression of CsPHYB in Arabidopsis indicated that CsPHYB delayed flowering. Y2H and bimolecular fluorescence complementation (BiFC) assays showed that CsPHYB interacted with CsPIF3/4, promoting or inhibiting the expression of downstream flowering-related genes, thereby affecting flowering time.
TFL1 is a mobile signal of the PEBP family, which controls Arabidopsis flowering [117,142,143]. Six PEBP family members have been reported in cucumber; Csa3G776350 has the highest similarity to AtTFL1, reaching 71.91%, and is named CsTFL1b [113]. The ectopic expression of CsTFL1b in Arabidopsis showed that CsTFL1b delayed the flowering in Arabidopsis, and the degree of delayed flowering in transgenic plants was correlated with the expression level of CsTFL1b [144]. In addition, the 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE (CsACS2) transferred into tobacco delayed flowering in tobacco [145]. Also, the heterotopic expression of ETHYLENE RESPONSE FACTOR31 (CsERF31) also leads to late-flowering phenotypes in Arabidopsis and tobacco [146]. Despite abundant evidence, many mysteries regarding the molecular regulatory mechanisms of early flowering traits in cucumbers remain unresolved.
Researchers have identified a large number of early flowering germplasm resources for cucumbers. Several molecular markers have been designed to search for genes that can regulate the early flowering traits in cucumbers and elucidate their molecular regulatory mechanisms. These molecular markers can be used to detect the phenotype of cucumber flowering time in early development and even in the seedling stage [93], thus significantly improving breeding efficiency and providing the theoretical basis for promoting the research on early flowering research in cucumbers.
In conclusion, the molecular regulation of flowering in cucumbers involves a complex network of conserved pathways (photoperiod, gibberellin, etc.) and key genes (CsFT, CsSPL13A, CsPHYB, etc.), many of which exhibit functional homology with Arabidopsis flowering regulators. Significant progress has been achieved in characterizing these genes by exploring overexpression, protein–protein interactions (e.g., CsSHP-CsSEPs), and transposon-mediated mutations (e.g., CsPHYB-LTR), revealing their roles in flowering control. However, further studies are needed to elucidate the crosstalk between pathways and exploit these findings for precision breeding of early flowering cucumber varieties.


Table 2.
Early flowering genes and their regulatory mechanisms in cucumbers.
Table 2.
Early flowering genes and their regulatory mechanisms in cucumbers.
| Gene Names | Regulation | Interaction Proteins or TFs | References |
|---|---|---|---|
| CsSPL13A | positive | CsFT and CsBAM | [6] |
| CsFT | positive | - | [4,93,119,120] |
| CsSHP | positive | CsSEPs | [72] |
| CsBCATs | positive | FT and SOC1 | [133] |
| CFL | positive | - | [138] |
| CsMADS02, 09 | positive | - | [129,130] |
| CsMADS08 | positive | - | [131] |
| CsPHYB | Negative | CsPIF3/4 | [48] |
| CsTFL1b | Negative | - | [144] |
| CsACS2 | Negative | - | [145] |
| CsERF31 | Negative | - | [146] |
6. Functional Genomics and Omics Insights into Early Flowering Traits in Cucumbers
Cucumber genome data (CLv1.0) were released in 2009, making it the first sequenced genome of vegetable crops worldwide and laying a solid foundation for functional genomics research [147]. Subsequently, CLv2.0, CLv3.0, and CLv4.0 were reassembled, significantly advancing the progress of functional genomics research in cucumbers [148,149,150]. The cucumber genome data can be queried in cucumber-DB (http://www.cucumberdb.com/ (accessed on 22 March 2025)). This bioinformatics or genomic database can provide researchers with cucumber genome and transcriptome data, along with cucumber gene annotation, thereby providing a foundation for cucumber omics research. Thus, omics analyses, such as transcriptomic and proteomic analyses, are widely used to identify early flowering genes in cucumbers. Transcriptomic analysis is used for exploring gene evolution and function. New genes involved in cucumber flowering and their interacting factors were identified by integrating the transcriptional expression profiles and predicting promoter elements [151]. For example, RNA sequencing (RNA-seq) and differentially expressed gene analysis were conducted on male cucumber flowers, revealing that the HD-ZIP IV transcription factor GL2-LIKE regulated male flowering time in cucumbers [67]. RNA-seq analysis was conducted in the initiation stage of floral primordia and the developmental stage of floral organs to examine the differential expression of specific genes [27]. They found that CsaNFYA1 integrated multiple types of genes to regulate the flowering of XIS cucumber. The transcriptome analysis was performed on plants of wild-type (“Hardwickei”) and cultivated (“9930”) cucumbers under SD and LD conditions. The results showed that the changes in a photoperiod-sensitive CsFT were associated with the day-neutral and early flowering of cultivated cucumbers [4]. The transcriptome analysis was conducted on “XIS49” cucumber under different light conditions. The results indicated that the FT gene, rather than its upstream circadian clock gene, regulated the SD flowering of XIS cucumber; also, the cis-regulation of FT might be due to TE insertion [120]. Proteomics research can be used to explore the mechanisms of cucumber flowering and the factors influencing it. Through proteomics research, Zhang et al. [138] demonstrated that CLF interacted with the LFY and MADS-box genes to regulate cucumber flowering time. A previous study showed that CsTFL1 competes with CsFT, interacts with CsNOT2a-CsFDP, and inhibits deterministic growth and terminal flower formation in cucumbers [152]. Moreover, CsSPL3A encoded a protein containing a nuclear localization signal and a highly conserved SBP domain, which, together with CsFT and CsBAM, mediated flowering in cucumbers [6].
In summary, the integration of functional genomics and multi-omics approaches has revolutionized the understanding of early flowering traits in cucumbers, from genome assembly to molecular networks. Transcriptomic studies have identified key flowering regulators such as CsFT and CsaNFYA1, whereas proteomic analyses have revealed critical protein interactions involving CsTFL1 and CsSPL3A. These findings highlight the power of omics technologies in uncovering both cis-regulatory mechanisms (e.g., TE-mediated FT regulation) and trans-acting factors governing flowering time. The established genomic resources and analytical frameworks provide a strong foundation for both fundamental research and precision breeding of early flowering cucumber varieties.
7. Challenges and Perspectives
Cucumber is an essential economic crop in China, with the highest yield and cultivation area worldwide. Early flowering is one of the main factors influencing critical agronomic traits such as cucumber growth period and yield. This is a key research direction in botany and an important goal for cucumber breeders. The phenomenon of early flowering in cucumbers is complex and is influenced by various factors such as temperature, light, plant hormones, and nutrients (Figure 1). Genetic research has clearly indicated that early flowering in cucumbers is a quantitative trait controlled by multiple genes. Numerous major and minor QTLs have been identified. Several genes have been identified as key regulators of early flowering in cucumbers. The development of high-precision, high-throughput phenotyping technology has significantly facilitated the investigation of these flowering traits. Furthermore, establishing comprehensive genetic resources, including overexpression libraries, mutant collections, and gene-edited populations, has provided both valuable germplasms and a solid theoretical foundation for advancing research on early flowering mechanisms in cucumbers.
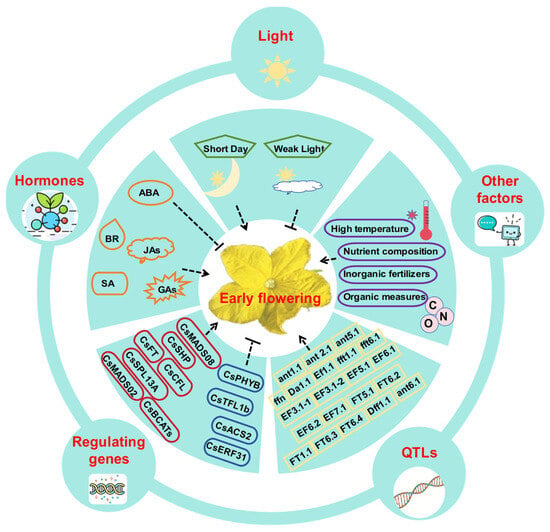
Figure 1.
Factors regulating early flowering trait in cucumbers. The green boxes represent different light conditions, orange boxes indicate different hormones, purple boxes represent other factors, yellow boxes indicate QTLs, and red and blue boxes represent positive and negative regulatory genes, respectively. The dashed arrows represent the promotion of early flowering, whereas the dashed horizontal lines indicate the inhibition of early flowering.
Despite considerable advances in research on early flowering in cucumber, significant challenges persist due to the inherent complexity of this polygenic trait. The genetic architecture of early flowering involves intricate interactions among multiple genes and their dynamic interplay with environmental factors. This makes both phenotypic stability and gene identification particularly challenging. Environmental variability further complicates research outcomes by influencing trait expression. Moreover, technical limitations in genetic transformation and functional validation, including background-dependent effects in transgenic studies, hinder the achievement of consistent results. Translating laboratory findings into practical applications involves additional obstacles, particularly in implementing molecular breeding techniques and ensuring trait stability under commercial production conditions. Addressing these multifaceted challenges may require continued innovation in research methodologies and closer integration between molecular studies and breeding practices to advance both the fundamental understanding and agricultural applications of early flowering traits in cucumbers.
Currently, the identification of cucumber early flowering genes and research on molecular mechanisms in cucumbers mainly depend on transcriptomics and functional genomics, with relatively few applications of technologies such as proteomics, cytomics, and metabolomics. Therefore, future studies should focus on enriching the resources of early flowering cucumber varieties, actively conducting research on genetic laws, and fully using modern molecular biology techniques (e.g., gene editing and molecular marker-assisted selection breeding) to explore the genes related to early flowering in cucumber, thereby providing abundant genetic resources for the cultivation of early flowering cucumber varieties. Technologies such as genomics, transcriptomics, proteomics, metabolomics, and cytomics should be used to elucidate the mechanisms of early flowering in cucumbers. This would provide a theoretical basis for improving cucumber yield and quality, meeting market demand, and cultivating new early flowering varieties.
Author Contributions
Funding acquisition, M.Z.; investigation, M.Z.; project administration, H.L.; supervision, M.M. and M.J.; visualization, M.M.; writing—original draft, M.Z.; writing—review and editing, H.L. and M.J. All authors have read and agreed to the published version of the manuscript.
Funding
The research was funded by The Doctoral Initiating Fund Project of Jilin Agricultural Science and Technology University, grant number No. (2022)716.
Data Availability Statement
All the data used in this review paper are available online.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Abbreviations
The following abbreviations are used in this manuscript:
| ABA | abscisic acid |
| BCAAs | branched-chain amino acids |
| BCATs | branched-chain amino acids and transferases |
| BiFC | bimolecular fluorescence complementation |
| BR | brassinosteroids |
| GAs | gibberellins |
| JA | jasmonic acid |
| LD | long-day |
| PAC | paclobutrazol |
| QTL | quantitative trait locus |
| RIL | recombinant inbred line |
| RNA-seq | RNA sequencing |
| SA | salicylic acid |
| SD | short-day |
| Y2H | yeast two-hybrid |
References
- Li, Y.H.; Wen, C.L.; Weng, Y.Q. Fine mapping of the pleiotropic locus B for black spine and orange mature fruit color in cucumber identifies a 50 kb region containing a R2R3-MYB transcription factor. Theor. Appl. Genet. 2013, 126, 2187–2196. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.L.; Liu, P.; Jin, Z.S.; Sun, J.; Weng, Y.Q.; Chen, P.; Du, S.L.; Wei, A.M.; Li, Y.H. A mutation in CsHY2 encoding a phytochromobilin (PΦB) synthase leads to an elongated hypocotyl 1 (elh1) phenotype in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2021, 134, 2639–2652. [Google Scholar] [CrossRef]
- Lv, J.; Qi, J.; Shi, Q.; Shen, D.; Zhang, S.; Shao, G.; Li, H.; Sun, Z.; Weng, Y.; Shang, Y.; et al. Genetic diversity and population structure of cucumber (Cucumis sativus L.). PLoS ONE 2012, 7, e46919. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Xu, Q.; Hong, Z.; Wang, X.; Zeng, K.; Yan, L.; Liu, Y.; Zhu, Z.; Wang, H.; Xu, Y. Modified photoperiod response of CsFT promotes day neutrality and early flowering in cultivated cucumber. Theor. Appl. Genet. 2022, 135, 2735–2746. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Liu, X.; Shen, D.; Miao, H.; Xie, B.; Li, X.; Zeng, P.; Wang, S.; Shang, Y.; Gu, X.; et al. A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat. Genet. 2013, 45, 1510–1515. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Deng, Q.; Xu, S.; Huang, Y.; Wei, D.; Wang, Z.; Zhang, H.; Wang, H.; Tang, Q. CsSPL13A directly binds and positively regulates CsFT and CsBAM to accelerate flowering in cucumber. Plant Physiol. Biochem. 2024, 207, 108395. [Google Scholar] [CrossRef]
- Robbins, M.D.; Staub, J.E. Comparative analysis of marker-assisted and phenotypic selection for yield components in cucumber. Theor. Appl. Genet. 2009, 119, 621–634. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, D.L. Preliminary report on the combination ability of early cucumber yield. J. Hebei Agric. Univ. 1991, 14, 84–88. [Google Scholar]
- Gu, X.F.; Fang, X.J.; Zhang, T.M.; Dong, X.H. Analysis of early yield coordination ability of cucumber in protected areas. North. Hortic. 2001, 5, 1–2. [Google Scholar]
- Miao, H.; Gu, X.F.; Zhang, S.P.; Zhang, Z.H.; Huang, S.W.; Wang, Y.; Fang, Z.Y. Mapping QTLs for Seedling-associated Traits in Cucumber. Acta Hortic. Sinica 2012, 39, 879–887. [Google Scholar]
- Soyk, S.; Müller, N.A.; Park, S.J.; Schmalenbach, I.; Jiang, K.; Hayama, R.; Zhang, L.; Van Eck, J.; Jiménez-Gómez, J.M.; Lippman, Z.B. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 2017, 49, 162–168. [Google Scholar] [CrossRef]
- Zhang, Y.Z. Genetic Analysis and QTL Mapping of Cucumber Flowering Traits Under Low Light Conditions. Master’s Thesis, Heilongjiang Bayi Agricultural University, Daqing, China, 1 June 2024. [Google Scholar]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Genet. 2012, 13, 627–639. [Google Scholar]
- Blümel, M.; Dally, N.; Jung, C. Flowering time regulation in crops—What did we learn from Arabidopsis? Curr. Opin. Biotechnol. 2015, 32, 121–129. [Google Scholar] [PubMed]
- Eshed, Y.; Lippman, Z.B. Revolutions in agriculture chart a course for targeted breeding of old and new crops. Science 2019, 366, eaax0025. [Google Scholar] [PubMed]
- Zhang, D.; Ai, G.; Ji, K.; Huang, R.; Chen, C.; Yang, Z.; Wang, J.; Cui, L.; Li, G.; Tahira, M.; et al. EARLY FLOWERING is a dominant gain-of-function allele of FANTASTIC FOUR 1/2c that promotes early flowering in tomato. Plant Biotechnol. J. 2024, 22, 698–711. [Google Scholar] [CrossRef]
- Albani, M.C.; Coupland, G. Comparative Analysis of Flowering in Annual and Perennial Plants. In Current Topics in Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2010; Volume 91, pp. 323–348. [Google Scholar]
- Kim, D.H.; Doyle, M.R.; Sung, S.; Amasino, R.M. Vernalization: Winter and the Timing of Flowering in Plants. Annu. Rev. Cell Dev. Biol. 2009, 25, 277–299. [Google Scholar] [CrossRef]
- Samach, A.; Wigge, P.A. Ambient temperature perception in plants. Curr. Opin. Plant Biol. 2005, 8, 483–486. [Google Scholar]
- Wang, J.W. Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 2014, 65, 4723–4730. [Google Scholar]
- Zhang, X.; Feng, Q.; Miao, J.; Zhu, J.; Zhou, C.; Fan, D.; Lu, Y.; Tian, Q.; Wang, Y.; Zhan, Q.; et al. The WD40 domain-containing protein Ehd5 positively regulates flowering in rice (Oryza sativa). Plant Cell 2023, 35, 4002–4019. [Google Scholar] [CrossRef]
- Wittern, L.; Steed, G.; Taylor, L.J.; Ramirez, D.C.; Pingarron-Cardenas, G.; Gardner, K.; Greenland, A.; Hannah, M.A.; Webb, A.A.R. Wheat EARLY FLOWERING 3 affects heading date without disrupting circadian oscillations. Plant Physiol. 2023, 191, 1383–1403. [Google Scholar] [PubMed]
- Wang, S.; Feng, D.; Zheng, Y.; Lu, Y.; Shi, K.; Yang, R.; Ma, W.; Li, N.; Liu, M.; Wang, Y.; et al. EARLY FLOWERING 3 alleles affect the temperature responsiveness of the circadian clock in Chinese cabbage. Plant Physiol. 2024, 197, kiae505. [Google Scholar]
- Odipio, J.; Getu, B.; Chauhan, R.D.; Alicai, T.; Bart, R.; Nusinow, D.A.; Taylor, N.J. Transgenic overexpression of endogenous FLOWERING LOCUS T-like gene MeFT1 produces early flowering in cassava. PLoS ONE 2020, 15, e0227199. [Google Scholar]
- Xu, P.; Lian, H.; Xu, F.; Zhang, T.; Wang, S.; Wang, W.; Du, S.; Huang, J.; Yang, H.Q. Phytochrome B and AGB1 Coordinately Regulate Photomorphogenesis by Antagonistically Modulating PIF3 Stability in Arabidopsis. Mol. Plant 2019, 12, 229–247. [Google Scholar]
- Yadav, A.; Singh, D.; Lingwan, M.; Yadukrishnan, P.; Masakapalli, S.K.; Datta, S. Light signaling and UV-B mediated plant growth regulation. J. Integr. Plant Biol. 2020, 62, 1270–1292. [Google Scholar]
- Tian, Z.; Jahn, M.; Qin, X.; Obel, H.O.; Yang, F.; Li, J.; Chen, J. Genetic and Transcriptomic Analysis Reveal the Molecular Basis of Photoperiod-Regulated Flowering in Xishuangbanna Cucumber (Cucumis sativus L. var. xishuangbannesis Qi et Yuan). Genes 2021, 12, 1064. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Koo, D.H.; Li, Y.; Zhang, X.; Luan, F.; Havey, M.J.; Jiang, J.; Weng, Y. Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J. 2012, 71, 895–906. [Google Scholar] [PubMed]
- Sheng, Y.; Pan, Y.; Li, Y.; Yang, L.; Weng, Y. Quantitative trait loci for fruit size and flowering time-related traits under domestication and diversifying selection in cucumber (Cucumis sativus). Plant Breed. 2020, 139, 176–191. [Google Scholar]
- Oh, J.; Park, E.; Song, K.; Bae, G.; Choi, G. PHYTOCHROME INTERACTING FACTOR8 Inhibits Phytochrome A-Mediated Far-Red Light Responses in Arabidopsis. Plant Cell 2019, 32, 186–205. [Google Scholar] [CrossRef] [PubMed]
- Kahle, N.; Sheerin, D.; Fischbach, P.; Koch, L.; Schwenk, P.; Lambert, D.; Rodriguez, R.; Kerner, K.; Hoecker, U.; Zurbriggen, M. COLD REGULATED 27 and 28 are targets of CONSTITUTIVELY PHOTOMORPHOGENIC 1 and negatively affect phytochrome B signalling. Plant J. 2020, 104, 1038–1053. [Google Scholar] [CrossRef]
- Miao, T.; Li, D.; Huang, Z.; Huang, Y.; Li, S.; Wang, Y. Gibberellin regulates UV-B-induced hypocotyl growth inhibition in Arabidopsis thaliana. Plant Signal. Behav. 2021, 16, 1966587. [Google Scholar]
- Miao, L.; Zhao, J.; Yang, G.; Xu, P.; Cao, X.; Du, S.; Xu, F.; Jiang, L.; Zhang, S.; Wei, X.; et al. Arabidopsis cryptochrome 1 undergoes COP1 and LRBs-dependent degradation in response to high blue light. New Phytol. 2021, 234, 1347–1362. [Google Scholar] [PubMed]
- Xu, P.; Chen, H.; Li, T.; Xu, F.; Mao, Z.; Cao, X.; Miao, L.; Du, S.; Hua, J.; Zhao, J.; et al. Blue light-dependent interactions of CRY1 with GID1 and DELLA proteins regulate gibberellin signaling and photomorphogenesis in Arabidopsis. Plant Cell 2021, 33, 2375–2394. [Google Scholar]
- Yan, B.; Yang, Z.; He, G.; Jing, Y.; Dong, H.; Ju, L.; Zhang, Y.; Zhu, Y.; Zhou, Y.; Sun, J. The blue light receptor CRY1 interacts with GID1 and DELLA proteins to repress gibberellin signaling and plant growth. Plant Commun. 2021, 2, 100245. [Google Scholar]
- Ichiro, I.S.; Eirini, K.; Xiang, Z.; Thomas, W.; Atsushi, T.; Masaki, O.; Hirotaka, T.; Motoaki, S.; Kazuo, S.; Yaeta, E.; et al. CIPK23 regulates blue light-dependent stomatal opening in Arabidopsis thaliana. Plant J. 2020, 104, 679–692. [Google Scholar]
- Rusaczonek, A.; Czarnocka, W.; Willems, P.; Sujkowska-Rybkowska, M.; Van Breusegem, F.; Karpiński, S. Phototropin 1 and 2 Influence Photosynthesis, UV-C Induced Photooxidative Stress Responses, and Cell Death. Cells 2021, 10, 200. [Google Scholar] [CrossRef]
- Min, G.X.; Xin, H.; Jun, Z.; Mei, H.Q.; Yuan, G.; Qi, L.Z.; Sha, L.; Min, H.J. UV RESISTANCE LOCUS8 mediates ultraviolet-B-induced stomatal closure in an ethylene-dependent manner. Plant Sci. 2020, 301, 110679. [Google Scholar]
- Hajdu, A.; Ádám, É.; Sheerin, D.J.; Dobos, O.; Bernula, P.; Hiltbrunner, A.; Kozma-Bognár, L.; Nagy, F. High-level expression and phosphorylation of phytochrome B modulates flowering time in Arabidopsis. Plant J. 2015, 83, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Fragoso, V.; Oh, Y.; Kim, S.G.; Gase, K.; Baldwin, I.T. Functional specialization of Nicotiana attenuata phytochromes in leaf development and flowering time. J. Integr. Plant Biol. 2017, 59, 205–224. [Google Scholar] [PubMed]
- Zou, Y.; Li, R.; Baldwin, I.T. ZEITLUPE is required for shade avoidance in the wild tobacco Nicotiana attenuata. J. Integr. Plant Biol. 2019, 62, 1341–1351. [Google Scholar]
- Liu, B.; Weng, J.; Guan, D.; Zhang, Y.; Niu, Q.; López-Juez, E.; Lai, Y.; Garcia-Mas, J.; Huang, D. A domestication-associated gene, CsLH, encodes a phytochrome B protein that regulates hypocotyl elongation in cucumber. Mol. Hortic. 2021, 1, 3. [Google Scholar]
- Liu, S.; Yang, L.; Li, J.; Tang, W.; Li, J.; Lin, R. FHY3 interacts with phytochrome B and regulates seed dormancy and germination. Plant Physiol. 2021, 187, 289–302. [Google Scholar] [PubMed]
- Wollenberg, A.C.; Strasser, B.; Cerdán, P.D.; Amasino, R.M. Acceleration of flowering during shade avoidance in Arabidopsis alters the balance between FLOWERING LOCUS C-mediated repression and photoperiodic induction of flowering. Plant Physiol. 2008, 148, 1681–1694. [Google Scholar] [PubMed]
- Iñigo, S.; Alvarez, M.J.; Strasser, B.; Califano, A.; Cerdán, P.D. PFT1, the MED25 subunit of the plant Mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. Plant J. 2012, 69, 601–612. [Google Scholar]
- Yasui, Y.; Mukougawa, K.; Uemoto, M.; Yokofuji, A.; Suzuri, R.; Nishitani, A.; Kohchi, T. The phytochrome-interacting vascular plant one-zinc finger1 and VOZ2 redundantly regulate flowering in Arabidopsis. Plant Cell 2012, 24, 3248–3263. [Google Scholar]
- Lazaro, A.; Mouriz, A.; Piñeiro, M.; Jarillo, J.A. Red Light-Mediated Degradation of CONSTANS by the E3 Ubiquitin Ligase HOS1 Regulates Photoperiodic Flowering in Arabidopsis. Plant Cell 2015, 27, 2437–2454. [Google Scholar]
- Hu, L.; Zhang, M.; Shang, J.; Liu, Z.; Weng, Y.; Yue, H.; Li, Y.; Chen, P. A 5.5-kb LTR-retrotransposon insertion inside phytochrome B gene (CsPHYB) results in long hypocotyl and early flowering in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2023, 136, 68. [Google Scholar] [PubMed]
- Sysoeva, M.I.; Marovskaia, E.F. Role of phytochrome B in organ formation processes in Cucumis sativus L. Russ. J. Dev. Biol. 2013, 44, 135–138. [Google Scholar]
- Halliday, K.J.; Koornneef, M.; Whitelam, G.C. Phytochrome B and at Least One Other Phytochrome Mediate the Accelerated Flowering Response of Arabidopsis thaliana L. to Low Red/Far-Red Ratio. Plant Physiol. 1994, 104, 1311–1315. [Google Scholar] [PubMed]
- Szymanski, D.B.; Jilk, R.A.; Pollock, S.M.; Marks, M.D. Control of GL2 expression in Arabidopsis leaves and trichomes. Development 1998, 125, 1161–1171. [Google Scholar]
- Vernoud, V.; Laigle, G.; Rozier, F.; Meeley, R.B.; Perez, P.; Rogowsky, P.M. The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J. 2009, 59, 883–894. [Google Scholar]
- Shi, L.; Katavic, V.; Yuanyuan, Y.; Ljerka, K.; George, H. Arabidopsis glabra2 mutant seeds deficient in mucilage biosynthesis produce more oil. Plant J. 2012, 69, 37–46. [Google Scholar]
- Fu, R.; Liu, W.; Li, Q.; Li, J.; Wang, L.; Ren, Z. Comprehensive analysis of the homeodomain-leucine zipper IV transcription factor family in Cucumis sativus. Genome 2013, 56, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.J.; Foster, K.R.; Morgan, P.W. Effect of Gibberellin Biosynthesis Inhibitors on Native Gibberellin Content, Growth and Floral Initiation in Sorghum bicolor. J. Plant Growth Regul. 1998, 17, 185–195. [Google Scholar] [PubMed]
- Lee, I.J.; Foster, K.R.; Morgan, P.W. Photoperiod Control of Gibberellin Levels and Flowering in Sorghum. Plant Physiol. 1998, 116, 1003–1011. [Google Scholar]
- Blázquez, M.A.; Weigel, D. Independent Regulation of Flowering by Phytochrome B and Gibberellins in Arabidopsis. Plant Physiol. 1999, 120, 1025–1032. [Google Scholar] [PubMed]
- Yu, H.; Ito, T.; Zhao, Y.; Peng, J.; Kumar, P.; Meyerowitz, E.M. Floral homeotic genes are targets of gibberellin signaling in flower development. Proc. Natl. Acad. Sci. USA 2004, 101, 7827–7832. [Google Scholar]
- Endo, M.; Nakamura, S.; Araki, T.; Mochizuki, N.; Nagatani, A. Phytochrome B in the Mesophyll Delays Flowering by Suppressing FLOWERING LOCUS T Expression in Arabidopsis Vascular Bundles. Plant Cell 2005, 17, 1941–1952. [Google Scholar]
- Fukazawa, J.; Ohashi, Y.; Takahashi, R.; Nakai, K.; Takahashi, Y. DELLA degradation by gibberellin promotes flowering via GAF1-TPR-dependent repression of floral repressors in Arabidopsis. Plant Cell 2021, 33, 2258–2272. [Google Scholar]
- Chini, A.; Gimenez-Ibanez, S.; Goossens, A.; Solano, R. Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 2016, 33, 147–156. [Google Scholar]
- Chung, H.S.; Koo, A.J.K.; Gao, X.; Jayanty, S.; Thines, B.; Jones, A.D.; Howe, G.A. Regulation and Function of Arabidopsis JASMONATE ZIM-Domain Genes in Response to Wounding and Herbivory. Plant Physiol. 2008, 146, 952–964. [Google Scholar]
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.-M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH Transcription Factors MYC3 and MYC4 Are Targets of JAZ Repressors and Act Additively with MYC2 in the Activation of Jasmonate Responses(C)(W). Plant Cell 2011, 23, 701–715. [Google Scholar] [PubMed]
- Schweizer, F.; Fernández-Calvo, P.; Zander, M.; Diez-Diaz, M.; Fonseca, S.; Glauser, G.; Lewsey, M.G.; Ecker, J.R.; Solano, R.; Reymond, P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 2013, 25, 3117–3132. [Google Scholar] [PubMed]
- Qi, T.; Huang, H.; Song, S.; Xie, D. Regulation of Jasmonate-Mediated Stamen Development and Seed Production by a bHLH-MYB Complex in Arabidopsis. Plant Cell 2015, 27, 1620–1633. [Google Scholar] [CrossRef]
- Wang, H.; Li, Y.; Pan, J.; Lou, D.; Hu, Y.; Yu, D. The bHLH Transcription Factors MYC2, MYC3, and MYC4 Are Required for Jasmonate-Mediated Inhibition of Flowering in Arabidopsis. Mol. Plant 2017, 10, 1461–1464. [Google Scholar]
- Cai, Y.; Bartholomew, E.S.; Dong, M.; Zhai, X.; Yin, S.; Zhang, Y.; Feng, Z.; Wu, L.; Liu, W.; Shan, N.; et al. The HD-ZIP IV transcription factor CsGL2-LIKE regulates male flowering time and fertility in cucumber. J. Exp. Bot. 2020, 71, 5425–5437. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Zapater, J.; Coupland, G.; Dean, C.; Koornneef, M. The transition to flowering in Arabidopsis. In Cold Spring Harbor Monograph Archive; Cold Spring Harbor Laboratory: New York, NY, USA, 1994; Volume 27, pp. 403–433. [Google Scholar]
- Yong, H.R.; Lee, S.Y.; Kim, K.S. ABA synthesis inhibitor inducing flowering of dormant peony without bud abortion. Acta Hort. 2020, 1291, 97–102. [Google Scholar]
- Hurr, B.M.; Huber, D.J.; Vallejos, C.E.; Talcott, S.T. Developmentally dependent responses of detached cucumber (Cucumis sativus L.) fruit to exogenous ethylene. Postharvest Biol. Technol. 2008, 52, 207–215. [Google Scholar]
- Wang, Y.; Wang, Y.; Ji, K.; Dai, S.; Hu, Y.; Sun, L.; Li, Q.; Chen, P.; Sun, Y.; Duan, C. The role of abscisic acid in regulating cucumber fruit development and ripening and its transcriptional regulation. Plant Physiol. Biochem. 2013, 64, 70–79. [Google Scholar] [PubMed]
- Cheng, Z.; Zhuo, S.; Liu, X.; Che, G.; Wang, Z.; Gu, R.; Shen, J.; Song, W.; Zhou, Z.; Han, D.; et al. The MADS-Box Gene CsSHP Participates in Fruit Maturation and Floral Organ Development in Cucumber. Front. Plant Sci. 2019, 10, 1781. [Google Scholar] [CrossRef]
- Pal, Y.N.; Vijay, B.; Gyanendra, S.; Vikram, S.N. Influence of Foliar Spray of Brassinosteroids (BR), Salicylic Acid (SA) and Gibberellic Acid (GA3) on Vegetative Growth and Flowering Parameters of Cucumber (Cucumis sativus L) cv. Arpit. Int. J. Environ. Clim. Change 2022, 12, 607–615. [Google Scholar]
- Hedhly, A.; Hormaza, J.I.; Herrero, M. Global warming and sexual plant reproduction. Trends Plant Sci. 2008, 14, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Thiruvengadam, M.; Chung, I. Optimization of factors influencing in vitro flowering of gherkin (Cucumis anguria L.). Acta Biol. Hung. 2014, 65, 72–84. [Google Scholar] [CrossRef]
- Sun, Y.L.; Zang, C.J.; Yao, T.T.; LI, X.L.; Yue, L.X. Research Advances on Heat Tolerance of Cucumber in China. China Fruit Veg. 2018, 38, 57–62. [Google Scholar]
- Prasad, P.H.; Mandal, A.R.; Sarkar, A.; Thapa, U.; Maity, T.K. Effect of bio-fertilizers and nitrogen on growth and yield attributes of bitter gourd (Momordica charantia L.). In Proceedings of the International Conference on Horticulture (ICH-2009), Bangalore, India, 9 December 2009. [Google Scholar]
- Anjanappa, M.; Kumara, B.S.; Indiresh, K.M. Growth, yield and quality attributes of cucumber (Cv. Hassan local) as influenced by integrated nutrient management grown under protected condition. Agric. Food Sci. 2012, 39, 47–50. [Google Scholar]
- Sahu, P.; Tripathy, P.; Sahu, G.S.; Dash, S.K.; Pattnayak, S.K.; Sarkar, S.; Nayak, R.K.; Nayak, N.J.; Mishra, S. Influence of Nutrient Management Practices on Growth, Flowering and Yield Attributes of Cucumber (Cucumis sativus). Int. J. Environ. Clim. Change 2022, 12, 493–503. [Google Scholar] [CrossRef]
- Zhang, S.P.; Miao, H.; Bo, K.L.; Dong, S.Y.; Gu, X.F. Research Progress on Cucumber Genetic Breeding During ‘The Thirteenth Five-year Plan’ in China. China Veg. 2021, 4, 16–26. [Google Scholar]
- Zhang, G.C.; Tong, W.J. “Jinchun No.4” Cucumber. Vegetable 2011, 4, 36–37. [Google Scholar]
- Cui, M.H. Zhongnong No.5 Cucumber. Jilin Veg. 2000, 4, 40. [Google Scholar]
- Liu, M.; Hsieh, C.; Chao, Y. Kaohsiung No. 3 Cucumber: An Early Flowering Variety Tolerant to Heat and Moisture. Hort. Sci. 2017, 52, 1435–1437. [Google Scholar] [CrossRef]
- Fang, X.J.; Wu, W.R.; Tang, J.L. Molecular Marker Assisted Selection; Science Press: Beijing, China, 2001; pp. 1–84. [Google Scholar]
- Qu, M.L. Analysis of Flower-Related Traits in Cucumber (Cucumis sativus L.). Master’s Thesis, Shanghai Jiao Tong University, Shanghai, China, 1 January 2016. [Google Scholar]
- Lu, H.; Lin, T.; Klein, J.; Wang, S.; Qi, J.; Zhou, Q.; Sun, J.; Zhang, Z.; Weng, Y.; Huang, S. QTL-seq identifies an early flowering QTL located near Flowering Locus T in cucumber. Theor. Appl. Genet. 2014, 127, 1491–1499. [Google Scholar] [CrossRef]
- Dijkhuizen, A.; Staub, J.E. QTL Conditioning Yield and Fruit Quality Traits in Cucumber (Cucumis sativus L.). J. New Seeds 2002, 4, 1–30. [Google Scholar]
- Fazio, G.; Staub, J.E.; Stevens, M.R. Genetic mapping and QTL analysis of horticultural traits in cucumber (Cucumis sativus L.) using recombinant inbred lines. Theor. Appl. Genet. 2003, 107, 864–874. [Google Scholar]
- Pan, J.S.; Wang, G.; Li, X.Z.; He, H.L.; Wu, A.Z.; Cai, R. Construction of a genetic map with SRAP markers and localization of the gene responsible for the first-flower-node trait in cucumber (Cucumis sativus L.). Prog. Nat. Sci. 2005, 15, 407–413. [Google Scholar]
- Bo, K.; Ma, Z.; Chen, J.; Weng, Y. Molecular mapping reveals structural rearrangements and quantitative trait loci underlying traits with local adaptation in semi-wild Xishuangbanna cucumber (Cucumis sativus L. var. xishuangbannanesis Qi et Yuan). Theor. Appl. Genet. 2015, 128, 25–39. [Google Scholar]
- Pan, Y.; Qu, S.; Bo, K.; Gao, M.; Haider, K.R.; Weng, Y. QTL mapping of domestication and diversifying selection related traits in round-fruited semi-wild Xishuangbanna cucumber (Cucumis sativus L. var. xishuangbannanesis). Theor. Appl. Genet. 2017, 130, 1531–1548. [Google Scholar]
- Pan, Y.; Wen, C.; Han, Y.; Wang, Y.; Li, Y.; Li, X.; Cheng, X.; Weng, Y. QTL for horticulturally important traits associated with pleiotropic andromonoecy and carpel number loci, and a paracentric inversion in cucumber. Theor. Appl. Genet. 2020, 133, 2271–2290. [Google Scholar]
- Wang, S.; Li, H.; Li, Y.; Li, Z.; Qi, J.; Lin, T.; Yang, X.; Zhang, Z.; Huang, S. FLOWERING LOCUS T Improves Cucumber Adaptation to Higher Latitudes. Plant Physiol. 2020, 182, 908–918. [Google Scholar] [PubMed]
- Nishioka, M.; Tamura, K.; Hayashi, M.; Fujimori, Y.; Ohkawa, Y.; Kuginuki, Y.; Harada, K. Mapping of QTLs for Bolting Time in Brassica rapa (syn. campestris) under Different Environmental Conditions. Breed. Sci. 2005, 55, 127–133. [Google Scholar]
- Kazan, K.; Lyons, R. The link between flowering time and stress tolerance. J. Exp. Bot. 2016, 67, 47–60. [Google Scholar]
- Fornara, F.; de Montaigu, A.; Coupland, G. SnapShot: Control of Flowering in Arabidopsis. Cell 2010, 141, 550–550.e2. [Google Scholar]
- Anusha, S.; Markus, S. Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar]
- Charles, W.; Caroline, D. The FLC Locus: A Platform for Discoveries in Epigenetics and Adaptation. Annu. Rev. Cell Dev. Biol. 2017, 33, 555–575. [Google Scholar]
- Simpson, G.G.; Dean, C. Arabidopsis, the Rosetta Stone of Flowering Time? Science 2002, 296, 285–289. [Google Scholar] [PubMed]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP Protein Mediating Signals from the Floral Pathway Integrator FT at the Shoot Apex. Science 2005, 309, 1052–1056. [Google Scholar]
- Liu, X.; Yang, Y.; Hu, Y.; Zhou, L.; Li, Y.; Hou, X. Temporal-Specific Interaction of NF-YC and CURLY LEAF during the Floral Transition Regulates Flowering. Plant Physiol. 2018, 177, 105–114. [Google Scholar]
- Yamaguchi, A.; Wu, M.F.; Yang, L.; Wu, G.; Poethig, R.S.; Wagner, D. The MicroRNA-Regulated SBP-Box Transcription Factor SPL3 Is a Direct Upstream Activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 2009, 17, 268–278. [Google Scholar]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996, 250, 7–16. [Google Scholar]
- Wang, S.; Yang, A.; Wang, H.; Xu, Y.M. Identification and expression analysis of miR156/157-SPL pathway genes in cucumber. Acta Hortic. 2021, 48, 2227–2238. [Google Scholar]
- Hong, Z.; Wang, X.; Fan, Z.; Wang, J.; Yang, A.; Yan, G.; He, Y.; Wang, H.; Zhu, Z.; Xu, X. The intrinsic developmental age signal defines an age-dependent climbing behavior in cucumber. Hortic. Plant J. 2024, 10, 797–808. [Google Scholar]
- Schmid, M.; Uhlenhaut, N.H.; Godard, F.; Demar, M.; Bressan, R.; Weigel, D.; Lohmann, J.U. Dissection of floral induction pathways using global expression analysis. Development 2003, 130, 6001–6012. [Google Scholar]
- Lee, J.; Oh, M.; Park, H.; Lee, I. SOC1 translocated to the nucleus by interaction with AGL24 directly regulates LEAFY. Plant J. 2008, 55, 832–843. [Google Scholar] [CrossRef] [PubMed]
- Preston, J.C.; Hileman, L.C. SQUAMOSA-PROMOTER BINDING PROTEIN 1 initiates flowering in Antirrhinum majus through the activation of meristem identity genes. Plant J. 2010, 62, 704–712. [Google Scholar] [CrossRef]
- Lal, S.; Pacis, L.B.; Smith, H.M.S. Regulation of the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE genes/microRNA156 Module by the Homeodomain Proteins PENNYWISE and POUND-FOOLISH in Arabidopsis. Mol. Plant 2011, 4, 1123–1132. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Ju, Y.; Seo, P.J.; Lee, J.H.; Park, C.M. The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 2012, 69, 577–588. [Google Scholar] [CrossRef]
- Teotia, S.; Tang, G. To Bloom or Not to Bloom: Role of MicroRNAs in Plant Flowering. Mol. Plant 2015, 8, 359–377. [Google Scholar] [CrossRef] [PubMed]
- Wickland, D.P.; Hanzawa, Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 Gene Family: Functional Evolution and Molecular Mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar]
- Sato, H.; Heang, D.; Sassa, H.; Koba, T. Identification and characterization of FT/TFL1 gene family in cucumber. Breed. Sci. 2009, 59, 3–11. [Google Scholar]
- Adeyemo, O.S.; Hyde, P.T.; Setter, T.L. Identification of FT family genes that respond to photoperiod, temperature and genotype in relation to flowering in cassava (Manihot esculenta, Crantz). Plant Reprod. 2019, 32, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Li, F.; Deng, Q.; Zhang, S.; Zhou, Q.; Chen, F.; Liu, B.; Bao, M.; Liu, G. Identification and Characterization of the FLOWERING LOCUS T/TERMINAL FLOWER 1 Gene Family in Petunia. DNA Cell Biol. 2019, 38, 982–995. [Google Scholar] [CrossRef]
- Yoshida, A.; Taoka, K.I.; Hosaka, A.; Tanaka, K.; Kobayashi, H.; Muranaka, T.; Toyooka, K.; Oyama, T.; Tsuji, H. Characterization of Frond and Flower Development and Identification of FT and FD Genes From Duckweed Lemna aequinoctialis Nd. Front. Plant Sci. 2021, 12, 697206. [Google Scholar] [CrossRef]
- Ratcliffe, O.J.; Bradley, D.J.; Coen, E.S. Separation of shoot and floral identity in Arabidopsis. Development 1999, 126, 1109–1120. [Google Scholar] [CrossRef]
- Shannon, S.; Meeks-Wagner, D.R. A Mutation in the Arabidopsis TFL1 Gene Affects Inflorescence Meristem Development. Plant Cell 1991, 3, 877–892. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, S.; Chai, S.; Yang, Z.; Zhang, Q.; Xin, H.; Xu, Y.; Lin, S.; Chen, X.; Yao, Z.; et al. Graph-based pan-genome reveals structural and sequence variations related to agronomic traits and domestication in cucumber. Nat. Commun. 2022, 13, 682. [Google Scholar] [CrossRef] [PubMed]
- Song, S.S.; Hao, Q.; Su, L.H.; Xia, S.W.; Zhang, R.J.; Liu, Y.J.; Li, Y.; Zhu, Y.Y.; Luo, Q.Y.; Lai, Y.S. FLOWERING LOCUS T (FT) gene regulates short-day flowering in low latitude Xishuangbanna cucumber (Cucumis sativus var. xishuangbannanesis). Veg. Res. 2023, 3, 15. [Google Scholar]
- Zhang, J.; Yan, S.S.; Zhao, W.S.; Zhang, X.L. Cloning and Functional Analysis of Cucumber CsFT Gene. Hortic. Plant J. 2013, 40, 2180–2188. [Google Scholar]
- Smaczniak, C.; Immink, R.G.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.S.; Yu, C.; Fan, P.P.; Bao, B.F.; Li, T.; Zhu, Z. Identification of Two Cucumber Putative Silicon Transporter Genes in Cucumis sativus. J. Plant Growth Regul. 2015, 34, 332–338. [Google Scholar] [CrossRef]
- Kater, M.M.; Colombo, L.; Franken, J.; Busscher, M.; Masiero, S.; Campagne, M.M.V.L.; Angenent, G.C. Multiple AGAMOUS homologs from cucumber and petunia differ in their ability to induce reproductive organ fate. Plant Cell 1998, 10, 171–182. [Google Scholar] [CrossRef]
- Kater, M.M.; Franken, J.; Carney, K.J.; Colombo, L.; Angenent, G.C. Sex determination in the monoecious species cucumber is confined to specific floral whorls. Plant Cell 2001, 13, 481–493. [Google Scholar] [CrossRef]
- Sun, J.J.; Li, F.; Wang, D.H.; Liu, X.F.; Li, X.; Liu, N.; Gu, H.T.; Zou, C.; Luo, J.C.; He, C.X.; et al. CsAP3: A Cucumber Homolog to Arabidopsis APETALA3 with Novel Characteristics. Front. Plant Sci. 2016, 7, 1181. [Google Scholar] [CrossRef]
- Wang, X.; Gao, D.; Sun, J.; Liu, M.; Lun, Y.; Zheng, J.; Wang, S.; Cui, Q.; Wang, X.; Huang, S. An exon skipping in a SEPALLATA-Like gene is associated with perturbed floral and fruits development in cucumber. J. Integr. Plant Biol. 2016, 58, 766–771. [Google Scholar]
- Hu, L.; Liu, S. Genome-wide analysis of the MADS-box gene family in cucumber. Genome 2012, 55, 245–256. [Google Scholar]
- Zhou, Y.; Hu, L.; Song, J.; Jiang, L.; Liu, S. Isolation and characterization of a MADS-box gene in cucumber (Cucumis sativus L.) that affects flowering time and leaf morphology in transgenic Arabidopsis. Biotechnol. Biotechnol. Equip. 2019, 33, 54–63. [Google Scholar]
- Zhou, Y.; Hu, L.; Ye, S.; Jiang, L.; Liu, S. Overexpression of an APETALA1 -like gene from cucumber (Cucumis sativus L.) induces earlier flowering and abnormal leaf development in transgenic Arabidopsis. Can. J. Plant Sci. 2019, 99, 210–220. [Google Scholar]
- Xu, S.; An, Y.; Wen, M.; Hu, K.; Yang, Y.; Gan, D. Function and expression analysis of cucumber CsMADS08 and its downstream regulatory genes. J. Northwest A F Univ. (Nat. Sci. Ed.) 2022, 50, 115–124. [Google Scholar]
- Thanda, W.K.; Chunying, Z.; Kihwan, S.; Hwan, L.J.; Sanghyeob, L. Development and characterization of a co-dominant molecular marker via sequence analysis of a genomic region containing the Female (F) locus in cucumber (Cucumis sativus L.). Mol. Breed. 2015, 35, 229. [Google Scholar]
- Hwan, L.J.; Young-Cheon, K.; Youjin, J.; Hoon, H.J.; Chunying, Z.; Cheol-Won, Y.; Sanghyeob, L. The overexpression of cucumber (Cucumis sativus L.) genes that encode the branched-chain amino acid transferase modulate flowering time in Arabidopsis thaliana. Plant Cell Rep. 2019, 38, 25–35. [Google Scholar]
- Engelhorn, J.; Moreau, F.; Fletcher, J.C.; Carles, C.C. ULTRAPETALA1 and LEAFY pathways function independently in specifying identity and determinacy at the Arabidopsis floral meristem. Ann. Bot. 2014, 114, 1497–1505. [Google Scholar] [PubMed]
- Hempel, F.D.; Welch, D.R.; Feldman, L.J. Floral induction and determination: Where is flowering controlled? Trends Plant Sci. 2000, 5, 17–21. [Google Scholar]
- Liu, F.Q.; Zhu, G.L.; Luo, D.; Wu, X.Y.; Xu, Z.H. Cloning and analysis of CFL-A LFY-like gene from cucumber. Acta Bot. Sin. 1999, 41, 813–819. [Google Scholar]
- Wang, L.L.; Pang, J.L.; Liang, H.M.; Zhu, M.Y. Expression of CFL gene during differentiation of floral and vegetative buds in cucumber cotyledonary nodes cultured in vitro. J. Plant Physiol. Mol. Biol. 2004, 30, 644–650. [Google Scholar]
- Zhang, M.Z.; Ye, D.; Wang, L.L.; Pang, J.L.; Zhang, Y.H.; Zheng, K.; Bian, H.W.; Han, N.; Pan, J.W.; Wang, J.H.; et al. Overexpression of the cucumber LEAFY homolog CFL and hormone treatments alter flower development in gloxinia (Sinningia speciosa). Plant Mol. Biol. 2008, 67, 419–427. [Google Scholar] [PubMed]
- Abhishek, K.; Anamika, S.; Madhusmita, P.; Kumar, S.P.; Panigrahi. Carbon nanoparticles influence photomorphogenesis and flowering time in Arabidopsis thaliana. Plant Cell Rep. 2018, 37, 901–912. [Google Scholar]
- Valverde, F.; Mouradov, A.; Soppe, W.; Ravenscroft, D.; Samach, A.; Coupland, G. Photoreceptor Regulation of CONSTANS Protein in Photoperiodic Flowering. Science 2004, 303, 1003–1006. [Google Scholar]
- Hun, S.Y.; Sung, S.J.; Kinmonth-Schultz, H.A.; Takato, I. Photoperiodic Flowering: Time Measurement Mechanisms in Leaves. Annu. Rev. Plant Biol. 2015, 66, 441–464. [Google Scholar]
- Bradley, D.; Ratcliffe, O.; Vincent, C.; Carpenter, R.; Coen, E. Inflorescence Commitment and Architecture in Arabidopsis. Science 1997, 275, 80–83. [Google Scholar] [CrossRef]
- Conti, L.; Bradley, D. TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell 2007, 19, 767–778. [Google Scholar]
- Zhao, W.; Gu, R.; Che, G.; Cheng, Z.; Zhang, X. CsTFL1b may regulate the flowering time and inflorescence architecture in cucumber (Cucumis sativus L.). Biochem. Biophys. Res. Commun. 2018, 2, 307–313. [Google Scholar] [CrossRef]
- Zheng, L.; Shu, W.; Qianyi, T.; Junsong, P.; Longting, S.; Zhenhui, G.; Run, C. A putative positive feedback regulation mechanism in CsACS2 expression suggests a modified model for sex determination in cucumber (Cucumis sativus L.). J. Exp. Bot. 2012, 63, 4475–4484. [Google Scholar]
- Pan, J.; Wen, H.; Chen, G.; Lin, W.H.; Du, H.; Chen, Y.; Zhang, L.; Lian, H.; Wang, G.; Cai, R.; et al. A positive feedback loop mediated by CsERF31 initiates female cucumber flower development. Plant Physiol. 2021, 186, 1088–1100. [Google Scholar]
- Huang, S.W.; Li, R.Q.; Zhang, Z.H.; Li, L.; Gu, X.F.; Fan, W.; Lucas, W.J.; Wang, X.W.; Xie, B.Y.; Nie, P.X.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Z.; Yan, P.; Huang, S.; Fei, Z.; Lin, K. RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genom. 2011, 12, 540. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, H.; Huang, W.; Xu, Y.; Zhou, Q.; Wang, S.; Ruan, J.; Huang, S.; Zhang, Z. A chromosome-scale genome assembly of cucumber (Cucumis sativus L.). GigaScience 2019, 8, giz072. [Google Scholar] [CrossRef]
- Guan, J.; Miao, H.; Zhang, Z.; Dong, S.; Zhou, Q.; Liu, X.; Beckles, D.M.; Gu, X.; Huang, S.; Zhang, S. A near-complete cucumber reference genome assembly and Cucumber-DB, a multi-omics database. Mol. Plant 2024, 17, 1178–1182. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Zhu, Z.; Lin, X.; Shen, X.; Yang, T.; Wang, H.; Zhou, X. Comparative Genomic Analysis of PEBP Genes in Cucurbits Explores the Interactors of Cucumber CsPEBPs Related to Flowering Time. Int. J. Mol. Sci. 2024, 25, 3815. [Google Scholar] [CrossRef]
- Wen, C.; Zhao, W.; Liu, W.; Yang, L.; Wang, Y.; Liu, X.; Xu, Y.; Ren, H.; Guo, Y.; Li, C.; et al. CsTFL1 inhibits determinate growth and terminal flower formation through interaction with CsNOT2a in cucumber. Development 2019, 146, dev180166. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).