Roles of WRKY Transcription Factors in Response to Sri Lankan Cassava Mosaic Virus Infection in Susceptible and Tolerant Cassava Cultivars
Abstract
1. Introduction
2. Results
2.1. Identification of WRKYs in SLCMV-Infected KU 50 and R 11 at 21, 32, and 67 dpi
2.2. Functions of Identified WRKYs and Phylogenetic Analysis
2.3. WRKY DEGs at 32 and 67 dpi in KU 50 and R 11
2.4. RT-qPCR Validation
3. Discussion
3.1. Exploring Functions of WRKYs
3.2. WRKY DEGs Determinations
3.3. Interacting Partners of WRKY Proteins
3.4. RT-qPCR Validation
4. Materials and Methods
4.1. Plant Materials, SLCMV Inoculation, and Leaf Sample Collection
4.2. RNA Extraction and cDNA Library Construction
4.3. RNA-Seq and WRKY Identification
4.4. WRKY Functional Annotation
4.5. Phylogenetic Tree Construction
4.6. WRKY Differentially Expressed Genes (DEGs)
4.7. RT-qPCR Validation
4.8. Interacting Partners of WRKYs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saunders, K.; Salim, N.; Mali, V.R.; Malathi, V.G.; Briddon, R.; Markham, P.G.; Stanley, J. Characterisation of Sri Lankan cassava mosaic virus and Indian cassava mosaic virus: Evidence for acquisition of a DNA B component by a monopartite Begomovirus. Virology 2002, 293, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A. ICTV virus taxonomy profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Pan, L.L.; Bouvaine, S.; Fan, Y.Y.; Liu, Y.Q.; Liu, S.S.; Seal, S.; Wang, X.W. Differential transmission of Sri Lankan cassava mosaic virus by three cryptic species of the whitefly Bemisia tabaci complex. Virology 2020, 540, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Saokham, K.; Hemniam, N.; Roekwan, S.; Hunsawattanakul, S.; Thawinampan, J.; Siriwan, W. Survey and molecular detection of Sri Lankan cassava mosaic virus in Thailand. PLoS ONE 2021, 16, e0252846. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Malik, A.I.; Sophearith, S.; Delaquis, E.; Cuellar, W.J.; Jimenez, J.; Newby, J.C. Susceptibility of cassava varieties to disease caused by Sri Lankan cassava mosaic virus and impacts on yield by use of asymptomatic and virus-free planting material. Agronomy 2022, 12, 1658. [Google Scholar] [CrossRef]
- Wang, H.L.; Cui, X.Y.; Wang, X.W.; Liu, S.S.; Zhang, Z.H.; Zhou, X.P. First report of Sri Lankan cassava mosaic virus infecting cassava in Cambodia. Plant Dis. 2016, 100, 1029. [Google Scholar] [CrossRef]
- Kishor, A.N.; Kumar, M.; Tarafdar, J. Occurrence of Sri Lankan cassava mosaic virus (SLCMV) and its characterization in West Bengal, India. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 2887–2893. [Google Scholar] [CrossRef][Green Version]
- Fraile, A.; García-Arenal, F. The coevolution of plants and viruses: Resistance and pathogenicity. Adv. Virus Res. 2010, 76, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, B.; Sanfaçon, H. Symptom recovery in virus-infected plants: Revisiting the role of RNA silencing mechanisms. Virology 2015, 479–480, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Molen, T.A. Host recovery and reduced virus level in the upper leaves after Potato virus Y infection occur in tobacco and tomato but not in potato plants. Viruses 2015, 7, 680–698. [Google Scholar] [CrossRef] [PubMed]
- Bengyella, L.; Waikhom, S.D.; Allie, F.; Rey, C. Virus tolerance and recovery from viral induced-symptoms in plants are associated with transcriptome reprograming. Plant Mol. Biol. 2015, 89, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Hemniam, N.; Saokham, K.; Roekwan, S.; Hunsawattanakul, S.; Thawinampan, J.; Siriwan, W. Severity of cassava mosaic disease in resistance and commercial varieties by grafting. In Proceedings of the 14th National Plant Protection Conference, Bangkok, Thailand, 12–14 November 2019. (In Thai). [Google Scholar]
- Barrera, L.O.; Ren, B. The transcriptional regulatory code of eukaryotic cells—Insights from genome-wide analysis of chromatin organization and transcription factor binding. Curr. Opin. Cell Biol. 2006, 18, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Seo, E.; Choi, D.; Choi. Functional studies of transcription factors involved in plant defenses in the genomics era. Brief. Funct. Genom. 2015, 14, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Meraj, T.A.; Fu, J.; Raza, M.A.; Zhu, C.; Shen, Q.; Xu, D.; Wang, Q. Transcriptional factors regulate plant stress responses through mediating secondary metabolism. Genes 2020, 11, 346. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Ng, D.W.K.; Abeysinghe, J.K.; Kamali, M. Regulating the regulators: The control of transcription factors in plant defense signalling. Int. J. Mol. Sci. 2018, 19, 3737. [Google Scholar] [CrossRef] [PubMed]
- Nicaise, V. Crop immunity against viruses: Outcomes and future challenges. Front. Plant Sci. 2014, 5, 660. [Google Scholar] [CrossRef] [PubMed]
- Ulker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signalling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Shi, H.; Xia, Z.; Tie, W.; Ding, Z.; Yan, Y.; Wang, W.; Hu, W.; Li, K. Genome-wide identification and expression analysis of the WRKY gene family in cassava. Front. Plant Sci. 2016, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Freeborough, W.; Gentle, N.; Rey, M.E.C. WRKY transcription factors in cassava contribute to regulation of tolerance and susceptibility to cassava mosaic disease through stress responses. Viruses 2021, 13, 1820. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.A.; Liu, Y.; Shen, Q.J. The WRKY gene family in rice (Oryza sativa). J. Integr. Plant Biol. 2007, 49, 827–842. [Google Scholar] [CrossRef]
- Wei, K.F.; Chen, J.; Chen, Y.F.; Wu, L.J.; Xie, D.X. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012, 19, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Gao, Y.; Liu, J. Genome-wide analysis of WRKY transcription factors in Solanum Lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Nakano, T.; Miyagawa, N.; Ishihama, N.; Yoshioka, M.; Katou, Y.; Yaeno, T.; Shirasu, K.; Yoshioka, H. WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 2015, 27, 2645–2663. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Chen, C.; Chen, Z. Expression profiles of the Arabidopsis. WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003, 51, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.Y.; Zhang, S. Activation of a mitogen-activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. Plant J. 2004, 38, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.; Brodersen, P.; Naested, H.; Andreasson, E.; Lindhart, U.; Johansen, B.; Nielsen, H.B.; Lacy, M.; Austin, M.J.; Parker, J.E.; et al. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 2000, 103, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef]
- Whiteham, S.; Dinesh-Kumar, S.P.; Choi, D.; Hehl, R.; Corr, C.; Baker, B. The product of the tobacco mosaic virus resistance gene N: Similarity to toll and the interleukin-1 receptor. Cell 1994, 78, 1101–1115. [Google Scholar] [CrossRef] [PubMed]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 cooperate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef] [PubMed]
- Geilen, K.; Heilmann, M.; Hillmer, S.; Böhmer, M. WRKY43 regulates polyunsaturated fatty acid content and seed germination under unfavourable growth conditions. Sci. Rep. 2017, 7, 14235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, L.; Ji, Y.; Jing, Y.; Li, L.; Chen, Y.; Wang, R.; Zhang, H.; Yu, D.; Chen, L. Arabidopsis SIGMA Factor BINDING PROTEIN1 (SIB1) and SIB2 inhibit WRKY75 function in abscisic acid-mediated leaf senescence and seed germination. J. Exp. Bot. 2022, 73, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Meng, X.; Zhang, J.; Xia, M.; Cao, S.; Tang, X.; Fan, T. AtWRKY1 negatively regulates the response of Arabidopsis thaliana to Pst. DC3000. Plant Physiol. Biochem. 2021, 166, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, X.; Jiang, M. CRISPR/Cas9-mediated mutagenesis of WRKY3 and WRKY4 function decreases salt and Me-JA stress tolerance in Arabidopsis thaliana. Mol. Biol. Rep. 2021, 48, 5821–5832. [Google Scholar] [CrossRef]
- Bhattarai, K.K.; Atamian, H.S.; Kaloshian, I.; Eulgem, T. WRKY72-type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene-for-gene resistance mediated by the tomato R gene Mi-1. Plant J. 2010, 63, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Kloth, K.J.; Wiegers, G.L.; Busscher-Lange, J.; van Haarst, J.C.; Kruijer, W.; Bouwmeester, H.J.; Dicke, M.; Jongsma, M.A. AtWRKY22 promotes susceptibility to aphids and modulates salicylic acid and jasmonic acid signalling. J. Exp. Bot. 2016, 67, 3383–3396. [Google Scholar] [CrossRef]
- Xing, D.H.; Lai, Z.B.; Zheng, Z.Y.; Vinod, K.M.; Fan, B.F.; Chen, Z.X. Stress- and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol. Plant 2008, 1, 459–470. [Google Scholar] [CrossRef]
- Yan, C.; Fan, M.; Yang, M.; Zhao, J.; Zhang, W.; Su, Y.; Xiao, L.; Deng, H.; Xie, D. Injury Activates Ca2+/Calmodulin-Dependent Phosphorylation of JAV1-JAZ8-WRKY51 Complex for Jasmonate Biosynthesis. Mol. Cell. 2018, 5, 136–149.e7. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.M.; Venugopal, S.; Navarre, D.; Kachroo, A. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 2011, 155, 464–476. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cui, X.; Yang, B.; Xu, S.; Wei, X.; Zhao, P.; Niu, F.; Sun, M.; Wang, C.; Cheng, H.; et al. WRKY55 transcription factor positively regulates leaf senescence and the defense response by modulating the transcription of genes implicated in the biosynthesis of reactive oxygen species and salicylic acid in Arabidopsis. Development 2020, 18, dev189647. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yu, D. The WRKY57 Transcription Factor Affects the Expression of Jasmonate ZIM-Domain Genes Transcriptionally to Compromise Botrytis cinerea Resistance. Plant Physiol. 2016, 171, 2771–2782. [Google Scholar] [CrossRef]
- Zhao, K.X.; Chu, S.S.; Zhang, X.D.; Wang, L.P.; Rono, J.K.; Yang, Z.M. AtWRKY21 negatively regulates tolerance to osmotic stress in Arabidopsis. Environ. Exp. Bot. 2019, 103920. [Google Scholar] [CrossRef]
- Pandey, S.P.; Roccaro, M.; Schön, M.; Logemann, E.; Somssich, I.E. Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J. 2010, 64, 912–923. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, W.; Karimi, M.; Wieczorek, K.; Van de Cappelle, E.; Wischnitzki, E.; Grundler, F.; Inzé, D.; Beeckman, T.; Gheysen, G. A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol. 2008, 148, 358–368. [Google Scholar] [CrossRef]
- Levée, V.; Major, I.; Levasseur, C.; Tremblay, L.; MacKay, J.; Séguin, A. Expression profiling and functional analysis of Populus WRKY23 reveals a regulatory role in defense. New Phytol. 2009, 184, 48–70. [Google Scholar] [CrossRef]
- Sheikh, A.H.; Sheikh, A.H.; Hussain, R.M.F.; Tabassum, N.; Badmi, R.; Marillonnet, S.; Scheel, D.; Lee, J.; Sinha, A. Possible role of WRKY transcription factors in regulating immunity in Oryza sativa ssp. indica. Physiol. Mol. Plant Pathol. 2021, 114, 101623. [Google Scholar] [CrossRef]
- Zheng, Z.; Qamar, S.A.; Chen, Z.; Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Fu, Q.; Chen, L.; Huang, W.; Yu, D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 2011, 233, 1237–1252. [Google Scholar] [CrossRef] [PubMed]
- Millyard, L. Investigating Transcription Factors in Wheat Defence against Zymoseptoria tritici Fungus. Ph.D. Thesis, Durham University, Durham, UK, 2019. [Google Scholar]
- Banerjee, A.; Roychoudhury, A. WRKY proteins: Signaling and regulation of expression during abiotic stress responses. Sci. World J. 2015, 807560. [Google Scholar]
- Hu, Y.; Dong, Q.; Yu, D. Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 2012, 185–186, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.P.; Elsässer, M.; Fuchs, P.; Fenske, R.; Schwarzländer, M.; Millar, A.H. The versatility of plant organic acid metabolism in leaves is underpinned by mitochondrial malate-citrate exchange. Plant Cell. 2021, 33, 3700–3720. [Google Scholar] [CrossRef]
- Li, S.; Zhou, X.; Chen, L.; Huang, W.; Yu, D. Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol. Cells. 2010, 29, 475–483. [Google Scholar] [CrossRef]
- Wang, L.; Yao, W.; Sun, Y.; Wang, J.; Jiang, T. Association of transcription factor WRKY56 gene from Populus simonii × P. nigra with salt tolerance in Arabidopsis thaliana. PeerJ 2019, 7, e7291. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cheng, X.; Yin, D.; Chen, D.; Luo, C.; Liu, H.; Huang, C. Advances in the research on plant WRKY transcription factors responsive to external stresses. Curr. Issues Mol. Biol. 2023, 45, 2861–2880. [Google Scholar] [CrossRef]
- Han, Z.; Xiong, D.; Schneiter, R.; Tian, C. The function of plant PR1 and other members of the CAP protein superfamily in plant–pathogen interactions. Mol. Plant Pathol. 2023, 24, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Allie, F.; Pierce, E.J.; Okoniewski, M.J.; Rey, C. Transcriptional analysis of South African cassava mosaic virus-infected susceptible and tolerant landraces of cassava highlights differences in resistance, basal defense and cell wall associated genes during infection. BMC Genom. 2014, 15, 1006. [Google Scholar] [CrossRef] [PubMed]
- Feys, B.J.; Parker, J.E. Interplay of signalling pathways in plant disease resistance. Trends Genet. 2000, 16, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Uknes, S.; Mauch, M.B.; Moyer, M.; Potter, S.; Williams, S.; Dincher, S.; Chandler, D.; Slusarenko, A.; Ward, E.; Ryals, J. Acquired resistance in Arabidopsis. Plant Cell 1992, 4, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Ryals, J.A.; Neuenschwander, U.H.; Willits, M.G.; Molina, A.; Steiner, H.Y.; Hunt, M.D. Systemic acquired resistance. Plant Cell 1996, 8, 1809–1819. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.D.; Liu, Y.; Klessig, D.F.; Dong, X. Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 1998, 10, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.D.; Volko, S.M.; Ledford, H.; Ausubel, F.M.; Dong, X. Roles of salicylic acid, jasmonic acid, and ethylene in cpr induced resistance in Arabidopsis. Plant Cell 2000, 12, 2175–2190. [Google Scholar] [CrossRef] [PubMed]
- Shah, J.; Kachroo, P.; Klessig, D.F. The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 1999, 11, 191–206. [Google Scholar] [CrossRef] [PubMed]
- Orek, C. A review of the functions of transcription factors and related genes involved in cassava (Manihot esculenta Crantz) response to drought stress. Trop. Plants 2023, 2, 14. [Google Scholar]
- Zhou, N.; Tootle, T.L.; Tsui, F.; Klessig, D.F.; Glazebrook, J. PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 1998, 10, 1021–1030. [Google Scholar] [CrossRef] [PubMed]
- Kalde, M.; Barth, M.; Somssich, I.E.; Lippok, B. Members of the Arabidopsis WRKY Group III transcription factors are part of different plant defense signalling pathways. Mol. Plant Microbe Interact. 2003, 16, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Lippok, B.; Birkenbihl, R.P.; Rivory, G.; Brümmer, J.; Schmelzer, E.; Logemann, E.; Somssich, I.E. Expression of AtWRKY33 encoding a pathogen- or PAMP-responsive WRKY transcription factor is regulated by a composite DNA motif containing W box elements. Mol. Plant Microbe Interact. 2007, 20, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Malichan, S.; Vannatim, N.; Chaowongdee, S. Comparative analysis of salicylic acid levels and gene expression in resistant, tolerant, and susceptible cassava varieties following whitefly mediated SLCMV infection. Sci. Rep. 2023, 13, 13610. [Google Scholar] [CrossRef] [PubMed]
- Yokotani, N.; Shikata, M.; Ichikawa, H. OsWRKY24, a blast-disease responsive transcription factor, positively regulates rice disease resistance. J. Gen. Plant Pathol. 2018, 84, 85–91. [Google Scholar] [CrossRef]
- Hernández, J.A.; Gullner, G.; Clemente-Moreno, M.J.; Künstler, A.; Juhasz, C.; Díaz-Vivancos, P.; Kiraly, L. Oxidative stress and antioxidative responses in plant–virus interactions. Physiol. Mol. Plant Pathol. 2016, 94, 134–148. [Google Scholar] [CrossRef]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signaling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Bali, S.; Gautam, A.; Dhiman, A.; Michael, R.; Dogra, V. Salicylate and jasmonate intertwine in ROS-triggered chloroplast-to-nucleus retrograde signalling. Physiol. Plant. 2023, 175, e14041. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.Y.; Spivey, N.W.; Zeng, W.; Liu, P.P.; Fu, Z.Q.; Klessig, D.F.; He, S.Y.; Dong, X. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signalling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 2012, 11, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The crosstalks between jasmonic acid and other plant hormone signalling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front. Plant Sci. 2019, 10, 1349. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, S.; Peng, A. CRISPR/Cas9-mediated editing of CsWRKY22 reduces susceptibility to Xanthomonas citri subsp. citri in Wanjincheng orange (Citrus sinensis (L.) Osbeck). Plant Biotechnol. Rep. 2019, 13, 501–510. [Google Scholar] [CrossRef]
- Long, Q.; Du, M.; Long, J.; Xie, Y.; Zhang, J.; Xu, L.; He, Y.; Li, Q.; Chen, S.; Zou, X. Transcription factor WRKY22 regulates canker susceptibility in sweet orange (Citrus sinensis Osbeck) by enhancing cell enlargement and CsLOB1 expression. Hortic. Res. 2021, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Jiang, Y.; Yu, D. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells 2011, 31, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Abbruscato, P.; Nepusz, T.; Mizzi, L.; Del Corvo, M.; Morandini, P.; Fumasoni, I.; Michel, C.; Paccanaro, A.; Guiderdoni, E.; Schaffrath, U.; et al. OsWRKY22, a monocot WRKY gene, plays a role in the resistance response to Blast. Mol. Plant Pathol. 2012, 13, 828–841. [Google Scholar] [CrossRef] [PubMed]
- Vanderauwera, S.; Vandenbroucke, K.; Inzé, A.; Cotte, B.; Mühlenbock, P.; De Rycke, R.; Naouar, N.; Van Gaever, T.; Van Montagu, M.C.E.; Van Breusegem, F. AtWRKY15 perturbation abolishes the mitochondrial stress response that steers osmotic stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 20113–20118. [Google Scholar] [CrossRef] [PubMed]
- Geilen, K.; Böhmer, M. Dynamic subnuclear relocalisation of WRKY40 in response to abscisic acid in Arabidopsis thaliana. Sci. Rep. 2015, 5, 13369. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Schuck, S.; Wu, J.; Yang, P.; Döring, A.C.; Zeier, J.; Tsuda, K. A MPK3/6-WRKY33-ALD1-Pipecolic acid regulatory loop contributes to systemic acquired resistance. Plant Cell 2018, 30, 2480–2494. [Google Scholar] [CrossRef] [PubMed]
- Lang, J.; Genot, B.; Bigeard, J.; Colcombet, J. MPK3 and MPK6 control salicylic acid signaling by up-regulating NLR receptors during pattern- and effector-triggered immunity. J. Exp. Bot. 2022, 73, 2190–2205. [Google Scholar] [CrossRef] [PubMed]
- Colcombet, J.; Berriri, S.; Hirt, H. Constitutively active MPK4 helps to clarify its role in plant immunity. Plant Signal. Behav. 2013, 8, e22991. [Google Scholar] [CrossRef] [PubMed]
- Siodmak, A.; Shahul Hameed, U.F.; Rayapuram, N.; Völz, R.; Boudsocq, M.; Alharbi, S.; Alhoraibi, H.; Lee, Y.-H.; Blilou, I.; Arold, S.T.; et al. Essential role of the CD docking motif of MPK4 in plant immunity, growth, and development. New Phytol. 2023, 239, 1112–1126. [Google Scholar] [CrossRef] [PubMed]
- Bittner, F.; Oreb, M.; Mendel, R.R. ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 40381–40384. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [PubMed]
- Oblessuc, P.R.; Obulareddy, N.; DeMott, L.; Matiolli, C.C.; Thompson, B.K.; Melotto, M. JAZ4 is involved in plant defense, growth, and development in Arabidopsis. Plant J. 2020, 101, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Wang, Y.; Qiu, L.; Han, X.; Zhu, Y.; Liu, L.; Man, M.; Li, F.; Ren, M.; Xing, Y. MYC2: A master switch for plant physiological processes and specialized metabolite synthesis. Int. J. Mol. Sci. 2023, 24, 3511. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Isozumi, N.; Mori, M.; Okuta, R.; Singkaravanit-Ogawa, S.; Imamura, T.; Kurita, J.I.; Gan, P.; Shirasu, K.; Ohki, S.; et al. Fungal effector SIB1 of Colletotrichum orbiculare has unique structural features and can suppress plant immunity in Nicotiana benthamiana. J. Biol. Chem. 2021, 297, 101370. [Google Scholar] [CrossRef] [PubMed]
- Jaspers, P.; Brosché, M.; Overmyer, K.; Kangasjärvi, J. The transcription factor interacting protein RCD1 contains a novel conserved domain. Plant Signal. Behav. 2010, 5, 78–80. [Google Scholar] [CrossRef] [PubMed]
- Hiltscher, H.; Rudnik, R.; Shaikhali, J.; Heiber, I.; Mellenthin, M.; Meirelles, D.I.; Schuster, G.; Kahmann, U.; Baier, M. The radical induced cell death protein 1 (RCD1) supports transcriptional activation of genes for chloroplast antioxidant enzymes. Front. Plant Sci. 2014, 5, 475. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Wu, F.; Wen, H.; Liu, X.; Luo, W.; Gao, L.; Jiang, Z.; Mo, B.; Chen, X.; Kong, W. RCD1 promotes salt stress tolerance in Arabidopsis by repressing ANAC017 activity. Int. J. Mol. Sci. 2023, 24, 9793. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.; Wakem, M.; Dijkman, G.; Alsarraj, M.; Nguyen, M. A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods 2010, 50, S1–S5. [Google Scholar] [CrossRef] [PubMed]
- De Keyser, E.; Desmet, L.; Van Bockstaele, E.; De Riek, J. How to perform RT-qPCR accurately in plant species? A case study on flower colour gene expression in an azalea (Rhododendron simsii hybrids) mapping population. BMC Mol. Biol. 2013, 14, 13. [Google Scholar] [CrossRef] [PubMed]
- Remans, T.; Keunen, E.; Bex, G.J.; Smeets, K.; Vangronsveld, J.; Cuypers, A. Reliable gene expression analysis by reverse transcription-quantitative PCR: Reporting and minimizing the uncertainty in data accuracy. Plant Cell 2014, 26, 3829–3837. [Google Scholar] [CrossRef] [PubMed]
- Gingrich, J.R.; Rubio, T.; Karlak, C. Effect of RNA degradation on data quality in quantitative PCR and microarray experiments. Bio-Rad Bull. 2006, 5452, 1106. [Google Scholar]
- Doyle, J.J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Behnam, B.; Bohorquez, C.A.; Castaneda, M.O.F.; Tsuji, H.; Ishitani, M.; Becerra, L.L.L.A. An optimized isolation protocol yields high-quality RNA from cassava tissues (Manihot esculenta Crantz). FEBS Open Bio 2019, 9, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt, A.O.; Mareuil, F.; Cohen, B.S.; Gascuel, O. NGPhylogeny.fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef] [PubMed]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Santos, A.; von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef] [PubMed]


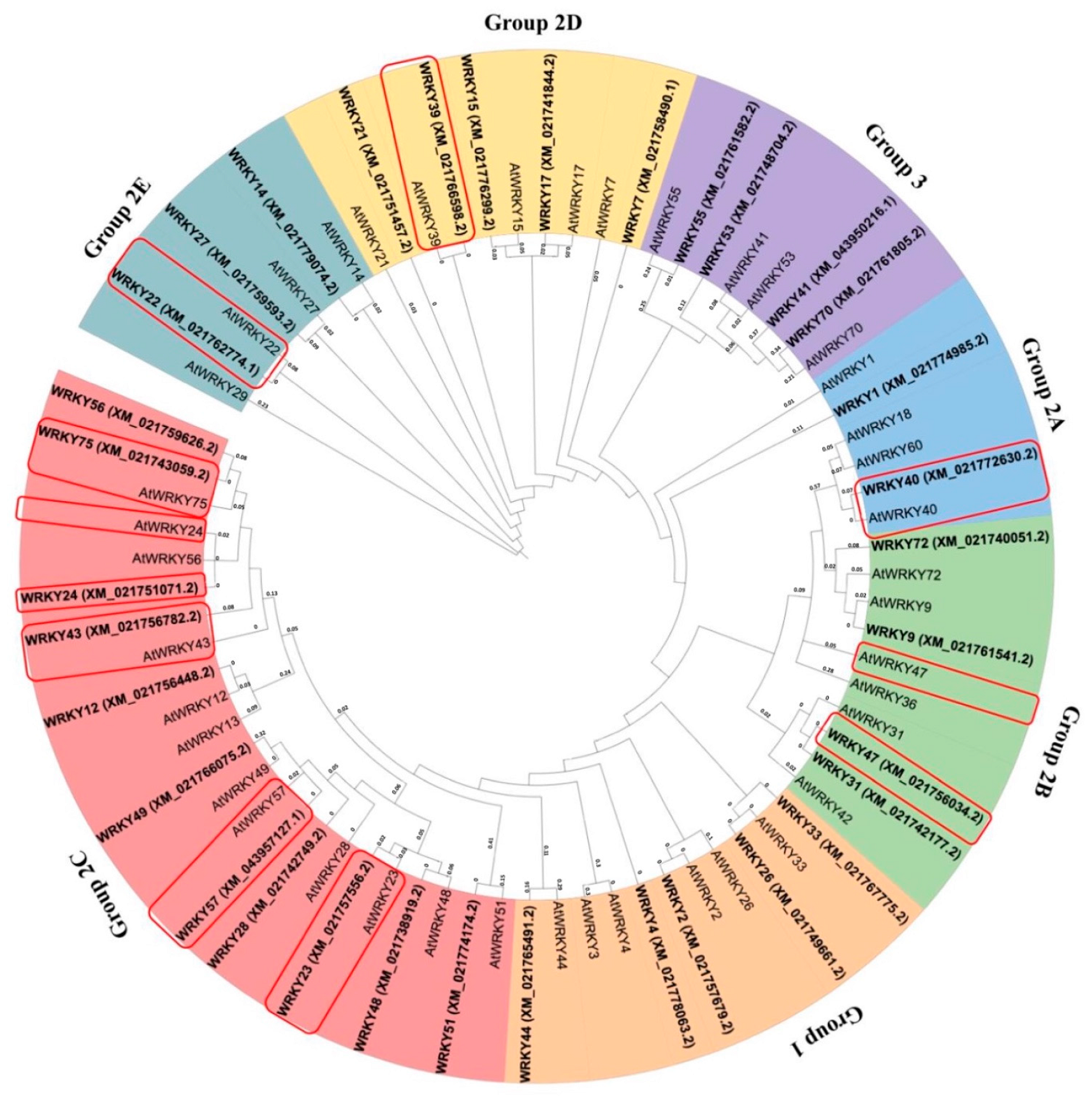

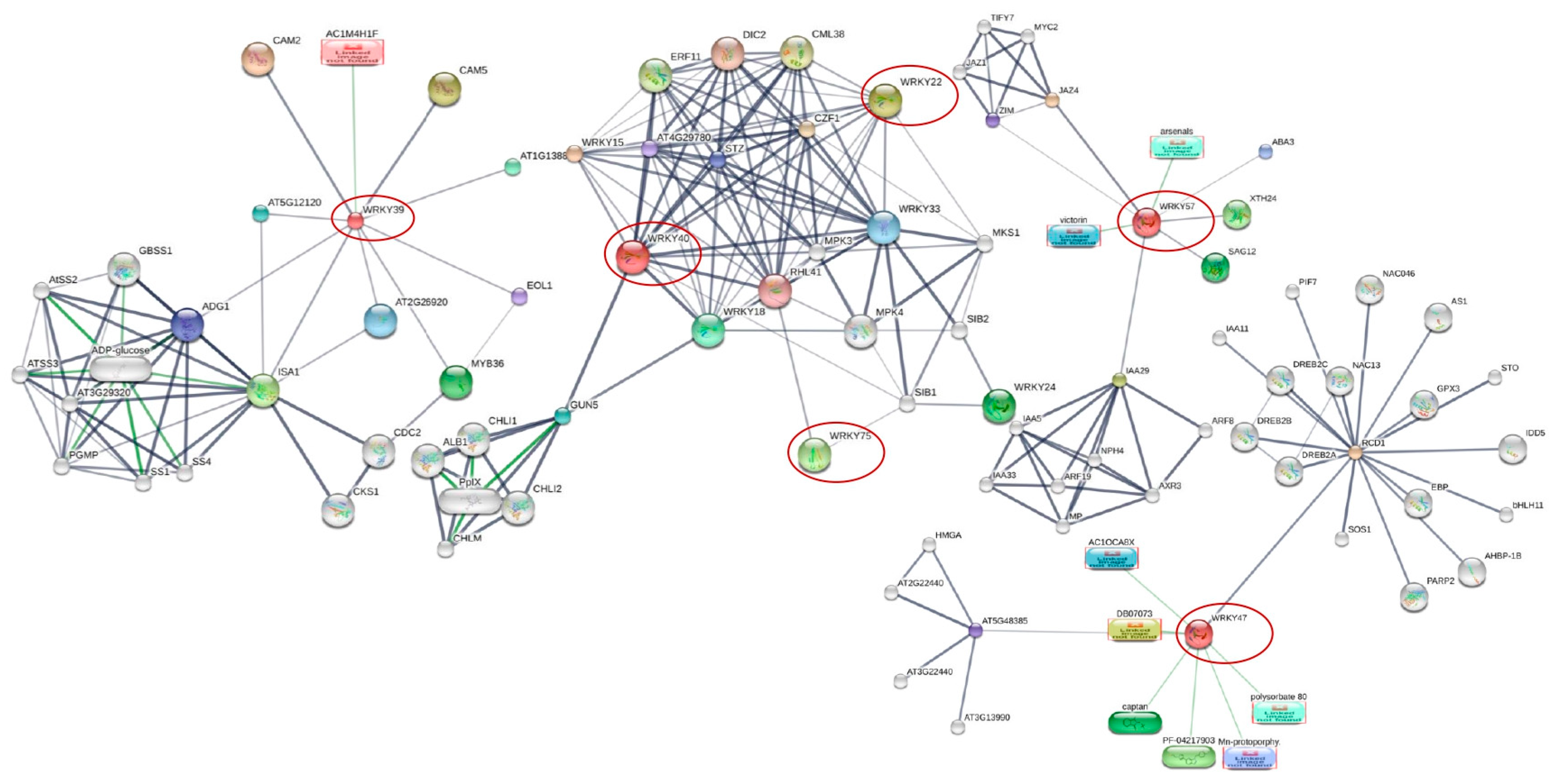

| NCBI Accession Numbers | Gene ID | WRKYs Family | AtWRKYs * (Arabidopsis thaliana Homolog) | Homolog Functions |
|---|---|---|---|---|
| KU 50 | ||||
| Plant defenses | ||||
| XM_021762774.1 | 110619370 | WRKY DNA-binding protein 22 | AtWRKY14, -22, -29 | Coordinating with the MAPK signaling pathway and related to H2O2 enhancement [31,32,33] |
| R 11 | ||||
| Other related hormones and further abiotic stress response | ||||
| XM_021756782.2 | 110615072 | WRKY DNA-binding protein 43 | AtWRKY24, -43, -56 | Regulation of the ABA-dependent gene expression [34] |
| XM_021743059.2 | 110604766 | WRKY DNA-binding protein 75 | AtWRKY24, -43, -56, -75 | Regulation of the ABA intermediate signaling pathway in Arabidopsis [35] |
| Plant developments | ||||
| XM_021743059.2 | 110604766 | WRKY DNA-binding protein 75 | AtWRKY24, -43, -56, -75 | Related to phosphate accumulation within plant growth and development [36] |
| KU 50 and R 11 | ||||
| Hormone signaling (abscisic acid, SA, and JA) | ||||
| XM_021774985.2 | 110628349 | WRKY DNA-binding protein 1 | AtWRKY1 | Suppressing SA signaling [37] |
| XM_021778063.2 | 110630526 | WRKY DNA-binding protein 4 | AtWRKY3, -4, -44 | Responding to JA stresses in Arabidopsis [38] |
| XM_021761541.2 | 110618414 | WRKY DNA-binding protein 9 | AtWRKY9, -72 | Utilizing SA-independent defense mechanisms [27,39] |
| XM_021779074.2 | 110631303 | WRKY DNA-binding protein 14 | AtWRKY14, -22, -27, -29 | Responding to SA and JA pathways in biotic and abiotic interruptions [40]. |
| XM_021759593.2 | 110617016 | WRKY DNA-binding protein 27 | AtWRKY22, -27, -29, -14 | Modulating the roles of the SA and JA pathways [40]. |
| XM_021742749.2 | 110604542 | WRKY DNA-binding protein 28 | AtWRKY28, -57 | Associated with JA signaling pathway [23] |
| XM_021742177.2 | 110604092 | WRKY DNA-binding protein 31 | AtWRKY31, -36, -42, -47 | Regulation through the modulation of SA signaling [33] |
| XM_021765491.2 | 110621275 | WRKY DNA-binding protein 44 | AtWRKY3, -4, -44 | Activating the tolerances within JA stresses in Arabidopsis [38] |
| XM_021756034.2 | 110614486 | WRKY DNA-binding protein 47 | AtWRKY31, -36, -42, -47 | Regulation through the modulation of SA signaling [33] |
| XM_021738919.2 | 110601688 | WRKY DNA-binding protein 48 | AtWRKY23, -28, -57, -48 | Associated with SA regulation by induce the PR1 in the bacterial pathogen infection [41] |
| XM_021774174.2 | 110627806 | WRKY DNA-binding protein 51 | AtWRKY24, -43, -51, -56, -75 | Intermediating the SA; otherwise, repressing JA signaling [42,43] |
| XM_021748704.2 | 110609254 | WRKY DNA-binding protein 53 | AtWRKY41, -53, -55 | Related to SA signaling induction of Arabidopsis [23] |
| XM_021761582.2 | 110618450 | WRKY DNA-binding protein 55 | AtWRKY41, -53, -55 | Regulating the SA signaling pathway [44] |
| XM_043957127.1 | 110614243 | WRKY DNA-binding protein 57 | AtWRKY28, -57 | Regulating the JA signaling pathway in case of fungal infection [45] |
| XM_021740051.2 | 110602513 | WRKY DNA-binding protein 72 | AtWRKY9, -72 | Utilizing SA-independent defense mechanisms [39] |
| Reactive Oxygen Species (ROS) | ||||
| XM_021742177.2 | 110604092 | WRKY DNA-binding protein 31 | AtWRKY31, -36, -42, -47 | Regulating of ROS synthesis [33] |
| XM_021756034.2 | 110614486 | WRKY DNA-binding protein 47 | AtWRKY31, -36, -42, -47 | Regulation through ROS synthesis [33] |
| XM_021761582.2 | 110618450 | WRKY DNA-binding protein 55 | AtWRKY41, -53, -55 | Regulation of ROS accumulation [44] |
| Basal immune mechanisms | ||||
| XM_021774985.2 | 110628349 | WRKY DNA-binding protein 1 | AtWRKY1 | Related to the pathogenesis-related (PR) proteins stimulated [37] |
| XM_021758490.1 | 110616151 | WRKY DNA-binding protein 7 | AtWRKY7, -15 | Regulating plant defense against bacterial pathogens and triggering the HR, which eventually induces cell death programming [27,29] |
| XM_021761541.2 | 110618414 | WRKY DNA-binding protein 9 | AtWRKY9, -72 | Contributing to the plant basal defense against bacteria and nematode pathogens and coordinating the elicited HR mechanism [39] |
| XM_021756448.2 | 110614786 | WRKY DNA-binding protein 12 | AtWRKY12, -13 | Regulated positively in the plant defense mechanism [23] |
| XM_021751457.2 | 110611253 | WRKY DNA-binding protein 21 | AtWRKY15, -17, -21, -39 | Controlling plant defense signaling against bacterial infection [46,47] |
| XM_021757556.2 | 110615593 | WRKY DNA-binding protein 23 | AtWRKY23, -48 | Accompanying an avirulent-to-bacterial infection [48,49] |
| XM_021751071.2 | 110610985 | WRKY DNA-binding protein 24 | AtWRKY24, -43, -51, -56, -75 | Has a role in the basal immunity (PTI) expression of the early defense response in Oryza sativa ssp. indica [50] |
| XM_021749661.2 | 110609837 | WRKY DNA-binding protein 26 | AtWRKY2, -26, -33 | Regulating resistance to necrotrophic pathogens [51,52] |
| XM_021759593.2 | 110617016 | WRKY DNA-binding protein 27 | AtWRKY22, -27, -29, -14 | Involved in pathogen-triggered immunity [40] |
| XM_021767775.2 | 110622985 | WRKY DNA-binding protein 33 | AtWRKY2, -26, -33 | Regulating plant-induced resistance to the necrotrophic pathogens [51,52] |
| XM_021738919.2 | 110601688 | WRKY DNA-binding protein 48 | AtWRKY23, -28, -57, -48 | Influencing the plant basal resistance associated with PR1 in the bacterial pathogen infection [41] |
| XM_021766075.2 | 110621777 | WRKY DNA-binding protein 49 | AtWRKY49 | Related to resistance and increasing defense gene expression [53] |
| XM_021761805.2 | 110618630 | WRKY DNA-binding protein 70 | AtWRKY70 | Corresponds to the NPR1 protein and is related to enhancing PR1 gene expression [54,55] |
| XM_021740051.2 | 110602513 | WRKY DNA-binding protein 72 | AtWRKY9, -72 | Utilizing plant basal immunity [39] |
| Other related hormones, metabolites, and abiotic stress responses | ||||
| XM_021757679.2 | 110615677 | WRKY DNA-binding protein 2 | AtWRKY2, -26, -33 | Enhanced during heat stress [51,52] |
| XM_021776299.2 | 110629365 | WRKY DNA-binding protein 15 | AtWRKY15, -17, -21, -39 | Enhancing plant metabolite pathways [56] |
| XM_021741844.2 | 110603865 | WRKY DNA-binding protein 17 | AtWRKY17, -15, -21, -39 | Responding to drought stress in bacterial infection [46,47] |
| XM_021757556.2 | 110615593 | WRKY DNA-binding protein 23 | AtWRKY23, -48 | Responding to auxin hormones in nematode resistance [48,49] |
| XM_021749661.2 | 110609837 | WRKY DNA-binding protein 26 | AtWRKY2, -26, -33 | Enhanced during heat stress [51,52] |
| XM_021742749.2 | 110604542 | WRKY DNA-binding protein 28 | AtWRKY28, -57 | Associated with ABA hormone transcriptional regulation [23] |
| XM_021767775.2 | 110622985 | WRKY DNA-binding protein 33 | AtWRKY2, -26, -33 | Enhanced during heat stress [51,52] |
| XM_021766598.2 | 110622177 | WRKY DNA-binding protein 39 | AtWRKY15, -17, -21, -39 | Functions in the ethylene hormone and heat tolerant in Arabidopsis thaliana [57] |
| XM_021772630.2 | 110626614 | WRKY DNA-binding protein 40 | AtWRKY18, -40, -60 | Regulating the intermediating of the ABA hormone signaling pathway [23] |
| XM_043950216.1 | 110631349 | WRKY DNA-binding protein 41 | AtWRKY41, -53, -55 | Regulating plant general hormone signaling and response to biotic stresses [23] |
| XM_021759626.2 | 110617045 | WRKY DNA-binding protein 56 | AtWRKY24, -43, -51, -56, -75 | Responding to salt stress in Arabidopsis thaliana [58] |
| NCBI Accession Numbers | Gene ID | WRKYs Families | KU 50 | R 11 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 32 dpi | 67 dpi | 32 dpi | 67 dpi | |||||||
| DEGs | p-Value | DEGs | p-Value | DEGs | p-Value | DEGs | p-Value | |||
| XM_021774985.2 | 110628349 | WRKY DNA-binding protein 1 | −0.88 | 0.58 | ND * | ND | −2.30 | 0.53 | ND | ND |
| XM_021757679.2 | 110615677 | WRKY DNA-binding protein 2 | ND | ND | −1.10 | 0.61 | 0.32 | 0.76 | −3.80 | 0.02 |
| XM_021778063.2 | 110630526 | WRKY DNA-binding protein 4 | −0.03 | 0.96 | −0.69 | 0.40 | 0.29 | 0.64 | −1.04 | 0.08 |
| XM_021758490.1 | 110616151 | WRKY DNA-binding protein 7 | −0.94 | 0.04 | −0.01 | 0.99 | −1.01 | 0.07 | −0.34 | 0.54 |
| XM_021761541.2 | 110618414 | WRKY DNA-binding protein 9 | ND | ND | ND | ND | ND | ND | ND | ND |
| XM_021756448.2 | 110614786 | WRKY DNA-binding protein 12 | ND | ND | ND | ND | ND | ND | ND | ND |
| XM_021779074.2 | 110631303 | WRKY DNA-binding protein 14 | ND | ND | ND | ND | ND | ND | ND | ND |
| XM_021776299.2 | 110629365 | WRKY DNA-binding protein 15 | −1.03 | 0.1 | 0.51 | 0.58 | −1.71 | 0.01 | 1.04 | 0.09 |
| XM_021741844.2 | 110603865 | WRKY DNA-binding protein 17 | ND | ND | 0.14 | 0.69 | ND | ND | −0.4 | 0.17 |
| XM_021751457.2 | 110611253 | WRKY DNA-binding protein 21 | 0.32 | 0.5 | 0.03 | 0.96 | 0.53 | 0.37 | −1.40 | 0.01 |
| XM_021762774.1 | 110619370 | WRKY DNA-binding protein 22 | −1.69 | 0.02 | −0.22 | 0.87 | ND | ND | ND | ND |
| XM_021757556.2 | 110615593 | WRKY DNA-binding protein 23 | 2.83 | 0.32 | 1.12 | 0.53 | 1.10 | 0.79 | −0.78 | 0.85 |
| XM_021751071.2 | 110610985 | WRKY DNA-binding protein 24 | 0.93 | 0.82 | 0.32 | 0.94 | −2.30 | 0.56 | 1.14 | 0.78 |
| XM_021749661.2 | 110609837 | WRKY DNA-binding protein 26 | ND | ND | −0.27 | 0.88 | ND | ND | −3.26 | 0.24 |
| XM_021759593.2 | 110617016 | WRKY DNA-binding protein 27 | ND | ND | ND | ND | ND | ND | ND | ND |
| XM_021742749.2 | 110604542 | WRKY DNA-binding protein 28 | ND | ND | ND | ND | ND | ND | ND | ND |
| XM_021742177.2 | 110604092 | WRKY DNA-binding protein 31 | ND | ND | −2.39 | 0.19 | −1.67 | 0.08 | −0.56 | 0.67 |
| XM_021767775.2 | 110622985 | WRKY DNA-binding protein 33 | ND | ND | −2.13 | 0.58 | ND | ND | −2.26 | 0.53 |
| XM_021766598.2 | 110622177 | WRKY DNA-binding protein 39 | 2.19 | 0.09 | −0.03 | 0.98 | −0.18 | 0.92 | 1.18 | 0.47 |
| XM_021772630.2 | 110626614 | WRKY DNA-binding protein 40 | ND | ND | 2.45 | 0.05 | ND | ND | −0.23 | 0.87 |
| XM_043950216.1 | 110631349 | WRKY DNA-binding protein 41 | ND | ND | ND | ND | ND | ND | ND | ND |
| XM_021756782.2 | 110615072 | WRKY DNA-binding protein 43 | ND | ND | ND | ND | 1.10 | 0.79 | −0.78 | 0.85 |
| XM_021765491.2 | 110621275 | WRKY DNA-binding protein 44 | ND | ND | 0.75 | 0.04 | 0.21 | 0.48 | ND | ND |
| XM_021756034.2 | 110614486 | WRKY DNA-binding protein 47 | 1.47 | 0.16 | −0.07 | 0.95 | 3.80 | 0.14 | 0.83 | 0.48 |
| XM_021738919.2 | 110601688 | WRKY DNA-binding protein 48 | 0.97 | 0.63 | 0.32 | 0.88 | −1.34 | 0.65 | −0.78 | 0.85 |
| XM_021766075.2 | 110621777 | WRKY DNA-binding protein 49 | ND | ND | ND | ND | ND | ND | ND | ND |
| XM_021774174.2 | 110627806 | WRKY DNA-binding protein 51 | 0.61 | 0.32 | −1.39 | 0.13 | −1.57 | 0.03 | −0.26 | 0.75 |
| XM_021748704.2 | 110609254 | WRKY DNA-binding protein 53 | ND | ND | −0.69 | 0.73 | ND | ND | −3.26 | 0.25 |
| XM_021761582.2 | 110618450 | WRKY DNA-binding protein 55 | ND | ND | ND | ND | ND | ND | ND | ND |
| XM_021759626.2 | 110617045 | WRKY DNA-binding protein 56 | ND | ND | ND | ND | ND | ND | ND | ND |
| XM_043957127.1 | 110614243 | WRKY DNA-binding protein 57 | 3.15 | 0.23 | −1.01 | 0.67 | −1.76 | 0.52 | −0.78 | 0.85 |
| XM_021761805.2 | 110618630 | WRKY DNA-binding protein 70 | 0.12 | 0.85 | −1.06 | 0.28 | ND | ND | −2.46 | 0.02 |
| XM_021740051.2 | 110602513 | WRKY DNA-binding protein 72 | ND | ND | ND | ND | ND | ND | ND | ND |
| XM_021743059.2 | 110604766 | WRKY DNA-binding protein 75 | ND | ND | ND | ND | 1.04 | 0.78 | −1.68 | 0.68 |
| NCBI Accession Numbers | Name of WRKYs Transcription Factors | R 11 | KU 50 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 32 dpi | 67 dpi | 32 dpi | 67 dpi | ||||||
| DEGs | log 2∆CQ | DEGs | log 2∆CQ | DEGs | log 2∆CQ | DEGs | log 2∆CQ | ||
| XM_021762774.1 | WRKY22 | ND * | −0.11 | ND | 0.01 | −1.69 | −0.54 | −0.22 | 0.13 |
| XM_021757556.2 | WRKY23 | 1.10 | 0.51 | −0.78 | 0.67 | 2.83 | −0.24 | 1.12 | −0.12 |
| XM_021751071.2 | WRKY24 | −2.30 | 1.34 | 1.14 | 0.61 | 0.93 | −0.35 | 0.31 | −0.01 |
| XM_021766598.2 | WRKY39 | −0.18 | 0.94 | 1.18 | 0.59 | 2.19 | −0.31 | −0.03 | −0.13 |
| XM_021772630.2 | WRKY40 | ND | 0.60 | −0.23 | 0.71 | ND | −1.24 | 2.45 | −0.30 |
| XM_021756782.2 | WRKY43 | 1.10 | 0.02 | −0.78 | 1.26 | ND | −0.42 | ND | 0.17 |
| XM_021756034.2 | WRKY47 | 3.80 | −0.01 | 0.83 | 0.01 | 1.47 | 0.09 | −0.07 | −0.03 |
| XM_043957127.1 | WRKY57 | −1.76 | 1.20 | −0.78 | 0.91 | 3.15 | −0.32 | −1.01 | −0.04 |
| XM_021743059.2 | WRKY75 | 1.04 | 0.71 | −1.68 | 0.50 | ND | −0.19 | ND | −0.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaowongdee, S.; Vannatim, N.; Malichan, S.; Kuncharoen, N.; Tongyoo, P.; Siriwan, W. Roles of WRKY Transcription Factors in Response to Sri Lankan Cassava Mosaic Virus Infection in Susceptible and Tolerant Cassava Cultivars. Plants 2025, 14, 1159. https://doi.org/10.3390/plants14081159
Chaowongdee S, Vannatim N, Malichan S, Kuncharoen N, Tongyoo P, Siriwan W. Roles of WRKY Transcription Factors in Response to Sri Lankan Cassava Mosaic Virus Infection in Susceptible and Tolerant Cassava Cultivars. Plants. 2025; 14(8):1159. https://doi.org/10.3390/plants14081159
Chicago/Turabian StyleChaowongdee, Somruthai, Nattachai Vannatim, Srihunsa Malichan, Nattakorn Kuncharoen, Pumipat Tongyoo, and Wanwisa Siriwan. 2025. "Roles of WRKY Transcription Factors in Response to Sri Lankan Cassava Mosaic Virus Infection in Susceptible and Tolerant Cassava Cultivars" Plants 14, no. 8: 1159. https://doi.org/10.3390/plants14081159
APA StyleChaowongdee, S., Vannatim, N., Malichan, S., Kuncharoen, N., Tongyoo, P., & Siriwan, W. (2025). Roles of WRKY Transcription Factors in Response to Sri Lankan Cassava Mosaic Virus Infection in Susceptible and Tolerant Cassava Cultivars. Plants, 14(8), 1159. https://doi.org/10.3390/plants14081159






