Identification of Genomic Regions Associated with Peanut Rust Resistance by Genome-Wide Association Studies
Abstract
:1. Introduction
2. Results
2.1. Resistance Evaluation for Peanut Rust
2.2. Genome-Wide Association Analysis of Peanut Rust Resistance
2.3. Candidate Gene Prediction
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Peanut Rust Resistance Evaluation
4.3. Trial Design and Phenotypic Identification
4.3.1. Field Trial Design and Management
4.3.2. Indoor Trial Design and Management
4.4. Data Analysis
4.5. Genome-Wide Association Study
4.6. Gene Predictions Within the Candidate Interval
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ojiewo, C.O.; Janila, P.; Bhatnagar-Mathur, P.; Pandey, M.K.; Desmae, H.; Okori, P.; Mwololo, J.; Ajeigbe, H.; Njuguna-Mungai, E.; Muricho, G.; et al. Advances in Crop Improvement and Delivery Research for Nutritional Quality and Health Benefits of Groundnut (Arachis hypogaea L.). Front. Plant Sci. 2020, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.J.; Liu, H.; Qin, L.; Qi, F.Y.; Sun, Z.Q.; Wu, J.H.; Dong, W.Z.; Huang, B.Y.; Zhang, X.Y. Identification of QTL for kernel weight and size and analysis of the pentatricopeptide repeat (PPR) gene family in cultivated peanut (Arachis hypogaea L.). BMC Genom. 2023, 24, 495. [Google Scholar] [CrossRef] [PubMed]
- Burow, M.D.; Simpson, C.E.; Starr, J.L.; Paterson, A.H. Transmission genetics of chromatin from a synthetic amphidiploid to cultivated peanut (Arachis hypogaea L.): Broadening the gene pool of a monophyletic polyploid species. Genetics 2001, 159, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Toomer, O.T. Nutritional chemistry of the peanut (Arachis hypogaea L.). Crit. Rev. Food Sci. Nutr. 2018, 58, 3042–3053. [Google Scholar] [CrossRef]
- Gong, J.J.; Liang, T.M. Occurrence characteristics and control measures of peanut rust disease. Mod. Agric. Sci. Technol. 2015, 6, 135. [Google Scholar] [CrossRef]
- Hou, H.M.; Liao, B.S.; Lei, Y.; Ren, X.P.; Wang, S.Y.; Li, D.; Jing, H.F.; Huang, J.Q.; Chen, B.Y. AFLP markers for resistance to peanut rust disease. Chin. J. Oil Crop Sci. 2007, 29, 195–198. [Google Scholar]
- Mondal, S.; Badigannavar, A.M. Peanut rust (Puccinia arachidis Speg.) disease: Its background and recent accomplishments towards disease resistance breeding. Protoplasma 2015, 252, 1409–1420. [Google Scholar] [CrossRef]
- Liao, B.S.; Wang, Y.Y.; Duan, N.X. Research on genetic improvement of rust resistance of peanut. Chin. J. Oil Crop Sci. 1991, 13, 92–94. [Google Scholar]
- Jiang, H.F.; Duan, N.X.; Tan, Y.J.; Hu, D.H. Preliminary identification of resistance to peanut rust in peanut germplasm resource. Chin. J. Oil Crop Sci. 1992, 45–47, 89. [Google Scholar]
- Chen, W.Y.; Ko, W.H. Flaxseed oil for control of peanut rust and its disease control mechanism. J. Phytopathol. 2015, 163, 271–278. [Google Scholar] [CrossRef]
- Hou, H.M. Studies on Genetic Diversity and Molecular Markers of Resistance to Rust Resistance in Peanut Germplasm. Master’s Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2007. [Google Scholar]
- Reddy, S.N.; Nigam, R.C.; Nageswara, R. Registration of ICGV87354 peanut germplasm with drought tolerance and rust resistance. Crop Sci. 2001, 41, 274–275. [Google Scholar] [CrossRef]
- Tang, Z.X.; Xu, R.R.; Lan, X.L.; Lin, Y.S.; Shi, G.Y. Breeding and cultivation of a new rust-resistance peanut variety, Fuhua 6. Fujian J. Agric. Sci. 2011, 26, 729–732. [Google Scholar]
- Zheng, Y.X.; Guo, D.D.; Chen, X.Y. Utilization of the rust-resistant and high-yield germplasm Shanyou 27 in peanut breeding. Sci. Agric. Sin. 2005, 38, 1755–1760. [Google Scholar]
- Zhao, M.Z.; Zhao, P.X.; Ma, G.R.; Xiao, Z.W.; Guo, T.Y.; Bai, X.H.; Li, J.H.; Zhou, H. Breeding and rust Resistance of a new coffee variety ‘Dere No.5’. Breed. Var. Sel. 2025, 1, 10–20, 26. [Google Scholar]
- Li, L.; Hu, M.X.; Zhang, W.T.; Wang, W.J.; Cao, S.Q.; Huang, J.; Zhang, B.; Sun, Z.Y.; Jia, Q.Z. Research progress on the application of biotechnology in wheat breeding against stripe rust. J. Cold-Arid. Agric. Sci. 2024, 3, 510–514. [Google Scholar]
- Shan, Z.H.; Zhou, X.A. The status of soybean rust research. Chin. J. Oil Crop Sci. 2007, 96–100. [Google Scholar]
- Wang, S.; Zhang, R.Y.; Wang, R.H.; Song, W.; Zhao, J.R. Research progress of southern corn rust and resistance breeding. Sci. Agric. Sin. 2024, 57, 2732–2743. [Google Scholar]
- Zhao, L.Q.; Pan, W.J.; Ma, J.F.; Weng, Q.Y.; Dong, L.; Quan, J.Z.; Xing, J.H.; Dong, Z.P.; Dong, J.G. Identification of AFLP markers linked to a novel rust resistance gene in foxtail millet. Sci. Agric. Sin. 2010, 43, 4349–4355. [Google Scholar]
- Subrahmanyam, P.; Williams, J.H.; McDonald, D.; Gibbons, R.W. The influence of foliar diseases and their control by selective fungicides on a range of groundnut (Arachis hypogaea L.) genotype. Ann. Appl. Biol. 1984, 104, 467–476. [Google Scholar] [CrossRef]
- Subrahmanyam, P.; Moss, J.P.; Rao, V.R. Resistance to peanut rust in wild arachis species. Plant Dis. 1983, 67, 209–212. [Google Scholar] [CrossRef]
- Pande, S.; Rao, N.J. Resistance of wild Arachis species to late leaf spot and rust in greenhouse trials. Plant Dis. 2001, 85, 851–855. [Google Scholar] [CrossRef] [PubMed]
- Khedikar, Y.P.; Gowda, M.V.C.; Sarvamangala, C.; Patgar, K.Y.; Upadhyaya, H.D.; Varshney, R.K. A QTL study on late leaf spot and rust revealed one major QTL for molecular breeding for rust resistance in groundnut (Arachis hypogaea L.). Theor. Appl. Genet. 2010, 121, 971–984. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.K.; Khan, A.W.; Singh, V.K.; Vishwakarma, M.K.; Varshney, R.K. QTL-seq approach identified genomic regions and diagnostic markers for rust and late leaf spot resistance in groundnut (Arachis hypogaea L.). Plant Biotechnol. J. 2017, 15, 927–941. [Google Scholar] [CrossRef] [PubMed]
- Sujay, V.; Gowda, M.V.C.; Pandey, M.K.; Bhat, R.S.; Khedikar, Y.P.; Nadaf, H.L.; Gautami, B.; Sarvamangala, C.; Lingaraju, S.; Radhakrishan, T.; et al. Quantitative trait locus analysis and construction of consensus genetic map for foliar disease resistance based on two recombinant inbred line populations in cultivated peanut (Arachis hypogaea L.). Mol. Breed. 2012, 30, 773–788. [Google Scholar] [CrossRef]
- Varman, P.V. A foliar disease resistant line developed through interspecific hybridization in groundnut (Arachis hypogaea). Indian J. Agric. Sci. 2013, 69, 67–68. [Google Scholar]
- Cheng, Y.J. Genome-Wide Association Analysis of Peanut Mesh Spot Resistance and Cytology of the Mechanism of Disease Resistance. Master’s Thesis, Zhengzhou University, Zhengzhou, China, 2021. [Google Scholar]
- Li, L. Linkage and Genome-Wide Association Analysis Were Used to Identify QTLs of Peanut Strain-Related Traits. Ph.D. Thesis, Hebei Agricultural University, Baoding, China, 2019. [Google Scholar]
- Zheng, Z.; Sun, Z.Q.; Qi, F.Y.; Fang, Y.J.; Lin, K.; Pavan, S.; Huang, B.Y.; Dong, W.Z.; Du, P.; Tian, M.D.; et al. DNA sequencing sheds light on the evolutionary history of peanut and identifies genes associated with phenotypic diversification. Nat. Genet. 2024, 56, 1975–1984. [Google Scholar] [CrossRef]
- Luo, D.D.; Shi, L.; Sun, Z.Q.; Qi, F.Y.; Liu, H.F.; Xue, L.L.; Li, X.N.; Liu, H.; Qu, P.Y.; Zhao, H.H.; et al. Genome-Wide Association Studies of Embryogenic Callus Induction Rate in Peanut (Arachis hypogaea L.). Genes 2024, 15, 160. [Google Scholar] [CrossRef]
- Sun, D.R.; Wang, Y.Y.; Wan, W.Z. Classification of Peanut Varieties Cultivated in China; Agricultural Press: Beijing, China, 1998. [Google Scholar]
- Krapovickas, A.G. Taxonomía del género Arachis (Leguminosae). Bonplandia 1994, 8, 1–186. [Google Scholar] [CrossRef]
- Yu, S.L. Chinese Peanut Varieties and Their Genealogies; Shanghai Science and Technology Press: Shanghai, China, 2008. [Google Scholar]
- Liao, B.S. Overview of the genetic breeding of peanut rust resistance at home and abroad. Peanut Technol. 1988, 33–35. [Google Scholar]
- Liao, B.S.; Wang, Y.; Xia, X.; Tang, G.; Tan, Y.; Sun, D. Genetic study of peanut rust resistance. China Oil. 1988, 14–19. [Google Scholar]
- Zhang, Y.M.; Xing, G.F.; Liu, M.T.; Liu, X.D.; Han, Y.H. Genome wide association study: Opportunities and challenges in genomic research. Biotechnol. Bull. 2013, 29, 1–6. [Google Scholar]
- Huang, Q.; Yin, J.Y.; Liu, Z.Q. Progress of genome-wide association studies in type 2 diabetes mellitus. Chin. J. Endocrinol. Metab. 2010, 26, 432–436. [Google Scholar]
- Shirasawa, K.; Bhat, R.S.; Khedikar, Y.P.; Sujay, V.; Kolekar, R.M.; Yeri, S.B.; Sukruth, M.; Cholin, S.; Asha, B.; Pandey, M.K.; et al. Sequencing analysis of genetic loci for resistance for late leaf spot and rust in peanut (Arachis hypogaea L.). Front. Plant Sci. 2018, 26, 1727. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Badigannavar, A.M. Mapping of a dominant rust resistance gene revealed two R genes around the major rust_QTL in cultivated peanut (Arachis hypogaea L.). Theor. Appl. Genet. Int. J. Breed. Res. Cell Genet. 2018, 131, 1671–1681. [Google Scholar] [CrossRef]
- Ahmad, S.; Nawade, B.; Sangh, C.; Mishra, G.P.; Bosamia, T.C.; Kumar, N.; Dobaria, J.R.; Gajera, H.P. Identification of novel QTLs for late leaf spot resistance and validation of a major rust QTL in peanut (Arachis hypogaea L.). 3 Biotech 2020, 10, 458. [Google Scholar]
- Qi, F.Y.; Sun, Z.Q.; Liu, H.; Zheng, Z.; Qin, L.; Shi, L.; Chen, Q.Z.; Liu, H.D.; Lin, X.F.; Miao, L.J.; et al. QTL identification, fine mapping, and marker development for breeding peanut (Arachis hypogaea L.) resistant to bacterial wilt. Theor. Appl. Genet. 2022, 135, 1319–1330. [Google Scholar] [CrossRef]
- Zhang, C.; Zhuang, W.J.; Chen, H.; Liu, H.; Wang, S.S.; Wei, T.; Zhuang, Y.H.; Cai, T.C.; Yang, Q.; Wang, L.H. A Peanut NBS-LRR Coding Gene AhRRS2 and Its Application in Plant Resistance Against Wilt Solanacearum. CN115109783B, 23 May 2023. [Google Scholar]
- Hu, Y.C.; Yao, L.P.; Zhang, T.; He, S.M.; Tang, X.B.; Chen, Q.X.; Wen, Z.F. FnCN gene and promoter cloning of the NB-ARC domain and FnCN gene expression analysis. J. Plant Resour. Environ. 2020, 29, 1–10. [Google Scholar]
- Collier, S.M.; Hamel, L.P.; Moffett, P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NBS-LRR protein. Molecular Plant-Microbe Interact. 2011, 24, 918–931. [Google Scholar] [CrossRef]
- Zhang, H.; Luo, H.Y.; Li, W.T.; Guo, J.B.; Chen, W.G.; Zhou, X.J.; Huang, L.; Liu, N.; Yan, L.Y.; Lei, Y. Identification of disease resistance genes and analysis of their response to B. green infection. J. Crop Sci. 2021, 47, 2314–2323. [Google Scholar]
- Gu, L.J.; Si, W.N.; Zhao, L.N.; Yang, S.H.; Zhang, X.H. Dynamic evolution of NBS-LRR genes in bread wheat and its progenitors. Mol. Genet. Genom. 2015, 290, 727–738. [Google Scholar] [CrossRef]
- Kang, Y.J.; Kim, K.H.; Shim, S.; Yoon, M.; Sun, S.; Kim, M.; Van, K.; Suk-Ha, L. Genome-wide mapping of NBS-LRR genes and their association with disease resistance in soybean. BMC Plant Biol. 2012, 9, 139. [Google Scholar] [CrossRef] [PubMed]
- Bai, J. Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res. 2002, 12, 1871–1884. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.P.; Yang, S.M.; Zhao, D.G.; Song, L. Bioinformatic analysis of the TIR-NBS gene family in tobacco. Plants Guangxi 2020, 40, 10. [Google Scholar]
- Lu, Y.; Liu, Y.; Song, Y.; Jing, L. Genome-wide analysis of the NBS-LRR disease resistance gene family in sunflower. Chin. J. Oil Crop Sci. 2020, 42, 441–452. [Google Scholar]
- Feng, Y.F.; Geng, L.L.; Han, R.; Zhang, J. Cloning and expression characteristics of NBS-LRR genes in peanut. Biotechnol. Bull. 2016, 32, 90–95. [Google Scholar]
- Zhang, C.; Chen, H.; Cai, T.; Deng, Y.; Zhuang, R.; Zhang, N.; Zeng, Y.H.; Zheng, Y.X.; Tang, R.R.; Pan, R.L.; et al. Overexpression of a novel peanut NBS-LRR gene AhRRS5 enhances disease resistance to ralstonia solanacearum in tobacco. Plant Biotechnol. J. 2017, 15, 39–55. [Google Scholar] [CrossRef]
- Zhao, X.B.; Zhang, T.T.; Yan, C.X.; Zhang, H.; Shan, S.H.; Chen, D.X. Cloning and expression analysis of a NBS-LRR class gene in peanut. Shandong Agric. Sci. 2015, 47, 1–5, 14. [Google Scholar]
- Yoon, Y.; Seo, D.H.; Shin, H. The role of stress responsive transcription factors in modulating abiotic stress tolerance in plants. Agronomy 2020, 10, 788. [Google Scholar] [CrossRef]
- SmallL, I.D.; Peeters, N. The PPR motif-a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000, 25, 45–47. [Google Scholar] [CrossRef]
- Su, H.G.; Li, B.; Song, X.Y.; Ma, J.; Ma, Y.Z. Genome-wide analysis of the DYW subgroup PPR gene family and identification of GmPPR4 responses to drought stress. Int. J. Mol. Sci. 2019, 20, 5667. [Google Scholar] [CrossRef]
- Chen, G.L.; Zou, Y.; Hu, J.H.; Ding, Y. Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments. BMC Genom. 2018, 19, 720. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kang, H. Roles of organellar RNA-binding proteins in plant growth, development, and abiotic stress responses. Int. J. Mol. Sci. 2020, 21, 4548. [Google Scholar] [CrossRef] [PubMed]
- Mergner, J.; Frejno, M.; List, M.; Papacek, M.; Chen, X.; Chaudhary, A.; Samaras, P.; Richter, S.; Shikata, H.; Messerer, M.; et al. Mass-spectrometrybased draft of the arabidopsis proteome. Nature 2020, 579, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.T.; Wang, Y.X.; Pei, L.L.; Xie, C.L.; Chen, M.; Chen, J.; Zhou, Y.B.; Ma, Y.Z.; Xu, Z.S. Plant protein kinases and abiotic stress resistance in crop plants. J. Plant Genet. Resour. 2017, 18, 763–770. [Google Scholar]
- Wang, G.F.; Peng, F.T.; Zhao, Y.F.; Luo, J.J.; Yu, W.; Xiao, Y.S.; Chen, X.L. Progress in plant SnRK 1 protein kinase studies. Shandong Agric. Sci. 2018, 50, 164–172. [Google Scholar]
- Zhao, Y.Q.; Zhou, Z.L.; Tang, J.; Yao, G.F.; Hu, K.D.; Zhang, H. Functional study of serine hydroxymethyltransferase gene family in sweetpotato, tomato and Arabidopsis. J. Hefei Univ. Technol. (Nat. Sci. Ed.) 2022, 45, 1705–1714. [Google Scholar]
- Ding, J.; Li, F.J.; Li, X.A.; Zhang, X.H. Regulation of E3 ubiquitin ligase and stress response. Food Sci. 2024, 1–18. [Google Scholar]
- Kamble, S.K.; Patil, B.J. Efficacy of foliar spray applications of plant extracts against groundnut rust. Curr. Opin. Environ. Sustain. 2019, 9, 113–121. [Google Scholar] [CrossRef]
- Shi, X.L.; Sun, Z.Q.; Qi, F.Y.; Han, S.Y.; Zheng, Y.X.; Dong, W.Z.; Zhang, X.Y. Effect of temperature on germination and infection of peanut rust fungus. J. Peanut 2023, 52, 1–6, 31. [Google Scholar]
- Yu, H.T.; Wang, L.P.; Lu, M.Y.; He, Y.H.; Yang, F.; Hu, C.Q.; Kang, Y.S.; Niu, W.W.; Yang, X.; Wang, Y.B. Method for Identifying Comprehensive Resistance to Broad Bean Rust. CN202010106381.7, 16 June 2020. [Google Scholar]
- Liu, H.; Sun, Z.Q.; Zhang, X.Y.; Dong, W.Z. QTL mapping of web blotch resistance in peanut by high-throughput genome-wide sequencing. BMC Plant Biol. 2020, 20, 249. [Google Scholar] [CrossRef]
- Wu, X.H.; Sun, Z.Q.; Qi, F.Y. Cytological and transcriptomic analysis to unveil the mechanism of web blotch resistance in Peanut. BMC Plant Biol. 2023, 23, 518. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.L.; Sun, Z.Q.; Xue, X.; Xu, H.M.; Wu, Y.; Zhang, Y.; Yang, Y.Q.; Han, S.Y.; Zhao, R.F.; Zhang, M.Y.; et al. Amelioration of hypothermia-induced damage on peanut by exogenous application of chitooligosaccharide. Agriculture 2023, 13, 217. [Google Scholar] [CrossRef]
- Sun, Z.Q.; Qi, F.Y.; Liu, H.; Xu, J.; Shi, L.; Zhang, Z.X.; Miao, L.J.; Huang, B.Y.; Dong, W.Z. QTL mapping of quality traits in peanut using whole-genome resequencing. Crop J. 2022, 10, 177–184. [Google Scholar] [CrossRef]
- Chen, K.M.; Luan, M.B.; Xiong, H.P.; Cheng, P.; Chen, J.K.; Gao, G.; Huang, K.Y.; Zhu, A.G.; Yu, C.M. Genome-wide association study discovered favorable single nucleotide polymorphisms and candidate genes associated with ramet number in ramie (Boehmeria nivea L.). BMC Plant Biol. 2018, 18, 345–354. [Google Scholar] [CrossRef]
- Li, F.; Chen, B.; Xu, K.; Wu, J.F.; Song, W.L.; Bancroft, I.; Harper, A.L.; Trick, M.; Liu, S.Y.; Gao, G.Z.; et al. Genome-wide association study dissects the genetic architecture of seed weight and seed quality in rapeseed (Brassica napus L.). DNA Res. 2014, 21, 355–368. [Google Scholar] [CrossRef]
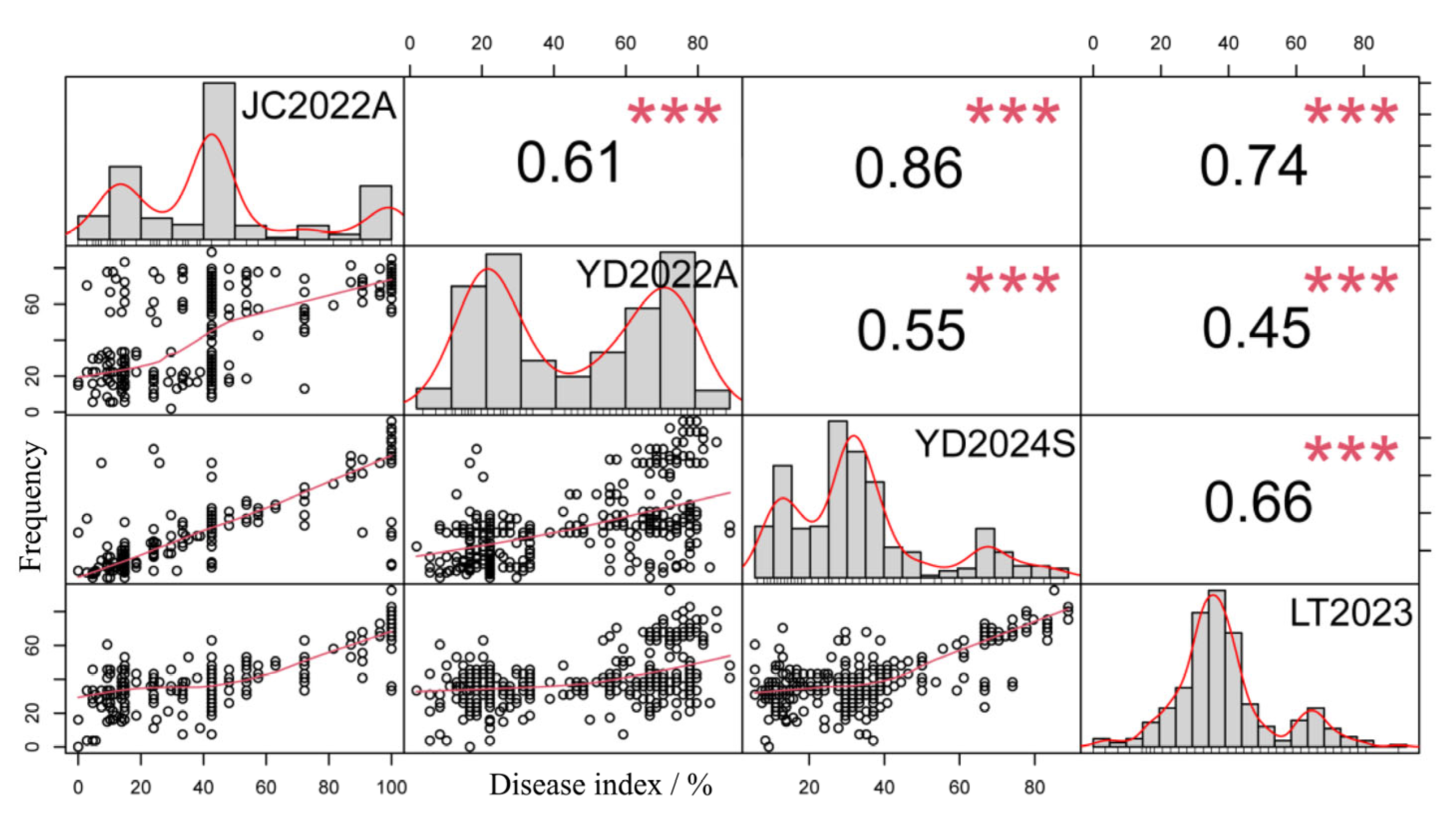


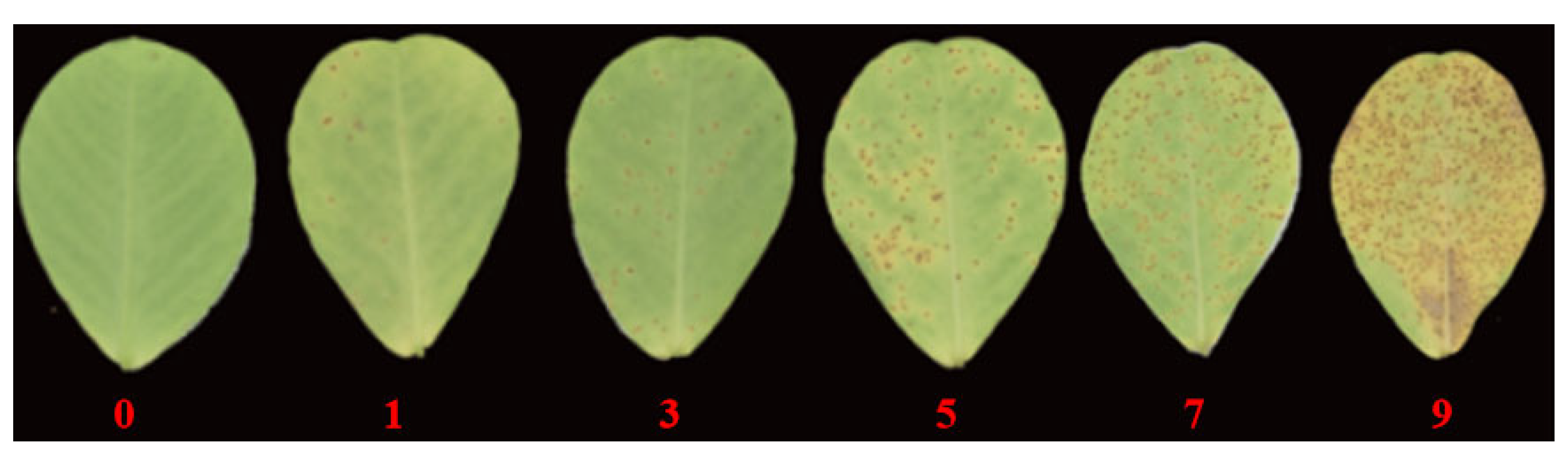
| Environment | Minimum | Maximum | Mid-Value | Average | Standard Error | Coefficient of Variation/% | Kurtosis | Skewness |
|---|---|---|---|---|---|---|---|---|
| JC2022A | 0.00 | 100.00 | 42.60 | 43.82 | 1.48 | 63.52 | −0.21 | 0.77 |
| YD2022A | 1.85 | 88.89 | 44.44 | 44.90 | 1.33 | 55.49 | −1.62 | 0.06 |
| YD2024S | 5.56 | 88.89 | 29.63 | 33.94 | 1.03 | 57.22 | 0.37 | 0.97 |
| LT2023 | 0.00 | 92.59 | 38.27 | 39.53 | 0.80 | 37.92 | 0.80 | 0.70 |
| Source | Degree of Freedom | Sum of Squares | Mean Squares | F-Value | p-Value | Heritability |
|---|---|---|---|---|---|---|
| Environment | 3 | 56,939.477 | 18,979.826 | 283.386 | 0.000 *** | 0.58 |
| Genotype | 352 | 1,024,795.125 | 2911.350 | 43.469 | 0.000 *** | |
| Environment × Genotype | 1056 | 454,018.125 | 429.941 | 6.419 | 0.000 *** | |
| Error | 1760 | 117,876.250 | 66.975 |
| SNP Marker | Chromosome | Location (bp) | Interval (bp) | Allele | −log10(P) |
|---|---|---|---|---|---|
| Arahy.05_93085395 | A05 | 93,085,395 | 92,885,395–93,285,395 | C/A | 7.67–8.32 |
| Arahy.05_93114354 | A05 | 93,114,354 | 92,914,354–93,314,354 | C/T | 7.72–8.12 |
| Arahy.12_4097252 | A12 | 4,097,252 | 3,897,252–4,297,252 | G/C | 7.95–8.14 |
| Number | Gene Name | Genn Location | Strand | Annotation |
|---|---|---|---|---|
| 1 | Arahy.66HKIQ | A12.3963819-3967481 | + | LRR and NB-ARC domain disease resistance protein |
| 2 | Arahy.NMJE7X | A12.3973701-3977692 | + | LRR and NB-ARC domain disease resistance protein |
| 3 | Arahy.1DE133 | A12.3983522-3989366 | + | LRR and NB-ARC domain disease resistance protein |
| 4 | Arahy.QREM2V | A12.3993406-3997397 | + | LRR and NB-ARC domain disease resistance protein |
| 5 | Arahy.CUPK9N | A12.4015364-4017374 | + | LRR and NB-ARC domain disease resistance protein |
| 6 | Arahy.KD5YG3 | A12.4024728-4028719 | + | LRR and NB-ARC domain disease resistance protein |
| 7 | Arahy.0C19HY | A12.4058399-4062049 | − | LRR and NB-ARC domain disease resistance protein |
| 8 | Arahy.V6I7WA | A12.4236131-4239737 | + | LRR and NB-ARC domain disease resistance protein |
| 9 | Arahy.0EHV1A | A12.4280257-4284123 | + | Disease resistance protein (TIR-NBS-LRR class) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, X.; Sun, Z.; Qi, F.; Han, S.; Zheng, Y.; Dong, W.; Zhang, M.; Zhang, X. Identification of Genomic Regions Associated with Peanut Rust Resistance by Genome-Wide Association Studies. Plants 2025, 14, 1219. https://doi.org/10.3390/plants14081219
Shi X, Sun Z, Qi F, Han S, Zheng Y, Dong W, Zhang M, Zhang X. Identification of Genomic Regions Associated with Peanut Rust Resistance by Genome-Wide Association Studies. Plants. 2025; 14(8):1219. https://doi.org/10.3390/plants14081219
Chicago/Turabian StyleShi, Xinlong, Ziqi Sun, Feiyan Qi, Suoyi Han, Yixiong Zheng, Wenzhao Dong, Maoning Zhang, and Xinyou Zhang. 2025. "Identification of Genomic Regions Associated with Peanut Rust Resistance by Genome-Wide Association Studies" Plants 14, no. 8: 1219. https://doi.org/10.3390/plants14081219
APA StyleShi, X., Sun, Z., Qi, F., Han, S., Zheng, Y., Dong, W., Zhang, M., & Zhang, X. (2025). Identification of Genomic Regions Associated with Peanut Rust Resistance by Genome-Wide Association Studies. Plants, 14(8), 1219. https://doi.org/10.3390/plants14081219






