Combating Root-Knot Nematodes (Meloidogyne spp.): From Molecular Mechanisms to Resistant Crops
Abstract
1. Introduction
2. RKN Infection Establishment: Hijacking the Plant Defense System
3. Common Strategies for Controlling RKN
3.1. Cultural Practices and Pesticides
3.2. Biopesticides
3.3. Host Plant Resistance
4. Omics Approaches to Understanding Plant–RKN Interactions and Resistance
Transcriptomic Efforts Toward Understanding Plant Resistance to RKN
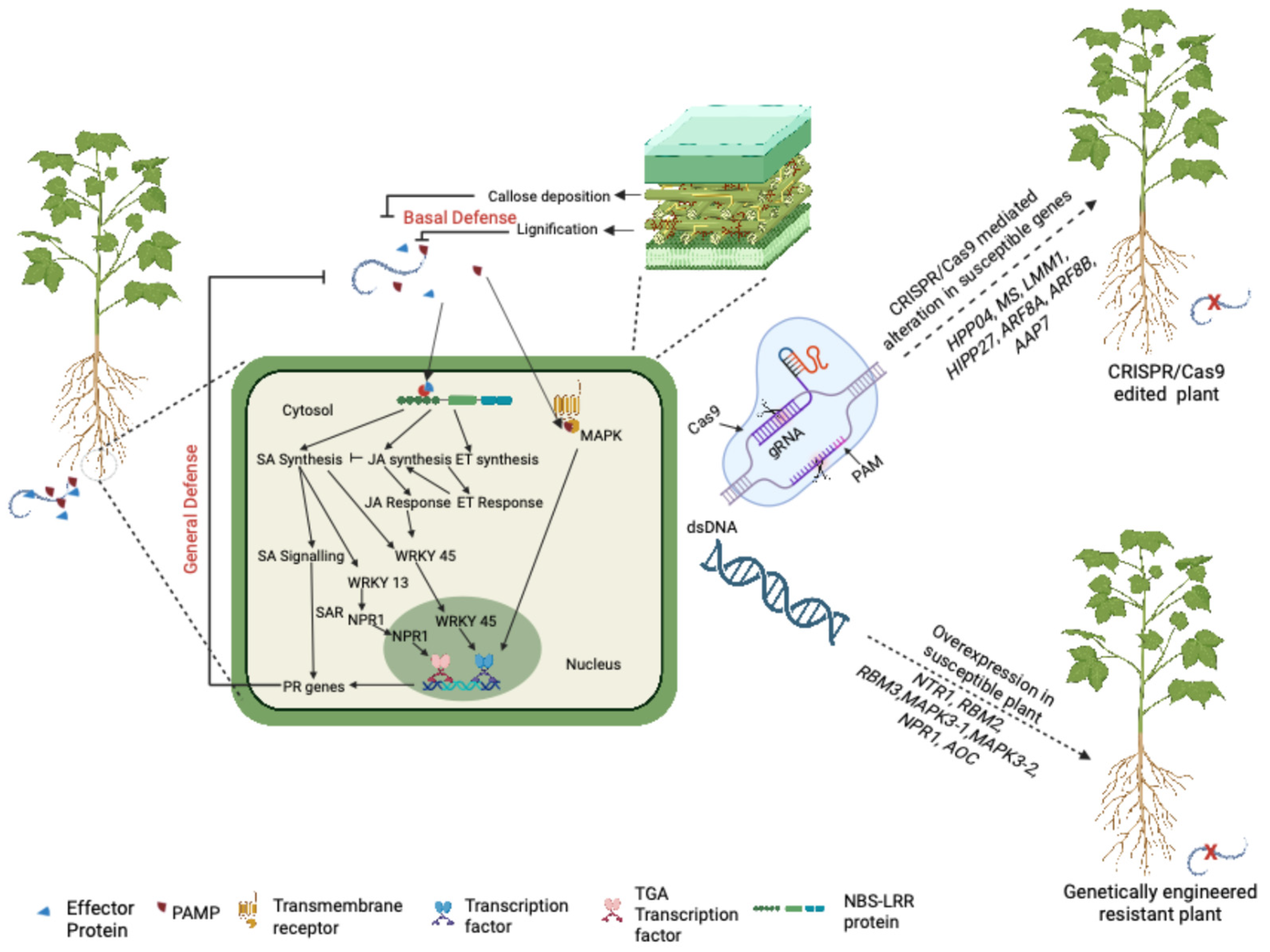
5. Genome Engineering Approaches to Improve RKN Resistance
5.1. Overexpression, Silencing, and Mutation of Genes to Develop Resistance to RKN
5.2. Genome Editing for Understanding and Developing Resistance to RKN
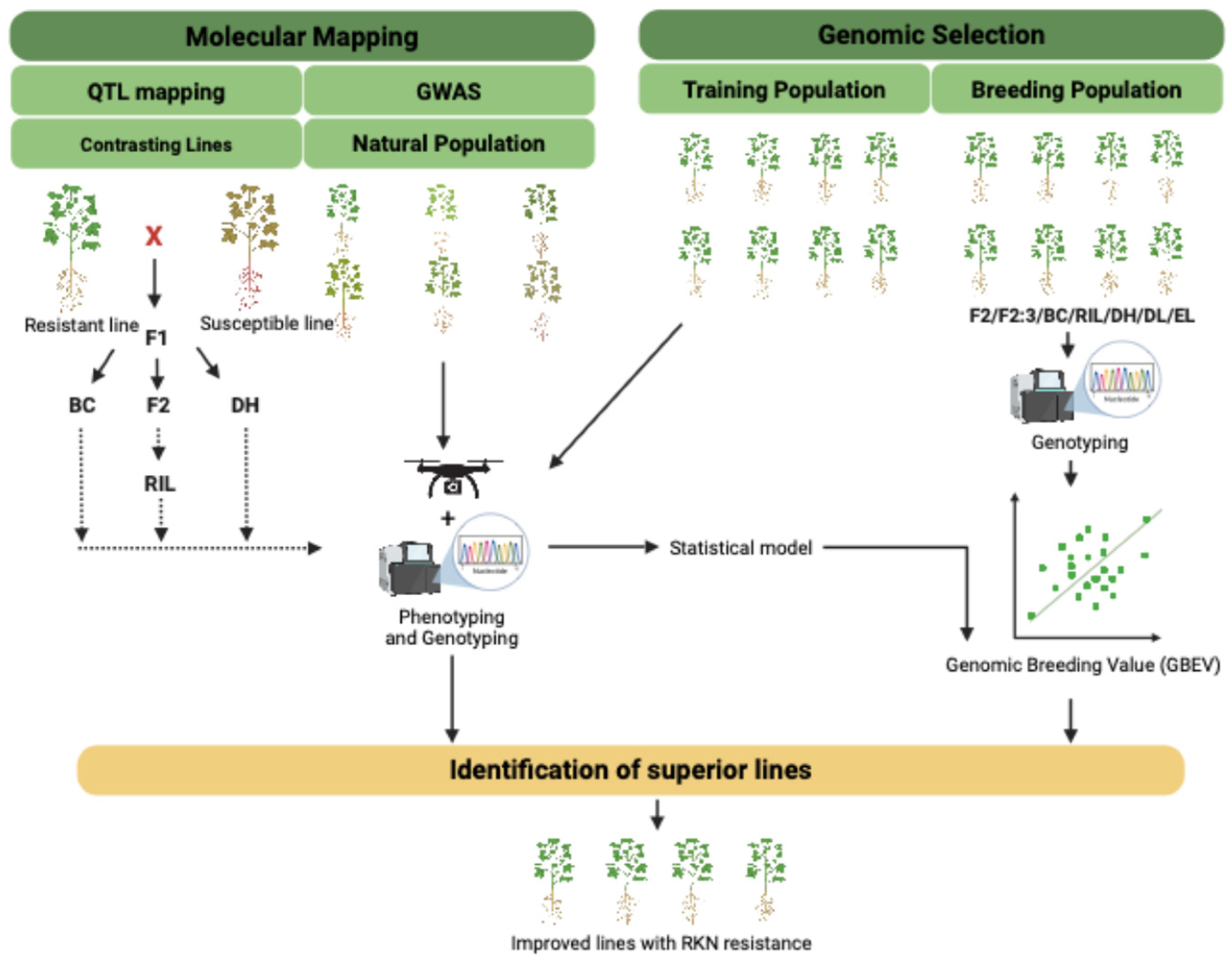
6. Genetic Approaches to Combat RKN Infestation
6.1. Marker Assisted Selection
6.2. QTL Mapping Efforts for RKN-Related Traits
| Crop | Population Type | No. of Lines Used | No. of Major QTLs Identified | Location of Identified QTLs | Reference |
|---|---|---|---|---|---|
| Cotton (Gossypium spp.) | RIL | 138 | 4 | Chr 3, 4, 11, 17 | [146] |
| (M120 × Pima S-6) F2 | 245 | 1 | Chr 14 | [159] | |
| Peanut (Arachis hypogaea) | RIL | 93 | 4 | LG02, 04,09 | [155] |
| Sorghum (Sorghum bicolor) | (PI 144,134 × Collier) F2 | 249 | 1 | Chr 5 | [160] |
| Cowpea (Vigna unguiculata) | RIL, F2:3 | 389 | 1 | VuLG11 | [161] |
| RIL | 264 | 2 | Vu01 and Vu04 | [74] | |
| Carrot (Daucus carota) | Two F2 mapping populations, (Br1091 × HM1) and (SFF × HM2), and one segregating HM3 population | - | 5 | Chr 1,2,4,8,9 | [152] |
| Pepper (Capsicum annuum) | (YW × DLL) F2:3 | 130 | 4 | Chr 1,9 | [160] |
| Sweet Potato | Tanzania × Beauregard | 240 | 9 (7 in Tanzania and 2 in beauregard) | T01.01, T05.26, T07.37, T07.38, T07.39, T07.41, T08.46 | [162] |
| TB population, F1 | 244 | 1 | IbLG07 | [154] | |
| Soybean (Glycine max) | RIL (Magellan × PI 567305) | 242 | 2 | Chr 10, 13 | [163] |
6.3. GWAS for RKN-Related Traits
6.4. GS: A Promising Tool to Improve RKN Resistance
6.5. Taking Advantage of Whole-Genome Resequencing to Track Down RKN Resistance Traits
7. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AFLPs | Amplified fragment length polymorphisms |
| AOC | Allene oxide cyclase |
| ARF | Auxin response factors |
| AtAAP6 | Arabidopsis thaliana Amino Acid Permease 6 |
| AtHIPP27 | Arabidopsis thaliana Heavy Metal-Associated Isoprenylated Plant Protein 27 |
| BZIP | Basic leucine zipper |
| BZIP60 | Basic leucine zipper 60 |
| CAPS | Cleaved amplified polymorphic sequence |
| CNVs | Copy number variants |
| COI-JAZ | Coronatine insensitive 1-jasmonate zim-domain |
| CRISPR/Cas9 | Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 |
| CsMS | Cucumis sativus malate synthase |
| DAMPs | Damage-associated molecular pattern |
| ddRAD-seq | Double-digested restriction site-association DNA sequencing |
| DEGs | Differentially expressed genes |
| DUF538 | Domain of unknown function538 |
| EMS | Ethyl methanesulfonate |
| ET | Ethylene |
| ETI | Effector-triggered immunity |
| GBS | Genotyping-by-sequencing |
| GEBVs | Genomic estimated breeding values |
| GhDIR4 | Gossypium hirsutum dirigent protein4 |
| GhPRXIIB | Gossypium hirsutum peroxiredoxin 11B |
| GmLMM1 | Glycine max lipid metabolism modulator 1 |
| GS | Genomic selection |
| GT-3A | Trihelix transcription factor GT-3a |
| GWAS | Genome-wide association mapping |
| JA | Jasmonic acid |
| J2 | Second stage RKN juveniles |
| MAPK | Mitogen-activated protein kinase |
| MAGIC | Multi-parent advanced generation intercross |
| MAS | Marker-assisted selection |
| MeJA | Methyl jasmonate |
| MiMsp40 | M. incognita esophageal gland cell secretory protrein40 |
| ML | Machine learning |
| MiPFN3 | Meloidogyne incognita profilin 3 |
| ML | Machine learning |
| MPKs | Mitogen-activated protein kinases |
| MYB | Myeloblastosis |
| NAM | Nested association mapping |
| NBS-LRR | Nucleotide-binding site—leucine-rich repeat |
| NGS | Next-generation sequencing |
| NLR | Nucleotide-binding leucine-rich repeat |
| NPR1 | Non-expressor of pathogenesis-related genes-1 |
| NtRK1 | Nicotiana tabacum Receptor Kinase 1 |
| OsHPP04 | Oryza sativa copper metallochaperone heavy metal-associated plant protein 04 |
| OsLOX7 | Oryza sativa lipoxygenase 7 |
| ONT | Oxford nanopore technologies |
| OsThion2 | Oryza sativa thionin2 |
| PacBio | Pacific Biosciences |
| PAL | Phenylalanine ammonia-lyase |
| PAMPs | Pathogen-associated molecular patterns |
| PBL | Plant bap-like |
| PCD | Programmed cell death protein |
| PI-II | Proteinase Inhibitor II |
| PLA2 | Phospholipase A2 |
| PR | Pathogenesis-related |
| PRR | Pattern recognition receptors |
| PR5K | Pathogenesis-related 5-like receptor kinase |
| PTI | Pattern-triggered immunity |
| QTL | Quantitative trait locus |
| RALFs | Rapid alkalinization factors |
| RALPs | Restriction amplified length polymorphisms |
| RAPD | Random amplified polymorphic DNA |
| RBM2 | RNA-binding motif protein 2 |
| RBM3 | RNA-binding motif protein 3 |
| RF | Random forest |
| RIL | Recombinant inbred lines |
| ROS | Reactive oxygen species |
| RKN | Root-knot nematode |
| RT-PCR | Reverse-transcription polymerase chain reaction |
| RNAi | RNA interference |
| SA | Salicylic acid |
| SCAR | Sequence characterized amplified region |
| SCL | SCARECROW-like |
| SlARF8A | Solanum lycopersicum auxin response factor 8A |
| SNPs | Single nucleotide polymorphisms |
| Spr2 | SPIRAL2 |
| SSR | Single sequence repeats |
| SVM | Support vector machine |
| TFs | Transcription factors |
| TIR-NBS-LRR | Toll/interleukin-1 receptor—nucleotide-binding site–leucine-rich repeat |
| TM-1 | Texas marker-1 |
| TUB-1 | Tubilin-1 |
| TS | Training set |
| VQ | Valine–glutamine |
| VS | Validation set |
| WGCNA | Weighted gene co-expression network analysis |
| WGR | Whole-genome resequencing |
| WMJ | Wild Mexican Jones |
| WRKY | Tryptophan–arginine–lysine–tyrosine (WRKY) |
References
- Somasekhar, N.; Prasad, J.S. Plant–nematode interactions: Consequences of climate change. In Crop Stress and Its Management: Perspectives and Strategies; Springer: Berlin/Heidelberg, Germany, 2011; pp. 547–564. [Google Scholar] [CrossRef]
- Mendy, B.; Wang’ombe, M.W.; Radakovic, Z.S.; Holbein, J.; Ilyas, M.; Chopra, D.; Holton, N.; Zipfel, C.; Grundler, F.M.; Siddique, S. Arabidopsis leucine-rich repeat receptor–like kinase NILR1 is required for induction of innate immunity to parasitic nematodes. PLoS Pathog. 2017, 13, e1006284. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, J.; Barcala, M.; Fenoll, C.; Escobar, C. The power of omics to identify plant susceptibility factors and to study resistance to root-knot nematodes. Curr. Issues Mol. Biol. 2016, 19, 53–72. [Google Scholar] [CrossRef] [PubMed]
- Forghani, F.; Hajihassani, A. Recent advances in the development of environmentally benign treatments to control root-knot nematodes. Front. Plant Sci. 2020, 11, 1125. [Google Scholar] [CrossRef]
- Wram, C.L.; Zasada, I.A. Short-term effects of sublethal doses of nematicides on Meloidogyne incognita. Phytopathology 2019, 109, 1605–1613. [Google Scholar] [CrossRef]
- Sikora, R.A.; Roberts, P.A. Management practices: An overview of integrated nematode management technologie. Plant Parasit. Nematodes Subtrop. Trop. Agric. 2018, 795–838. [Google Scholar] [CrossRef]
- Rodiuc, N.; Vieira, P.; Banora, M.Y.; de Almeida Engler, J. On the track of transfer cell formation by specialized plant-parasitic nematodes. Front. Plant Sci. 2014, 5, 160. [Google Scholar] [CrossRef] [PubMed]
- Rutter, W.B.; Franco, J.; Gleason, C. Rooting out the mechanisms of root-knot nematode–plant interactions. Annu. Rev. Phytopathol. 2022, 60, 43–76. [Google Scholar] [CrossRef]
- Gheysen, G.; Mitchum, M.G. Phytoparasitic nematode control of plant hormone pathways. Plant Physiol. 2019, 179, 1212–1226. [Google Scholar] [CrossRef]
- Karczmarek, A.; Overmars, H.; Helder, J.; Goverse, A. Feeding cell development by cyst and root-knot nematodes involves a similar early, local and transient activation of a specific auxin-inducible promoter element. Mol. Plant Pathol. 2004, 5, 343–346. [Google Scholar] [CrossRef]
- Grunewald, W.; Cannoot, B.; Friml, J.; Gheysen, G. Parasitic nematodes modulate PIN-mediated auxin transport to facilitate infection. PLoS Pathog. 2009, 5, e1000266. [Google Scholar] [CrossRef]
- Lohar, D.P.; Schaff, J.E.; Laskey, J.G.; Kieber, J.J.; Bilyeu, K.D.; Bird, D.M. Cytokinins play opposite roles in lateral root formation, and nematode and rhizobial symbioses. Plant J. 2004, 38, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Medina, A.; Appels, F.V.; van Wees, S.C. Impact of salicylic acid-and jasmonic acid-regulated defences on root colonization by Trichoderma harzianum T-78. Plant Signal. Behav. 2017, 12, e1345404. [Google Scholar] [CrossRef] [PubMed]
- Molinari, S.; Fanelli, E.; Leonetti, P. Expression of tomato salicylic acid (SA)-responsive pathogenesis-related genes in Mi-1-mediated and SA-induced resistance to root-knot nematodes. Mol. Plant Pathol. 2014, 15, 255–264. [Google Scholar] [CrossRef]
- Sanz-Alférez, S.; Mateos, B.; Alvarado, R.; Sánchez, M. SAR induction in tomato plants is not effective against root-knot nematode infection. Eur. J. Plant Pathol. 2008, 120, 417–425. [Google Scholar] [CrossRef]
- Shukla, N.; Yadav, R.; Kaur, P.; Rasmussen, S.; Goel, S.; Agarwal, M.; Jagannath, A.; Gupta, R.; Kumar, A. Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-infected tomato (Solanum lycopersicum) roots reveals complex gene expression profiles and metabolic networks of both host and nematode during susceptible and resistance responses. Mol. Plant Pathol. 2018, 19, 615–633. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, T.; Tomitaka, Y.; Abe, H.; Tsuda, S.; Futai, K.; Mizukubo, T. Expression profile of jasmonic acid-induced genes and the induced resistance against the root-knot nematode (Meloidogyne incognita) in tomato plants (Solanum lycopersicum) after foliar treatment with methyl jasmonate. J. Plant Physiol. 2011, 168, 1084–1097. [Google Scholar] [CrossRef]
- Gleason, C.; Leelarasamee, N.; Meldau, D.; Feussner, I. OPDA has key role in regulating plant susceptibility to the root-knot nematode Meloidogyne hapla in Arabidopsis. Front. Plant Sci. 2016, 7, 1565. [Google Scholar] [CrossRef]
- Fan, J.; Hu, C.; Zhang, L.; Li, Z.; Zhao, F.; Wang, S. Jasmonic acid mediates tomato’s response to root knot nematodes. J. Plant Growth Regul. 2015, 34, 196–205. [Google Scholar] [CrossRef]
- Kyndt, T.; Nahar, K.; Haeck, A.; Verbeek, R.; Demeestere, K.; Gheysen, G. Interplay between carotenoids, abscisic acid and jasmonate guides the compatible rice-Meloidogyne graminicola interaction. Front. Plant Sci. 2017, 8, 951. [Google Scholar] [CrossRef]
- Wang, G.; Hu, C.; Zhou, J.; Liu, Y.; Cai, J.; Pan, C.; Wang, Y.; Wu, X.; Shi, K.; Xia, X.; et al. Systemic Root-Shoot Signaling Drives Jasmonate-Based Root Defense against Nematodes. Curr. Biol. 2019, 29, 3430–3438.e3434. [Google Scholar] [CrossRef]
- Huang, H.; Ma, X.; Sun, L.; Wang, Y.; Ma, J.; Hong, Y.; Zhao, M.; Zhao, W.; Yang, R.; Song, S. SlVQ15 recruits SlWRKY30IIc to link with jasmonate pathway in regulating tomato defence against root-knot nematodes. Plant Biotechnol. J. 2025, 23, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Gahoi, S.; Gautam, B. Genome-wide analysis of excretory/secretory proteins in root-knot nematode, Meloidogyne incognita provides potential targets for parasite control. Comput. Biol. Chem. 2017, 67, 225–233. [Google Scholar] [CrossRef]
- Ranty-Roby, S. Identification and functional analysis of plant targets of effectors of the root-knot nematode Meloidogyne incognita. Ph.D. Thesis, Université Côte d’Azur, Nice, France, 2024. [Google Scholar]
- Wan, J.; Vuong, T.; Jiao, Y.; Joshi, T.; Zhang, H.; Xu, D.; Nguyen, H.T. Whole-genome gene expression profiling revealed genes and pathways potentially involved in regulating interactions of soybean with cyst nematode (Heterodera glycines Ichinohe). BMC Genom. 2015, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Liu, P.; Liu, Q.; Chen, C.; Guo, Q.; Yin, J.; Yang, G.; Jian, H. Msp40 effector of root-knot nematode manipulates plant immunity to facilitate parasitism. Sci. Rep. 2016, 6, 19443. [Google Scholar] [CrossRef]
- Xue, B.; Hamamouch, N.; Li, C.; Huang, G.; Hussey, R.S.; Baum, T.J.; Davis, E.L. The 8D05 parasitism gene of Meloidogyne incognita is required for successful infection of host roots. Phytopathology 2013, 103, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Soulé, S.; Huang, K.; Mulet, K.; Mejias, J.; Bazin, J.; Truong, N.M.; Kika, J.L.; Jaubert, S.; Abad, P.; Zhao, J. The root-knot nematode effector MiEFF12 targets the host ER quality control system to suppress immune responses and allow parasitism. Mol. Plant Pathol. 2024, 25, e13491. [Google Scholar] [CrossRef]
- Leelarasamee, N.; Zhang, L.; Gleason, C. The root-knot nematode effector MiPFN3 disrupts plant actin filaments and promotes parasitism. PLoS Pathog. 2018, 14, e1006947. [Google Scholar] [CrossRef]
- Huang, G.; Allen, R.; Davis, E.L.; Baum, T.J.; Hussey, R.S. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. USA 2006, 103, 14302–14306. [Google Scholar] [CrossRef]
- Huang, G.; Dong, R.; Allen, R.; Davis, E.L.; Baum, T.J.; Hussey, R.S. A root-knot nematode secretory peptide functions as a ligand for a plant transcription factor. Mol. Plant-Microbe Interact. 2006, 19, 463–470. [Google Scholar] [CrossRef]
- Jianlong, Z.; Kaiwei, H.; Rui, L.; Yuqing, L.; Pierre, A.; Bruno, F.; Heng, J.; Jian, L.; Yan, L.; Yuhong, Y.; et al. The root-knot nematode effector Mi2G02 hijacks a host plant trihelix transcription factor to promote nematode parasitism. Plant Commun. 2024, 5, 100723. [Google Scholar] [CrossRef]
- Nguyen, C.; Perfus-Barbeoch, L.; Quentin, M.; Zhao, J.; Magliano, M.; Marteu, N.; Da Rocha, M.; Nottet, N.; Abad, P.; Favery, B. A root-knot nematode small glycine and cysteine-rich secreted effector, MiSGCR1, is involved in plant parasitism. New Phytol. 2018, 217, 687–699. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lin, B.; Huang, Q.; Hu, L.; Zhuo, K.; Liao, J. A novel Meloidogyne graminicola effector, MgGPP, is secreted into host cells and undergoes glycosylation in concert with proteolysis to suppress plant defenses and promote parasitism. PLoS Pathog. 2017, 13, e1006301. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, K.; Chen, J.; Lin, B.; Wang, J.; Sun, F.; Hu, L.; Liao, J. A novel Meloidogyne enterolobii effector MeTCTP promotes parasitism by suppressing programmed cell death in host plants. Mol. Plant Pathol. 2017, 18, 45–54. [Google Scholar] [CrossRef]
- Navarrete, M.; Djian-Caporalino, C.; Mateille, T.; Palloix, A.; Sage-Palloix, A.-M.; Lefèvre, A.; Fazari, A.; Marteu, N.; Tavoillot, J.; Dufils, A. A resistant pepper used as a trap cover crop in vegetable production strongly decreases root-knot nematode infestation in soil. Agron. Sustain. Dev. 2016, 36, 68. [Google Scholar] [CrossRef]
- Ferris, H.; Griffiths, B.S.; Porazinska, D.L.; Powers, T.O.; Wang, K.-H.; Tenuta, M. Reflections on plant and soil nematode ecology: Past, present and future. J. Nematol. 2012, 44, 115. [Google Scholar] [PubMed]
- Desaeger, J.; Dickson, D.W.; Locascio, S. Methyl Bromide Alternatives for Control of Root-knot Nematode (spp.) in Tomato Production in Florida. J. Nematol. 2017, 49, 140–149. [Google Scholar] [CrossRef]
- Nelson, S.; Locascio, S.; Allen, L.; Dickson, D.; Mitchell, D. Soil flooding and fumigant alternatives to methyl bromide in tomato and eggplant production. HortScience 2002, 37, 1057–1060. [Google Scholar] [CrossRef]
- Osteen, C.D. Economic Implications of the Methyl Bromide Phaseout; United States Department of Agriculture, Economic Research Service, Agriculture Information Bulletin Numbere 756: Washington, DC, USA, 2000. [CrossRef]
- Su, L.; Ruan, Y.; Yang, X.; Wang, K.; Li, R.; Shen, Q. Suppression on plant-parasitic nematodes using a soil fumigation strategy based on ammonium bicarbonate and its effects on the nematode community. Sci. Rep. 2015, 5, 17597. [Google Scholar] [CrossRef]
- Behzadian, S.; Sahebani, N.; Karimi, S. Effectiveness of Plant-Induced Resistance Against Root-Knot Nematode Depends on the Policy of Using Inducer on the Host Plant. Curr. Microbiol. 2025, 82, 88. [Google Scholar] [CrossRef]
- Meyer, S.L.F.; Zasada, I.A.; Rupprecht, S.M.; VanGessel, M.J.; Hooks, C.R.R.; Morra, M.J.; Everts, K.L. Mustard Seed Meal for Management of Root-knot Nematode and Weeds in Tomato Production. HortTechnology Hortte 2015, 25, 192–202. [Google Scholar] [CrossRef]
- Tranier, M.-S.; Pognant-Gros, J.; Quiroz, R.D.l.C.; González, C.N.A.; Mateille, T.; Roussos, S. Commercial biological control agents targeted against plant-parasitic root-knot nematodes. Braz. Arch. Biol. Technol. 2014, 57, 831–841. [Google Scholar] [CrossRef]
- Goswami, B.K.; Pandey, R.K.; Rathour, K.S.; Bhattacharya, C.; Singh, L. Integrated application of some compatible biocontrol agents along with mustard oil seed cake and furadan on Meloidogyne incognita infecting tomato plants. J. Zhejiang Univ. Sci. B 2006, 7, 873–875. [Google Scholar] [CrossRef]
- Huang, X.; Zhao, N.; Zhang, K. Extracellular enzymes serving as virulence factors in nematophagous fungi involved in infection of the host. Res. Microbiol. 2004, 155, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, I. Effect of some biocides and entomopathogenic nematodes on suppressing root-knot nematode. Al-Azhar J. Agric. Res. 2023, 48, 319–330. [Google Scholar] [CrossRef]
- Ibrahim, H.M.M.; Ahmad, E.M.; Martínez-Medina, A.; Aly, M.A.M. Effective approaches to study the plant-root knot nematode interaction. Plant Physiol. Biochem. 2019, 141, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, R. Transgressive Segregation for Root-Knot Nematode Resistance in Cotton 1. Crop Sci. 1974, 14, 872–875. [Google Scholar] [CrossRef]
- Shepherd, R. Registration of Auburn 623 RNR cotton germplasm. (Reg. No. GP 20). Crop. Sci. 1974, 14, 911. [Google Scholar] [CrossRef]
- Kirkpatrick, T.L.; Rockroth, C. Compendium of Cotton Diseases; American Phytopathological Society (APS Press): St Paul, MN, USA, 2001. [Google Scholar]
- Wang, C.; Ulloa, M.; Roberts, P. Identification and mapping of microsatellite markers linked to a root-knot nematode resistance gene (rkn1) in Acala NemX cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 2006, 112, 770–777. [Google Scholar] [CrossRef]
- Ulloa, M.; Wang, C.; Saha, S.; Hutmacher, R.; Stelly, D.; Jenkins, J.; Burke, J.; Roberts, P. Analysis of root-knot nematode and fusarium wilt disease resistance in cotton (Gossypium spp.) using chromosome substitution lines from two alien species. Genetica 2016, 144, 167–179. [Google Scholar] [CrossRef]
- Wang, C.; Ulloa, M.; Roberts, P.A. A transgressive segregation factor (RKN2) in Gossypium barbadense for nematode resistance clusters with gene rkn1 in G. hirsutum. Mol. Genet. Genom. 2008, 279, 41–52. [Google Scholar] [CrossRef]
- Wang, C.; Ulloa, M.; Nichols, R.L.; Roberts, P.A. Sequence composition of bacterial chromosome clones in a transgressive root-knot nematode resistance chromosome region in tetraploid cotton. Front. Plant Sci. 2020, 11, 574486. [Google Scholar] [CrossRef] [PubMed]
- Roberts, P.A.; Ulloa, M.; Wang, C. Host plant resistance to root-knot nematode in cotton. In Proceedings of the Fourth World Cotton Research Conference 2007, Lubbock, TX, USA, 10–14 September 2007. [Google Scholar]
- Hu, H.; Yu, F. Embracing the Omics Era for Plant Breeding. Crop Breed. Genet. Genom. 2025, 7, e250002. [Google Scholar] [CrossRef]
- Lamichhane, S.; Thapa, S. Advances from conventional to modern plant breeding methodologies. Plant Breed. Biotechnol. 2022, 10, 1–4. [Google Scholar] [CrossRef]
- Ritchie, M.D.; Holzinger, E.R.; Li, R.; Pendergrass, S.A.; Kim, D. Methods of integrating data to uncover genotype–phenotype interactions. Nat. Rev. Genet. 2015, 16, 85–97. [Google Scholar] [CrossRef]
- Lappalainen, T.; Sammeth, M.; Friedländer, M.R.; ‘t Hoen, P.A.; Monlong, J.; Rivas, M.A.; Gonzalez-Porta, M.; Kurbatova, N.; Griebel, T.; Ferreira, P.G. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013, 501, 506–511. [Google Scholar] [CrossRef]
- Kumar, V.; Vats, S.; Kumawat, S.; Bisht, A.; Bhatt, V.; Shivaraj, S.; Padalkar, G.; Goyal, V.; Zargar, S.; Gupta, S. Omics advances and integrative approaches for the simultaneous improvement of seed oil and protein content in soybean (Glycine max L.). Crit. Rev. Plant Sci. 2021, 40, 398–421. [Google Scholar] [CrossRef]
- Morabito, A.; De Simone, G.; Pastorelli, R.; Brunelli, L.; Ferrario, M. Algorithms and tools for data-driven omics integration to achieve multilayer biological insights: A narrative review. J. Transl. Med. 2025, 23, 425. [Google Scholar] [CrossRef]
- Sanches, P.H.G.; de Melo, N.C.; Porcari, A.M.; de Carvalho, L.M. Integrating Molecular Perspectives: Strategies for Comprehensive Multi-Omics Integrative Data Analysis and Machine Learning Applications in Transcriptomics, Proteomics, and Metabolomics. Biology 2024, 13, 848. [Google Scholar] [CrossRef]
- Wang, M.; Li, R.; Zhao, Q. Multi-Omics Techniques in Genetic Studies and Breeding of Forest Plants. Forests 2023, 14, 1196. [Google Scholar] [CrossRef]
- Dimitriu, M.A.; Lazar-Contes, I.; Roszkowski, M.; Mansuy, I.M. Single-cell multiomics techniques: From conception to applications. Front. Cell Dev. Biol. 2022, 10, 854317. [Google Scholar] [CrossRef]
- Postnikova, O.A.; Hult, M.; Shao, J.; Skantar, A.; Nemchinov, L.G. Transcriptome analysis of resistant and susceptible alfalfa cultivars infected with root-knot nematode Meloidogyne incognita. PLoS ONE 2015, 10, e0118269. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yu, Y.; Haberer, G.; Marri, P.R.; Fan, C.; Goicoechea, J.L.; Zuccolo, A.; Song, X.; Kudrna, D.; Ammiraju, J.S. The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat. Genet. 2014, 46, 982–988. [Google Scholar] [CrossRef]
- Jammes, F.; Lecomte, P.; de Almeida-Engler, J.; Bitton, F.; Martin-Magniette, M.L.; Renou, J.P.; Abad, P.; Favery, B. Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis a. Plant J. 2005, 44, 447–458. [Google Scholar] [CrossRef]
- Petitot, A.-S.; Kyndt, T.; Haidar, R.; Dereeper, A.; Collin, M.; de Almeida Engler, J.; Gheysen, G.; Fernandez, D. Transcriptomic and histological responses of African rice (Oryza glaberrima) to Meloidogyne graminicola provide new insights into root-knot nematode resistance in monocots. Ann. Bot. 2017, 119, 885–899. [Google Scholar] [CrossRef]
- Ojeda-Rivera, J.O.; Ulloa, M.; Roberts, P.A.; Kottapalli, P.; Wang, C.; Nájera-González, H.-R.; Payton, P.; Lopez-Arredondo, D.; Herrera-Estrella, L. Root-Knot Nematode Resistance in Gossypium hirsutum Determined by a Constitutive Defense-Response Transcriptional Program Avoiding a Fitness Penalty. Front. Plant Sci. 2022, 13, 858313. [Google Scholar] [CrossRef] [PubMed]
- Khanal, S.; Kumar, P.; da Silva, M.B.; Singh, R.; Suassuna, N.; Jones, D.C.; Davis, R.F.; Chee, P.W. Time-course RNA-seq analysis of upland cotton (Gossypium hirsutum L.) responses to Southern root-knot nematode (Meloidogyne incognita) during compatible and incompatible interactions. BMC Genom. 2025, 26, 183. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.R.P.; Ndeve, A.D.; Huynh, B.-L.; Matthews, W.C.; Roberts, P.A. QTL mapping and transcriptome analysis of cowpea reveals candidate genes for root-knot nematode resistance. PLoS ONE 2018, 13, e0189185. [Google Scholar] [CrossRef]
- Ndeve, A.D.; Matthews, W.C.; Santos, J.R.; Huynh, B.L.; Roberts, P.A. Broad-based root-knot nematode resistance identified in cowpea gene-pool two. J. Nematol. 2018, 50, 545. [Google Scholar] [CrossRef]
- Ndeve, A.D.; Santos, J.R.; Matthews, W.C.; Huynh, B.L.; Guo, Y.-N.; Lo, S.; Muñoz-Amatriaín, M.; Roberts, P.A. A novel root-knot nematode resistance QTL on chromosome Vu01 in cowpea. G3 Genes Genomes Genet. 2019, 9, 1199–1209. [Google Scholar] [CrossRef]
- Das, S.; DeMason, D.A.; Ehlers, J.D.; Close, T.J.; Roberts, P.A. Histological characterization of root-knot nematode resistance in cowpea and its relation to reactive oxygen species modulation. J. Exp. Bot. 2008, 59, 1305–1313. [Google Scholar] [CrossRef]
- Das, S.; Ehlers, J.D.; Close, T.J.; Roberts, P.A. Transcriptional profiling of root-knot nematode induced feeding sites in cowpea (Vigna unguiculata L. Walp.) using a soybean genome array. BMC Genom. 2010, 11, 480. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Sakure, A.A.; Kumar, S.; Mishra, A.; Ahmad, S.; Rojasara, Y.M.; Vaja, M.B.; Patel, D.A. The Mi-1 gene is a key regulator of defence mechanisms and cellular gene dynamics in response to root-knot nematodes. Plant Cell Rep. 2025, 44, 96. [Google Scholar] [CrossRef] [PubMed]
- Amrine, K.C.; Blanco-Ulate, B.; Cantu, D. Discovery of core biotic stress responsive genes in Arabidopsis by weighted gene co-expression network analysis. PLoS ONE 2015, 10, e0118731. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, H.; Tan, J.; Huang, S.; Chen, X.; Jiang, D.; Xiao, X. Transcriptome analysis of eggplant root in response to root-knot nematode infection. Pathogens 2021, 10, 470. [Google Scholar] [CrossRef]
- Hu, W.; Kingsbury, K.; Mishra, S.; DiGennaro, P. A comprehensive transcriptional profiling of pepper responses to root-knot nematode. Genes 2020, 11, 1507. [Google Scholar] [CrossRef]
- Lee, I.H.; Shim, D.; Jeong, J.C.; Sung, Y.W.; Nam, K.J.; Yang, J.-W.; Ha, J.; Lee, J.J.; Kim, Y.-H. Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-resistant and susceptible sweetpotato cultivars. Planta 2019, 249, 431–444. [Google Scholar] [CrossRef]
- Li, X.; Xing, X.; Tian, P.; Zhang, M.; Huo, Z.; Zhao, K.; Liu, C.; Duan, D.; He, W.; Yang, T. Comparative transcriptome profiling reveals defense-related genes against Meloidogyne incognita invasion in tobacco. Molecules 2018, 23, 2081. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Li, X.; Zhang, M.; Wang, Y.; Liu, B.; Xi, Q.; Zhao, K.; Wu, Y.; Yang, T. Transcriptome analysis of resistant and susceptible tobacco (Nicotiana tabacum) in response to root-knot nematode Meloidogyne incognita infection. Biochem. Biophys. Res. Commun. 2017, 482, 1114–1121. [Google Scholar] [CrossRef]
- Arraes, F.B.; Vasquez, D.D.; Tahir, M.; Pinheiro, D.H.; Faheem, M.; Freitas-Alves, N.S.; Moreira-Pinto, C.E.; Moreira, V.J.; Paes-de-Melo, B.; Lisei-de-Sa, M.E. Integrated Omic Approaches Reveal Molecular Mechanisms of Tolerance during Soybean and Meloidogyne incognita Interactions. Plants 2022, 11, 2744. [Google Scholar] [CrossRef]
- Cao, K.; Li, H.; Wang, Q.; Zhao, P.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wang, L. Comparative transcriptome analysis of genes involved in the response of resistant and susceptible peach cultivars to nematode infection. Sci. Hortic. 2017, 215, 20–27. [Google Scholar] [CrossRef]
- Kumar, P.; Khanal, S.; Da Silva, M.; Singh, R.; Davis, R.F.; Nichols, R.L.; Chee, P.W. Transcriptome analysis of a nematode resistant and susceptible upland cotton line at two critical stages of Meloidogyne incognita infection and development. PLoS ONE 2019, 14, e0221328. [Google Scholar] [CrossRef] [PubMed]
- Kumari, C.; Dutta, T.K.; Banakar, P.; Rao, U. Comparing the defence-related gene expression changes upon root-knot nematode attack in susceptible versus resistant cultivars of rice. Sci. Rep. 2016, 6, 22846. [Google Scholar] [CrossRef] [PubMed]
- Klink, V.P.; Alkharouf, N.W.; Lawrence, K.S.; Lawaju, B.R.; Sharma, K.; Niraula, P.M.; McNeece, B.T. The heterologous expression of conserved Glycine max (soybean) mitogen activated protein kinase 3 (MAPK3) paralogs suppresses Meloidogyne incognita parasitism in Gossypium hirsutum (upland cotton). Transgenic Res. 2022, 31, 457–487. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Zhao, W.; Qiao, H.; Li, C.; Sun, L.; Yang, R.; Ma, X.; Ma, J.; Song, S.; Wang, S. SlWRKY45 interacts with jasmonate-ZIM domain proteins to negatively regulate defense against the root-knot nematode Meloidogyne incognita in tomato. Hortic. Res. 2022, 9, uhac197. [Google Scholar] [CrossRef]
- Dutta, T.K.; Rupinikrishna, K.; Akhil, V.S.; Vashisth, N.; Phani, V.; Pankaj; Sirohi, A.; Chinnusamy, V. CRISPR/Cas9-induced knockout of an amino acid permease gene (AAP6) reduced Arabidopsis thaliana susceptibility to Meloidogyne incognita. BMC Plant Biol. 2024, 24, 515. [Google Scholar] [CrossRef]
- Milligan, S.B.; Bodeau, J.; Yaghoobi, J.; Kaloshian, I.; Zabel, P.; Williamson, V.M. The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 1998, 10, 1307–1319. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, Y.; Wang, J.; Ling, X.; Hu, Z.; Liu, T.; Chen, T.; Zhang, W. A CC-NBS-LRR type gene GHNTR1 confers resistance to southern root-knot nematode in Nicotiana. benthamiana and Nicotiana. tabacum. Eur. J. Plant Pathol. 2015, 142, 715–729. [Google Scholar] [CrossRef]
- Xiao, K.; Zhu, H.; Zhu, X.; Liu, Z.; Wang, Y.; Pu, W.; Guan, P.; Hu, J. Overexpression of PsoRPM3, an NBS-LRR gene isolated from myrobalan plum, confers resistance to Meloidogyne incognita in tobacco. Plant Mol. Biol. 2021, 107, 129–146. [Google Scholar] [CrossRef]
- Zhu, X.; Xiao, K.; Cui, H.; Hu, J. Overexpression of the Prunus sogdiana NBS-LRR Subgroup Gene PsoRPM2 Promotes Resistance to the Root-Knot Nematode Meloidogyne incognita in Tobacco. Front. Microbiol. 2017, 8, 2113. [Google Scholar] [CrossRef]
- Goggin, F.L.; Jia, L.; Shah, G.; Hebert, S.; Williamson, V.M.; Ullman, D.E. Heterologous expression of the Mi-1.2 gene from tomato confers resistance against nematodes but not aphids in eggplant. Mol. Plant-Microbe Interact. 2006, 19, 383–388. [Google Scholar] [CrossRef]
- Williamson, V.M.; Kumar, A. Nematode resistance in plants: The battle underground. Trends Genet. 2006, 22, 396–403. [Google Scholar] [CrossRef]
- Pant, S.R.; McNeece, B.T.; Sharma, K.; Niruala, P.; Burson, H.E.; Lawrence, G.W.; Klink, V.P. The heterologous expression of a Glycine max homolog of NONEXPRESSOR OF PR1 (NPR1) and α-hydroxynitrile glucosidase suppresses parasitism by the root pathogen Meloidogyne incognita in Gossypium hirsutum. J. Plant Interact. 2016, 11, 41–52. [Google Scholar] [CrossRef]
- Priya, D.B.; Somasekhar, N.; Prasad, J.; Kirti, P. Transgenic tobacco plants constitutively expressing Arabidopsis NPR1 show enhanced resistance to root-knot nematode, Meloidogyne incognita. BMC Res. Notes 2011, 4, 231. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, Z.; Jiang, Z.; Chen, X.; Li, B.; Xu, L.; Zhang, Z. Cloning and functional analysis of the root-knot nematode resistance gene NtRk1 in tobacco. Physiol. Plant. 2023, 175, e13894. [Google Scholar] [CrossRef]
- Chinnapandi, B.; Bucki, P.; Fitoussi, N.; Kolomiets, M.; Borrego, E.; Braun Miyara, S. Tomato SlWRKY3 acts as a positive regulator for resistance against the root-knot nematode Meloidogyne javanica by activating lipids and hormone-mediated defense-signaling pathways. Plant Signal. Behav. 2019, 14, 1601951. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, C.; Carmo, L.S.; Távora, F.T.; Lima, R.; da Nobrega Mendes, P.; de Sá, M.E.L.; Grossi-de-Sa, M.F.; Mehta, A. Overexpression of cotton genes GhDIR4 and GhPRXIIB in Arabidopsis thaliana improves plant resistance to root-knot nematode (Meloidogyne incognita) infection. 3 Biotech 2022, 12, 211. [Google Scholar] [CrossRef] [PubMed]
- Araujo, A.C.G.; Guimaraes, P.M.; Mota, A.P.Z.; Guimaraes, L.A.; Pereira, B.M.; Vinson, C.C.; Lacerda, A.L.; Martins, A.C.Q.; Brasileiro, A.C.M. Overexpression of DUF538 from wild Arachis enhances plant resistance to Meloidogyne spp. Agronomy 2021, 11, 559. [Google Scholar] [CrossRef]
- Lilley, C.J.; Urwin, P.E.; Johnston, K.A.; Atkinson, H.J. Preferential expression of a plant cystatin at nematode feeding sites confers resistance to Meloidogyne incognita and Globodera pallida. Plant Biotechnol. J. 2004, 2, 3–12. [Google Scholar] [CrossRef]
- Chan, Y.-L.; Yang, A.-H.; Chen, J.-T.; Yeh, K.-W.; Chan, M.-T. Heterologous expression of taro cystatin protects transgenic tomato against Meloidogyne incognita infection by means of interfering sex determination and suppressing gall formation. Plant Cell Rep. 2010, 29, 231–238. [Google Scholar] [CrossRef]
- Chan, Y.-L.; He, Y.; Hsiao, T.-T.; Wang, C.-J.; Tian, Z.; Yeh, K.-W. Pyramiding taro cystatin and fungal chitinase genes driven by a synthetic promoter enhances resistance in tomato to root-knot nematode Meloidogyne incognita. Plant Sci. 2015, 231, 74–81. [Google Scholar] [CrossRef]
- Papolu, P.K.; Dutta, T.K.; Tyagi, N.; Urwin, P.E.; Lilley, C.J.; Rao, U. Expression of a Cystatin Transgene in Eggplant Provides Resistance to Root-knot Nematode, Meloidogyne incognita. Front. Plant Sci. 2016, 7, 1122. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, M.; Zhang, Y.; Peng, L.; Dai, D.; Zhang, F.; Zhang, J. Efficient control of root-knot nematodes by expressing Bt nematicidal proteins in root leucoplasts. Mol. Plant 2024, 17, 1504–1519. [Google Scholar] [CrossRef]
- Li, Y.; Wu, X.; Zhang, Y.; Zhang, Q. CRISPR/Cas genome editing improves abiotic and biotic stress tolerance of crops. Front. Genome Ed. 2022, 4, 987817. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.S.-E.; Mukhtar, M.S.; Mansoor, S. Genome editing: Targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 2018, 36, 898–906. [Google Scholar] [CrossRef]
- Huang, Q.; Lin, B.; Cao, Y.; Zhang, Y.; Song, H.; Huang, C.; Sun, T.; Long, C.; Liao, J.; Zhuo, K. CRISPR/Cas9-mediated mutagenesis of the susceptibility gene OsHPP04 in rice confers enhanced resistance to rice root-knot nematode. Front. Plant Sci. 2023, 14, 1134653. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, D.; Chen, J.; Wu, D.; Feng, X.; Yu, F. Nematode RALF-like 1 targets soybean malectin-like receptor kinase to facilitate parasitism. Front. Plant Sci. 2021, 12, 775508. [Google Scholar] [CrossRef]
- Zhang, X.; Li, S.; Li, X.; Song, M.; Ma, S.; Tian, Y.; Gao, L. Peat-based hairy root transformation using Rhizobium rhizogenes as a rapid and efficient tool for easily exploring potential genes related to root-knot nematode parasitism and host response. Plant Methods 2023, 19, 22. [Google Scholar] [CrossRef]
- Noureddine, Y.; da Rocha, M.; An, J.; Médina, C.; Mejias, J.; Mulet, K.; Quentin, M.; Abad, P.; Zouine, M.; Favery, B. AUXIN RESPONSIVE FACTOR8 regulates development of the feeding site induced by root-knot nematodes in tomato. J. Exp. Bot. 2023, 74, 5752–5766. [Google Scholar] [CrossRef] [PubMed]
- Dutta, T.K.; Vashisth, N.; Ray, S.; Phani, V.; Chinnusamy, V.; Sirohi, A. Functional analysis of a susceptibility gene (HIPP27) in the Arabidopsis thaliana–Meloidogyne incognita pathosystem by using a genome editing strategy. BMC Plant Biol. 2023, 23, 390. [Google Scholar] [CrossRef]
- Saini, H.; Devrani, A.; Synrem, G.; Priyanka. Application of CRISPR Technology in Plant Improvement: An Update Review. Adv. Agric. 2025, 2025, 4578877. [Google Scholar] [CrossRef]
- El-Sappah, A.H.; M., I.M.; El-Awady, H.H.; Yan, S.; Qi, S.; Liu, J.; Cheng, G.-T.; Liang, Y. Tomato Natural Resistance Genes in Controlling the Root-Knot Nematode. Genes 2019, 10, 925. [Google Scholar] [CrossRef]
- Takeda, S.; Matsuoka, M. Genetic approaches to crop improvement: Responding to environmental and population changes. Nat. Rev. Genet. 2008, 9, 444–457. [Google Scholar] [CrossRef]
- Anand, A.; Subramanian, M.; Kar, D. Breeding techniques to dispense higher genetic gains. Front. Plant Sci. 2022, 13, 1076094. [Google Scholar] [CrossRef]
- Abd-Elgawad, M.M.M. Understanding Molecular Plant–Nematode Interactions to Develop Alternative Approaches for Nematode Control. Plants 2022, 11, 2141. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-S.; Vuong, T.D.; Qiu, D.; Robbins, R.T.; Grover Shannon, J.; Li, Z.; Nguyen, H.T. Advancements in breeding, genetics, and genomics for resistance to three nematode species in soybean. Theor. Appl. Genet. 2016, 129, 2295–2311. [Google Scholar] [CrossRef]
- Zheng, W.; Li, S.; Liu, Z.; Zhou, Q.; Feng, Y.; Chai, S. Molecular marker assisted gene stacking for disease resistance and quality genes in the dwarf mutant of an elite common wheat cultivar Xiaoyan22. BMC Genet. 2020, 21, 45. [Google Scholar] [CrossRef]
- Luo, W.; Guo, T.; Yang, Q.; Wang, H.; Liu, Y.; Zhu, X.; Chen, Z. Stacking of five favorable alleles for amylase content, fragrance and disease resistance into elite lines in rice (Oryza sativa) by using four HRM-based markers and a linked gel-based marker. Mol. Breed. 2014, 34, 805–815. [Google Scholar] [CrossRef]
- Eagles, H.A.; Bariana, H.S.; Ogbonnaya, F.C.; Rebetzke, G.J.; Hollamby, G.; Henry, R.J.; Henschke, P.; Carter, M. Implementation of markers in Australian wheat breeding. Aust. J. Agric. Res. 2001, 52, 1349–1356. [Google Scholar] [CrossRef]
- Simko, I.; Jia, M.; Venkatesh, J.; Kang, B.-C.; Weng, Y.; Barcaccia, G.; Lanteri, S.; Bhattarai, G.; Foolad, M.R. Genomics and marker-assisted improvement of vegetable crops. Crit. Rev. Plant Sci. 2021, 40, 303–365. [Google Scholar] [CrossRef]
- Banu, J.G.; Sankari Meena, K.; Selvi, C.; Manickam, S. Molecular marker-assisted selection for nematode resistance in crop plants. J. Entomol. Zool. Stud. 2017, 5, 1307–1311. [Google Scholar]
- Seifi, A.; Kaloshian, I.; Vossen, J.; Che, D.; Bhattarai, K.K.; Fan, J.; Naher, Z.; Goverse, A.; Tjallingii, W.F.; Lindhout, P. Linked, if not the same, Mi-1 homologues confer resistance to tomato powdery mildew and root-knot nematodes. Mol. Plant-Microbe Interact. 2011, 24, 441–450. [Google Scholar] [CrossRef]
- El-Mehrach, K.; Hatimi, A.; Chouchane, S.; Salus, M.; Martin, C.; Maxwell, D.; Mejia, L.; Williamson, V.; Vidavski, F. PCR-based methods for tagging the Mi-1 locus for resistance to root-knot nematode in begomovirus-resistant tomato germplasm. Acta Hortic. 2005, 695, 263–270. [Google Scholar] [CrossRef]
- Jablonska, B.; Ammiraju, J.S.; Bhattarai, K.K.; Mantelin, S.; de Ilarduya, O.M.; Roberts, P.A.; Kaloshian, I. The Mi-9 gene from Solanum arcanum conferring heat-stable resistance to root-knot nematodes is a homolog of Mi-1. Plant Physiol. 2007, 143, 1044–1054. [Google Scholar] [CrossRef] [PubMed]
- Ynturi, P.; Jenkins, J.N.; McCarty, J.C., Jr.; Gutierrez, O.A.; Saha, S. Association of root-knot nematode resistance genes with simple sequence repeat markers on two chromosomes in cotton. Crop Sci. 2006, 46, 2670–2674. [Google Scholar] [CrossRef][Green Version]
- Wang, C.; Roberts, P.A. Development of AFLP and derived CAPS markers for root-knot nematode resistance in cotton. Euphytica 2006, 152, 185–196. [Google Scholar] [CrossRef]
- Gutiérrez, O.A.; Jenkins, J.N.; McCarty, J.C.; Wubben, M.J.; Hayes, R.W.; Callahan, F.E. SSR markers closely associated with genes for resistance to root-knot nematode on chromosomes 11 and 14 of Upland cotton. Theor. Appl. Genet. 2010, 121, 1323–1337. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, J.N.; McCarty, J.C.; Wubben, M.J.; Hayes, R.; Gutierrez, O.A.; Callahan, F.; Deng, D. SSR markers for marker assisted selection of root-knot nematode (Meloidogyne incognita) resistant plants in cotton (Gossypium hirsutum L). Euphytica 2012, 183, 49–54. [Google Scholar] [CrossRef]
- DEVRAN, Z.; Sogut, M. Response of heat-stable tomato genotypes to Mi-1 virulent root-knot nematode populations. Turk. Entomoloji Derg.-Turk. J. Entomol. 2014, 38, 229–238. [Google Scholar] [CrossRef][Green Version]
- Devran, Z.; Süğüt, M.; Goezel, U.; Toer, M.; Elekcioglu, I.H. Analysis of genetic variation between populations of Meloidogyne spp. from Turkey. Russ. J. Nematol. 2008, 16, 143–149. [Google Scholar]
- Williamson, V.; Ho, J.-Y.; Wu, F.; Miller, N.; Kaloshian, I. A PCR-based marker tightly linked to the nematode resistance gene, Mi, in tomato. Theor. Appl. Genet. 1994, 87, 757–763. [Google Scholar] [CrossRef]
- Devran, Z.; Firat, A.F.; Tör, M.; Mutlu, N.; Elekçioğlu, I.H. AFLP and SRAP markers linked to the mj gene for root-knot nematode resistance in cucumber. Sci. Agric. 2011, 68, 115–119. [Google Scholar] [CrossRef]
- Mohanta, S.; Swain, P.; Sial, P.; Rout, G. Morphological and molecular screening of turmeric (Curcuma longa L.) cultivars for resistance against parasitic nematode, Meloidogyne incognita. J. Plant Pathol. Microbiol. 2015, 6, 1. [Google Scholar] [CrossRef]
- Kumar, P.; He, Y.; Singh, R.; Davis, R.F.; Guo, H.; Paterson, A.H.; Peterson, D.G.; Shen, X.; Nichols, R.L.; Chee, P.W. Fine mapping and identification of candidate genes for a QTL affecting Meloidogyne incognita reproduction in Upland cotton. BMC Genom. 2016, 17, 567. [Google Scholar] [CrossRef]
- Carpentieri-Pípolo, V.; Gallo-Meagher, M.; Dickson, D.W.; Gorbet, D.W.; de Lurdes Mendes, M.; de Souza, S.H. Molecular marker screening of peanut (Arachis hypogaea L.) germplasm for Meloidogyne arenaria resistance. Afr. J. Biotechnol. 2014, 13, 2608–2612. [Google Scholar] [CrossRef]
- Chu, Y.; Wu, C.; Holbrook, C.; Tillman, B.; Person, G.; Ozias-Akins, P. Marker-assisted selection to pyramid nematode resistance and the high oleic trait in peanut. Plant Genome 2011, 4, 110–117. [Google Scholar] [CrossRef]
- Ramzan, M.; Ahmed, R.Z.; Khanum, T.A.; Akram, S.; Jabeen, S. Survey of root knot nematodes and RMi resistance to Meloidogyne incognita in soybean from Khyber Pakhtunkhwa, Pakistan. Eur. J. Plant Pathol. 2021, 160, 1–13. [Google Scholar] [CrossRef]
- Arunakumar, G.S.; Gnanesh, B.N.; Manojkumar, H.B.; Doss, S.G.; Mogili, T.; Sivaprasad, V.; Tewary, P. Genetic diversity, identification, and utilization of novel genetic resources for resistance to Meloidogyne incognita in mulberry (Morus spp.). Plant Dis. 2021, 105, 2919–2928. [Google Scholar] [CrossRef]
- Boiteux, L.; Hyman, J.; Bach, I.C.; Fonseca, M.; Matthews, W.; Roberts, P.; Simon, P. Employment of flanking codominant STS markers to estimate allelic substitution effects of a nematode resistance locus in carrot. Euphytica 2004, 136, 37–44. [Google Scholar] [CrossRef]
- Manisha; Padmini, K.; Umamaheswari, R.; Reddy, D.C.L.; Dhananjaya, M.V.; Rao, V.K. Evaluation of a resistant line of tropical carrot to root-knot nematode Meloidogyne incognita using conventional method and molecular markers. Eur. J. Plant Pathol. 2024, 168, 363–371. [Google Scholar] [CrossRef]
- Djian-Caporalino, C.; Pijarowski, L.; Fazari, A.; Samson, M.; Gaveau, L.; O’byrne, C.; Lefebvre, V.; Caranta, C.; Palloix, A.; Abad, P. High-resolution genetic mapping of the pepper (Capsicum annuum L.) resistance loci Me3 and Me4 conferring heat-stable resistance to root-knot nematodes (Meloidogyne spp.). Theor. Appl. Genet. 2001, 103, 592–600. [Google Scholar] [CrossRef]
- Wang, C.; Ulloa, M.; Mullens, T.R.; Yu, J.Z.; Roberts, P.A. QTL analysis for transgressive resistance to root-knot nematode in interspecific cotton (Gossypium spp.) progeny derived from susceptible parents. PLoS ONE 2012, 7, e34874. [Google Scholar] [CrossRef][Green Version]
- Shen, X.; Van Becelaere, G.; Kumar, P.; Davis, R.F.; May, O.L.; Chee, P. QTL mapping for resistance to root-knot nematodes in the M-120 RNR Upland cotton line (Gossypium hirsutum L.) of the Auburn 623 RNR source. Theor. Appl. Genet. 2006, 113, 1539–1549. [Google Scholar] [CrossRef] [PubMed]
- Roberts, P.; Ulloa, M. Introgression of Root-Knot Nematode Resistance into Tetraploid Cottons. Crop Sci. 2010, 50, 940–951. [Google Scholar] [CrossRef]
- Wubben, M.J.; Callahan, F.E.; Jenkins, J.N.; Deng, D.D. Coupling of MIC-3 overexpression with the chromosomes 11 and 14 root-knot nematode (RKN) (Meloidogyne incognita) resistance QTLs provides insights into the regulation of the RKN resistance response in Upland cotton (Gossypium hirsutum). Theor. Appl. Genet. 2016, 129, 1759–1767. [Google Scholar] [CrossRef] [PubMed]
- Gaudin, A.G.; Wubben, M.J. Genotypic and phenotypic evaluation of wild cotton accessions previously identified as resistant to root-knot (Meloidogyne incognita) or reniform nematode (Rotylenchulus reniformis). Euphytica 2021, 217, 207. [Google Scholar] [CrossRef]
- Ali, A.; Matthews, W.C.; Cavagnaro, P.F.; Iorizzo, M.; Roberts, P.A.; Simon, P.W. Inheritance and mapping of Mj-2, a new source of root-knot nematode (Meloidogyne javanica) resistance in carrot. J. Hered. 2014, 105, 288–291. [Google Scholar] [CrossRef] [PubMed]
- Parsons, J.; Matthews, W.; Iorizzo, M.; Roberts, P.; Simon, P. Meloidogyne incognita nematode resistance QTL in carrot. Mol. Breed. 2015, 35, 114. [Google Scholar] [CrossRef]
- Davis, R.F.; Harris-Shultz, K.; Knoll, J.E.; Krakowsky, M.; Scully, B. A Quantitative Trait Locus on Maize Chromosome 5 Is Associated with Root-Knot Nematode Resistance. Phytopathology® 2024, 114, 1657–1663. [Google Scholar] [CrossRef]
- Oloka, B.M.; da Silva Pereira, G.; Amankwaah, V.A.; Mollinari, M.; Pecota, K.V.; Yada, B.; Olukolu, B.A.; Zeng, Z.-B.; Craig Yencho, G. Discovery of a major QTL for root-knot nematode (Meloidogyne incognita) resistance in cultivated sweetpotato (Ipomoea batatas). Theor. Appl. Genet. 2021, 134, 1945–1955. [Google Scholar] [CrossRef]
- Leal-Bertioli, S.C.; Moretzsohn, M.C.; Roberts, P.A.; Ballén-Taborda, C.; Borba, T.C.; Valdisser, P.A.; Vianello, R.P.; Araújo, A.C.G.; Guimarães, P.M.; Bertioli, D.J. Genetic mapping of resistance to Meloidogyne arenaria in Arachis stenosperma: A new source of nematode resistance for peanut. G3 Genes Genomes Genet. 2016, 6, 377–390. [Google Scholar] [CrossRef]
- Xie, X.; Ling, J.; Lu, J.; Mao, Z.; Zhao, J.; Zheng, S.; Yang, Q.; Li, Y.; Visser, R.G.; Bai, Y. Genetic dissection of Meloidogyne incognita resistance genes based on VIGS functional analysis in Cucumis metuliferus. BMC Plant Biol. 2024, 24, 964. [Google Scholar] [CrossRef] [PubMed]
- Harris-Shultz, K.R.; Davis, R.F.; Wallace, J.; Knoll, J.E.; Wang, H. A novel QTL for root-knot nematode resistance is identified from a South African sweet sorghum line. Phytopathology 2019, 109, 1011–1017. [Google Scholar] [CrossRef]
- Changkwian, A.; Venkatesh, J.; Lee, J.-H.; Han, J.-W.; Kwon, J.-K.; Siddique, M.I.; Solomon, A.M.; Choi, G.-J.; Kim, E.; Seo, Y. Physical localization of the root-knot nematode (Meloidogyne incognita) resistance locus Me7 in pepper (Capsicum annuum). Front. Plant Sci. 2019, 10, 886. [Google Scholar] [CrossRef]
- He, Y.; Kumar, P.; Shen, X.; Davis, R.F.; Van Becelaere, G.; May, O.L.; Nichols, R.L.; Chee, P.W. Re-evaluation of the inheritance for root-knot nematode resistance in the Upland cotton germplasm line M-120 RNR revealed two epistatic QTLs conferring resistance. Theor. Appl. Genet. 2014, 127, 1343–1351. [Google Scholar] [CrossRef]
- Barbary, A.; Djian-Caporalino, C.; Marteu, N.; Fazari, A.; Caromel, B.; Castagnone-Sereno, P.; Palloix, A. Plant genetic background increasing the efficiency and durability of major resistance genes to root-knot nematodes can be resolved into a few resistance QTLs. Front. Plant Sci. 2016, 7, 632. [Google Scholar] [CrossRef]
- Huynh, B.-L.; Matthews, W.C.; Ehlers, J.D.; Lucas, M.R.; Santos, J.R.; Ndeve, A.; Close, T.J.; Roberts, P.A. A major QTL corresponding to the Rk locus for resistance to root-knot nematodes in cowpea (Vigna unguiculata L. Walp.). Theor. Appl. Genet. 2016, 129, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Flores, J.C.; Yencho, G.C.; Pecota, K.V.; Sosinski, B.; Mwanga, R.O. Detection of quantitative trait loci and inheritance of root-knot nematode resistance in sweetpotato. J. Am. Soc. Hortic. Sci. 2008, 133, 844–851. [Google Scholar] [CrossRef]
- Vuong, T.; Sonah, H.; Patil, G.; Meinhardt, C.; Usovsky, M.; Kim, K.; Belzile, F.; Li, Z.; Robbins, R.; Shannon, J. Identification of genomic loci conferring broad-spectrum resistance to multiple nematode species in exotic soybean accession PI 567305. Theor. Appl. Genet. 2021, 134, 3379–3395. [Google Scholar] [CrossRef]
- Maranna, S.; Kumawat, G.; Nataraj, V.; Gireesh, C.; Gupta, S.; Satpute, G.K.; Ratnaparkhe, M.B.; Yadav, D.P. NAM population–a novel genetic resource for soybean improvement: Development and characterization for yield and attributing traits. Plant Genet. Resour. 2019, 17, 545–553. [Google Scholar] [CrossRef]
- Qi, Z.; Zhang, Z.; Wang, Z.; Yu, J.; Qin, H.; Mao, X.; Jiang, H.; Xin, D.; Yin, Z.; Zhu, R. Meta-analysis and transcriptome profiling reveal hub genes for soybean seed storage composition during seed development. Plant Cell Environ. 2018, 41, 2109–2127. [Google Scholar] [CrossRef]
- Van, K.; McHale, L.K. Meta-analyses of QTLs associated with protein and oil contents and compositions in soybean [Glycine max (L.) Merr.] seed. Int. J. Mol. Sci. 2017, 18, 1180. [Google Scholar] [CrossRef]
- Ma, C.-X.; Casella, G.; Shen, Z.-J.; Osborn, T.C.; Wu, R. A unified framework for mapping quantitative trait loci in bivalent tetraploids using single-dose restriction fragments: A case study from alfalfa. Genome Res. 2002, 12, 1974–1981. [Google Scholar] [CrossRef] [PubMed]
- Mollinari, M.; Garcia, A.A.F. Linkage analysis and haplotype phasing in experimental autopolyploid populations with high ploidy level using hidden Markov models. G3 Genes Genomes Genet. 2019, 9, 3297–3314. [Google Scholar] [CrossRef]
- Sonah, H.; O’Donoughue, L.; Cober, E.; Rajcan, I.; Belzile, F. Identification of loci governing eight agronomic traits using a GBS-GWAS approach and validation by QTL mapping in soya bean. Plant Biotechnol. J. 2015, 13, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, R.; Sonah, H.; Patil, G.; Chen, W.; Prince, S.; Mutava, R.; Vuong, T.; Valliyodan, B.; Nguyen, H.T. Integrating omic approaches for abiotic stress tolerance in soybean. Front. Plant Sci. 2014, 5, 244. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Boyse, J.F.; Song, Q.; Cregan, P.B.; Wang, D. Genomic consequences of selection and genome-wide association mapping in soybean. BMC Genom. 2015, 16, 671. [Google Scholar] [CrossRef]
- Yu, J.; Jung, S.; Cheng, C.H.; Lee, T.; Zheng, P.; Buble, K.; Crabb, J.; Humann, J.; Hough, H.; Jones, D.; et al. CottonGen: The Community Database for Cotton Genomics, Genetics, and Breeding Research. Plants 2021, 10, 2805. [Google Scholar] [CrossRef]
- Bayer, M.M.; Rapazote-Flores, P.; Ganal, M.; Hedley, P.E.; Macaulay, M.; Plieske, J.; Ramsay, L.; Russell, J.; Shaw, P.D.; Thomas, W.; et al. Development and Evaluation of a Barley 50k iSelect SNP Array. Front. Plant Sci. 2017, 8, 1792. [Google Scholar] [CrossRef]
- Song, Q.; Hyten, D.L.; Jia, G.; Quigley, C.V.; Fickus, E.W.; Nelson, R.L.; Cregan, P.B. Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS ONE 2013, 8, e54985. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Amatriaín, M.; Cuesta-Marcos, A.; Endelman, J.B.; Comadran, J.; Bonman, J.M.; Bockelman, H.E.; Chao, S.; Russell, J.; Waugh, R.; Hayes, P.M. The USDA barley core collection: Genetic diversity, population structure, and potential for genome-wide association studies. PLoS ONE 2014, 9, e94688. [Google Scholar] [CrossRef]
- Dimkpa, S.O.; Lahari, Z.; Shrestha, R.; Douglas, A.; Gheysen, G.; Price, A.H. A genome-wide association study of a global rice panel reveals resistance in Oryza sativa to root-knot nematodes. J. Exp. Bot. 2016, 67, 1191–1200. [Google Scholar] [CrossRef] [PubMed]
- Hada, A.; Dutta, T.K.; Singh, N.; Singh, B.; Rai, V.; Singh, N.K.; Rao, U. A genome-wide association study in Indian wild rice accessions for resistance to the root-knot nematode Meloidogyne graminicola. PLoS ONE 2020, 15, e0239085. [Google Scholar] [CrossRef]
- Alekcevetch, J.C.; de Lima Passianotto, A.L.; Ferreira, E.G.C.; Dos Santos, A.B.; da Silva, D.C.G.; Dias, W.P.; Belzile, F.; Abdelnoor, R.V.; Marcelino-Guimarães, F.C. Genome-wide association study for resistance to the Meloidogyne javanica causing root-knot nematode in soybean. Theor. Appl. Genet. 2021, 134, 777–792. [Google Scholar] [CrossRef] [PubMed]
- Warmerdam, S.; Sterken, M.G.; van Schaik, C.; Oortwijn, M.E.; Sukarta, O.C.; Lozano-Torres, J.L.; Dicke, M.; Helder, J.; Kammenga, J.E.; Goverse, A. Genome-wide association mapping of the architecture of susceptibility to the root-knot nematode Meloidogyne incognita in Arabidopsis thaliana. New Phytol. 2018, 218, 724–737. [Google Scholar] [CrossRef]
- Passianotto, A.L.d.L.; Sonah, H.; Dias, W.P.; Marcelino-Guimarães, F.C.; Belzile, F.; Abdelnoor, R.V. Genome-wide association study for resistance to the southern root-knot nematode (Meloidogyne incognita) in soybean. Mol. Breed. 2017, 37, 148. [Google Scholar] [CrossRef]
- Canella Vieira, C.; Zhou, J.; Usovsky, M.; Vuong, T.; Howland, A.D.; Lee, D.; Li, Z.; Zhou, J.; Shannon, G.; Nguyen, H.T.; et al. Exploring Machine Learning Algorithms to Unveil Genomic Regions Associated With Resistance to Southern Root-Knot Nematode in Soybeans. Front. Plant Sci. 2022, 13, 883280. [Google Scholar] [CrossRef]
- Giordani, W.; Gama, H.C.; Chiorato, A.F.; Marques, J.P.R.; Huo, H.; Benchimol-Reis, L.L.; Camargo, L.E.A.; Garcia, A.A.F.; Vieira, M.L.C. Genetic mapping reveals complex architecture and candidate genes involved in common bean response to Meloidogyne incognita infection. Plant Genome 2022, 15, e20161. [Google Scholar] [CrossRef]
- Obata, N.; Tabuchi, H.; Kurihara, M.; Yamamoto, E.; Shirasawa, K.; Monden, Y. Mapping of nematode resistance in hexaploid sweetpotato using a next-generation sequencing-based association study. Front. Plant Sci. 2022, 13, 858747. [Google Scholar] [CrossRef]
- Bastien, M.; Sonah, H.; Belzile, F. Genome wide association mapping of Sclerotinia sclerotiorum resistance in soybean with a genotyping-by-sequencing approach. Plant Genome 2014, 7, plantgenome2013.2010.0030. [Google Scholar] [CrossRef]
- Tardivel, A.; Sonah, H.; Belzile, F.; O’Donoughue, L.S. Rapid identification of alleles at the soybean maturity gene E3 using genotyping by sequencing and a haplotype-based approach. Plant Genome 2014, 7, plantgenome2013.2010.0034. [Google Scholar] [CrossRef]
- Szymczak, S.; Holzinger, E.; Dasgupta, A.; Malley, J.D.; Molloy, A.M.; Mills, J.L.; Brody, L.C.; Stambolian, D.; Bailey-Wilson, J.E. r2VIM: A new variable selection method for random forests in genome-wide association studies. BioData Min. 2016, 9, 7. [Google Scholar] [CrossRef]
- Nicholls, H.L.; John, C.R.; Watson, D.S.; Munroe, P.B.; Barnes, M.R.; Cabrera, C.P. Reaching the end-game for GWAS: Machine learning approaches for the prioritization of complex disease loci. Front. Genet. 2020, 11, 521712. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. GigaScience 2018, 8, giy154. [Google Scholar] [CrossRef] [PubMed]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Stewart-Brown, B.B.; Song, Q.; Vaughn, J.N.; Li, Z. Genomic selection for yield and seed composition traits within an applied soybean breeding program. G3 Genes Genomes Genet. 2019, 9, 2253–2265. [Google Scholar] [CrossRef]
- Abed, A.; Belzile, F. Exploring the Realm of Possibilities: Trying to Predict Promising Crosses and Successful Offspring through Genomic Mating in Barley. Crop Breed. Genet. Genom. 2019, 1, e190019. [Google Scholar] [CrossRef]
- Mohammadi, M.; Tiede, T.; Smith, K.P. PopVar: A genome-wide procedure for predicting genetic variance and correlated response in biparental breeding populations. Crop Sci. 2015, 55, 2068–2077. [Google Scholar] [CrossRef]
- Huynh, B.L.; Ehlers, J.D.; Huang, B.E.; Muñoz-Amatriaín, M.; Lonardi, S.; Santos, J.R.; Ndeve, A.; Batieno, B.J.; Boukar, O.; Cisse, N. A multi-parent advanced generation inter-cross (MAGIC) population for genetic analysis and improvement of cowpea (Vigna unguiculata L. Walp.). Plant J. 2018, 93, 1129–1142. [Google Scholar] [CrossRef]
- Huynh, B.-L.; Stangoulis, J.C.; Vuong, T.D.; Shi, H.; Nguyen, H.T.; Duong, T.; Boukar, O.; Kusi, F.; Batieno, B.J.; Cisse, N. Quantitative trait loci and genomic prediction for grain sugar and mineral concentrations of cowpea [Vigna unguiculata (L.) Walp.]. Sci. Rep. 2024, 14, 4567. [Google Scholar] [CrossRef]
- Muñoz-Amatriaín, M.; Mirebrahim, H.; Xu, P.; Wanamaker, S.I.; Luo, M.; Alhakami, H.; Alpert, M.; Atokple, I.; Batieno, B.J.; Boukar, O. Genome resources for climate-resilient cowpea, an essential crop for food security. Plant J. 2017, 89, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Ratnaparkhe, M.B.; Marmat, N.; Kumawat, G.; Shivakumar, M.; Kamble, V.G.; Nataraj, V.; Ramesh, S.V.; Deshmukh, M.P.; Singh, A.K.; Sonah, H. Whole genome re-sequencing of soybean accession EC241780 providing genomic landscape of candidate genes involved in rust resistance. Curr. Genom. 2020, 21, 504–511. [Google Scholar] [CrossRef]
- Lee, S.; Van, K.; Sung, M.; Nelson, R.; LaMantia, J.; McHale, L.K.; Mian, M.R. Genome-wide association study of seed protein, oil and amino acid contents in soybean from maturity groups I to IV. Theor. Appl. Genet. 2019, 132, 1639–1659. [Google Scholar] [CrossRef] [PubMed]
- Barabaschi, D.; Tondelli, A.; Desiderio, F.; Volante, A.; Vaccino, P.; Valè, G.; Cattivelli, L. Next generation breeding. Plant Sci. 2016, 242, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef]
- Esposito, S.; Aiese Cigliano, R.; Cardi, T.; Tripodi, P. Whole-genome resequencing reveals genomic footprints of Italian sweet and hot pepper heirlooms giving insight into genes underlying key agronomic and qualitative traits. BMC Genom. Data 2022, 23, 21. [Google Scholar] [CrossRef]
- Lim, J.-H.; Yang, H.-J.; Jung, K.-H.; Yoo, S.-C.; Paek, N.-C. Quantitative trait locus mapping and candidate gene analysis for plant architecture traits using whole genome re-sequencing in rice. Mol. Cells 2014, 37, 149–160. [Google Scholar] [CrossRef]
- Varshney, R.K.; Saxena, R.K.; Upadhyaya, H.D.; Khan, A.W.; Yu, Y.; Kim, C.; Rathore, A.; Kim, D.; Kim, J.; An, S.; et al. Whole-genome resequencing of 292 pigeonpea accessions identifies genomic regions associated with domestication and agronomic traits. Nat. Genet. 2017, 49, 1082–1088. [Google Scholar] [CrossRef]
- Xu, X.; Zeng, L.; Tao, Y.; Vuong, T.; Wan, J.; Boerma, R.; Noe, J.; Li, Z.; Finnerty, S.; Pathan, S.M. Pinpointing genes underlying the quantitative trait loci for root-knot nematode resistance in palaeopolyploid soybean by whole genome resequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 13469–13474. [Google Scholar] [CrossRef]
- Cheng, C.; Wang, X.; Liu, X.; Yang, S.; Yu, X.; Qian, C.; Li, J.; Lou, Q.; Chen, J. Candidate genes underlying the quantitative trait loci for root-knot nematode resistance in a Cucumis hystrix introgression line of cucumber based on population sequencing. J. Plant Res. 2019, 132, 813–823. [Google Scholar] [CrossRef]
- Lee, J.-D.; Kim, H.-J.; Robbins, R.T.; Wrather, J.A.; Bond, J.; Nguyen, H.T.; Shannon, J.G. Reaction of soybean cyst nematode resistant plant introductions to root-knot and reniform nematodes. Plant Breed. Biotechnol. 2015, 3, 346–354. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, Y.; Liu, J.; Zhang, H.; Wang, T. The regulatory roles of plant miRNAs in biotic stress responses. Biochem. Biophys. Res. Commun. 2025, 755, 151568. [Google Scholar] [CrossRef] [PubMed]

| Attributes | Conventional Breeding | Omics-Based Breeding |
|---|---|---|
| Example of approach | GS, MAS, GWAS, QTL mapping | Transcriptomics, genomics, metabolomics, lipidomics, proteomics |
| Labor intensity | Highly labor extensive | Moderately labor extensive |
| Cost | Low to moderate | Moderate to low |
| Precision | Low, since it is based primarily on phenotype | More accurate because it is based on genotype |
| Time requirement | Dependent on crop cycle; 8–12 years to release an improved variety | From weeks to a few months to generate data; candidate gene(s) responsible for the trait(s) is then identified and assessed through overexpression, silencing, and gene editing approaches; generation of transgenic crops can take from a few months to 1–2 years based on genotype |
| Regulation | Flexible regulation and release of germplasm; dependent on country regulations | Strict regulatory frame depending on the country, e.g., under regulation and approval from USDA, APHIS, and FDA in the USA |
| Variability | Highly variable, as it is created by hybridization; low number of replicates | Low variability; most approaches are high-throughput and allow a high number of replicates |
| Accessibility | Widely practiced as no special equipment is required | Requires sophisticated instrumentations and expertise |
| Reliability | Less reliable because it is based on phenotype and breeder’s subjective analysis | Highly reliable, though dependent on genotype |
| Other | Provide potential benefits to consumers, farmers, and the environment | Provide potential benefits to consumers, farmers, and the environment Provide acknowledgement and resources for marker-assisted selection and GS, MAS, GWAS, and QTL mapping Provide acknowledgement that helps develop more effective and safer strategies/technologies to control pests and diseases, and allows conceptual advances in plant biology/physiology and other related fields |
| Crop | Platform | Total No. of DEGs | Key Findings * | Reference | |
|---|---|---|---|---|---|
| Susceptible Line | Resistant Line | ||||
| Alfalfa (Medicago sativa) | Illumina Hi-Seq 2000 | 1143 | 319 | R genes, signaling pathways, oxidative stress, chemical stimulus, antioxidant activity, oxidoreductase and peroxidase activity | [66] |
| Cowpea (Vigna unguiculata) | Affymetrix GeneChip expression array | 1060 | 552 | Genes related to ROS, toxins, and defense | [76] |
| Eggplant (Solanum melongena) | Illumina Hi-Seq 4000 | 8148 | 4761 | Genes related to cell wall biogenesis/organization, stimulus, hormone, plant hormone signal Transduction, and plant–pathogen interaction | [79] |
| Pepper (Capsicum annuum) | Illumina Hi-Seq | 2057 | 1217 | Genes located on chromosome 9 (NBS-LRR resistance gene, genes belonging to transcription factors or kinases) | [80] |
| Tomato (Solanum lycopersicum) | Illumina Hi-Seq 2000 | 1827 | 25 | Cell wall structure, development, primary and secondary metabolism, defense signaling pathway, hormone- mediated defense response | [16] |
| Sweetpotato (Ipomoea batatas) | Illumina Hi-Seq 2000 | 881 | 929 | Genes related to hormone signaling-related transcription factors, PR genes | [81] |
| Tobaco (Nicotiana tabacum) | Illumina Hi-Seq 2000 | 545 | 4354 | Genes related to cell wall modification, toxic compound synthesis, ROS, salicylic acid signal transduction and metabolites | [82] |
| Illumina Hi-Seq 2000 | 545 | 2623 | Auxin-related proteins, cell wall modifying proteins, ROS | [83] | |
| Soybean (Glycine max) | Illumina Hi-Seq 4000 | 5842 | 7041 | Genes related to mTOR, OI3K-Akt, thermogenesis, relaxin and phenylpropanoid pathway | [84] |
| Peach (Prunus kansuensis) | Illumina Hi-Seq 2000 | 1476 | 2107 | Genes related to phytohormone metabolism | [85] |
| Cotton (Gossypium hirsutum) | Illumina Hi-Seq 300 | 1355 | 1250 | Cell wall organization, defense response, phytohormones, protein serine/threonine kinase activity | [86] |
| Illumina Hi-Seq 2500 | 8247 | 1093 | Phytohormone signaling (particularly salicylic and jasmonic acid), cell surface-related receptors | [70] | |
| Crop | Nematode Species | Marker Type | Resistance Gene | References |
|---|---|---|---|---|
| Tomato (Solanum lycopersicum) | M. incognita, M. Javanica | CAPS | Mi-1 | [133] |
| M. incognita, M. arenaria, M. javanica | RAPD | Mi1.1, Mi1.2 | [134] | |
| M. incognita | RAPD, RFLP | Mi 3 | [135] | |
| Cucumber (Cucumis metuliferus) | M. Javanica | AFLP and SRAP | mj | [136] |
| Turmeric (Curcuma longa) | M. incognita | ISSR | - | [137] |
| Cotton (Gossypium hirsutum) | M. incognita | SSR | qMi-C14 | [138] |
| M. incognita | AFLP and derived CAPS | rkn1 | [130] | |
| M. incognita | SSR | - | [132] | |
| Peanut (Arachis hypogaea) | M. arenaria | RFLP | - | [139] |
| M. arenaria | CAPS, SSR, AFLP | Rma | [140] | |
| Soybean (Glycine max) | M. incognita | SSR | Rmi | [141] |
| Mulberry (Morus spp.) | M. incognita | SSR | - | [142] |
| Carrot (Daucus carota) | M. javanica | RAPD and STS | Mj-1 | [143] |
| M. incognita | SSR | Mj-1 | [144] | |
| Eggplant (Solanum melongena) | M. javanica | RT-PCR | Mi-1.2 | [95] |
| Pepper (Capsicum annuum) | M. incognita, M. arenaria, M. javanica | RAPD, RFLP | Me3 and Me4 | [145] |
| Crop | Platform/Technique | No. of Genotypes | No. of Loci Tested | Reference |
|---|---|---|---|---|
| Arabidopsis thaliana | Association mapping | 340 | 214,051 | [179] |
| Indian wild rice (Oryza spp.) | 50K “OsSNPnks” genic Affymetrix chip | 272 | 50,051 | [177] |
| Asian Rice (Oryza sativa) | 44K Affymetrix SNP chip | 332 | 44,100 | [176] |
| Soybean (Glycine max) | GBS | 317 | 44,992 | [178] |
| GBS | 193 | 46,196 | [180] | |
| BARCSoySNP6K BeadChip | 717 | 4974 | [181] | |
| Common bean (Phaseolus vulgaris) | Association mapping | 180 | 10,362 | [182] |
| Sweetpotato (Ipomoea batatas) | GBS | 107 | 46,982 | [183] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadav, H.; Roberts, P.A.; Lopez-Arredondo, D. Combating Root-Knot Nematodes (Meloidogyne spp.): From Molecular Mechanisms to Resistant Crops. Plants 2025, 14, 1321. https://doi.org/10.3390/plants14091321
Yadav H, Roberts PA, Lopez-Arredondo D. Combating Root-Knot Nematodes (Meloidogyne spp.): From Molecular Mechanisms to Resistant Crops. Plants. 2025; 14(9):1321. https://doi.org/10.3390/plants14091321
Chicago/Turabian StyleYadav, Himanshu, Philip A. Roberts, and Damar Lopez-Arredondo. 2025. "Combating Root-Knot Nematodes (Meloidogyne spp.): From Molecular Mechanisms to Resistant Crops" Plants 14, no. 9: 1321. https://doi.org/10.3390/plants14091321
APA StyleYadav, H., Roberts, P. A., & Lopez-Arredondo, D. (2025). Combating Root-Knot Nematodes (Meloidogyne spp.): From Molecular Mechanisms to Resistant Crops. Plants, 14(9), 1321. https://doi.org/10.3390/plants14091321






