Unveiling Novel Genetic Loci and Superior Alleles for Nickel Accumulation in Wheat via Genome-Wide Association Study
Abstract
1. Introduction
2. Results
2.1. Phenotypic Variation in Ni Content in Wheat Grains in Natural Population
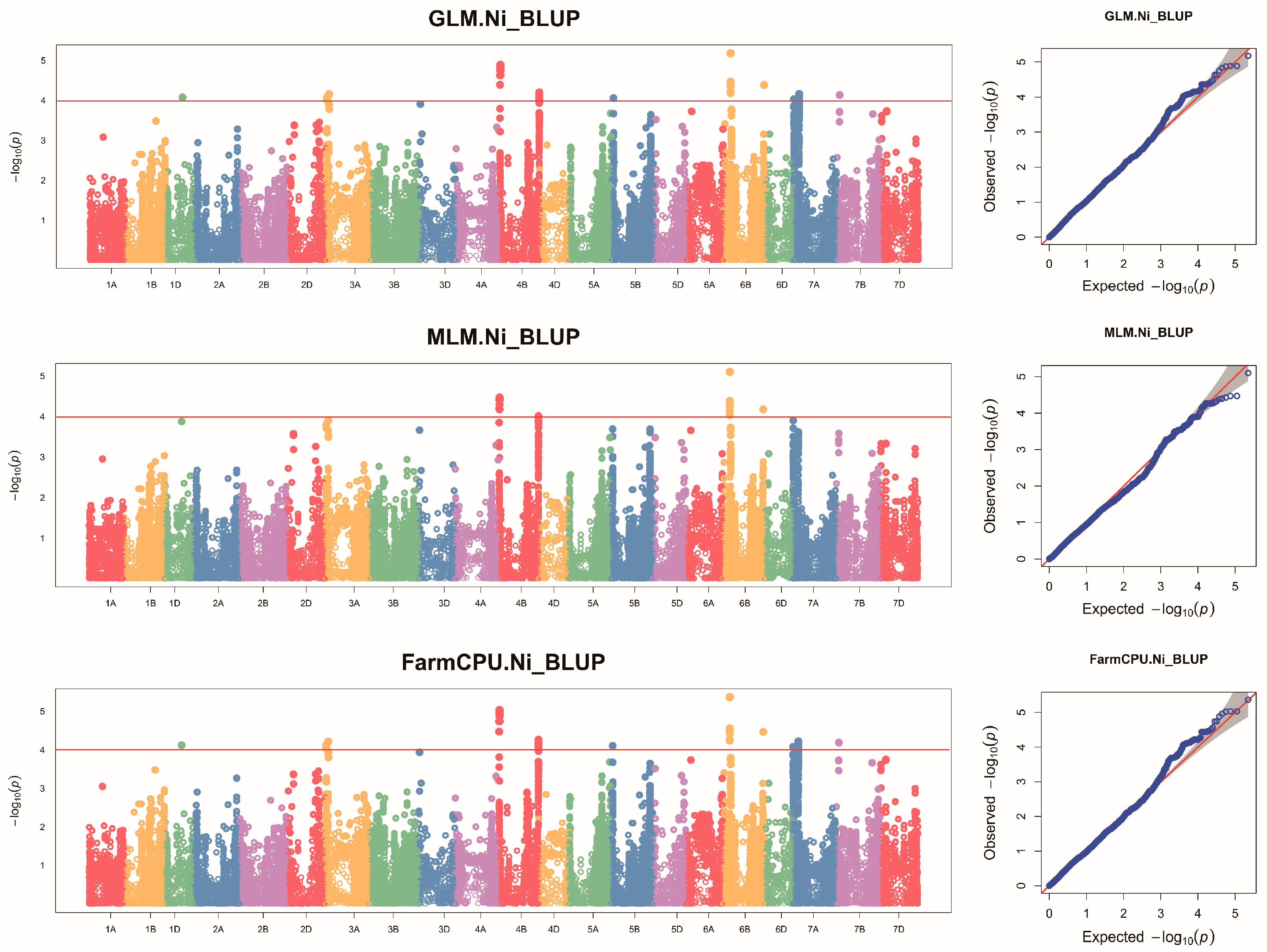
2.2. GWAS of Ni Content in Wheat Grains
2.3. Prediction of the Candidate Genes for Genetic Loci Controlling Ni Accumulation
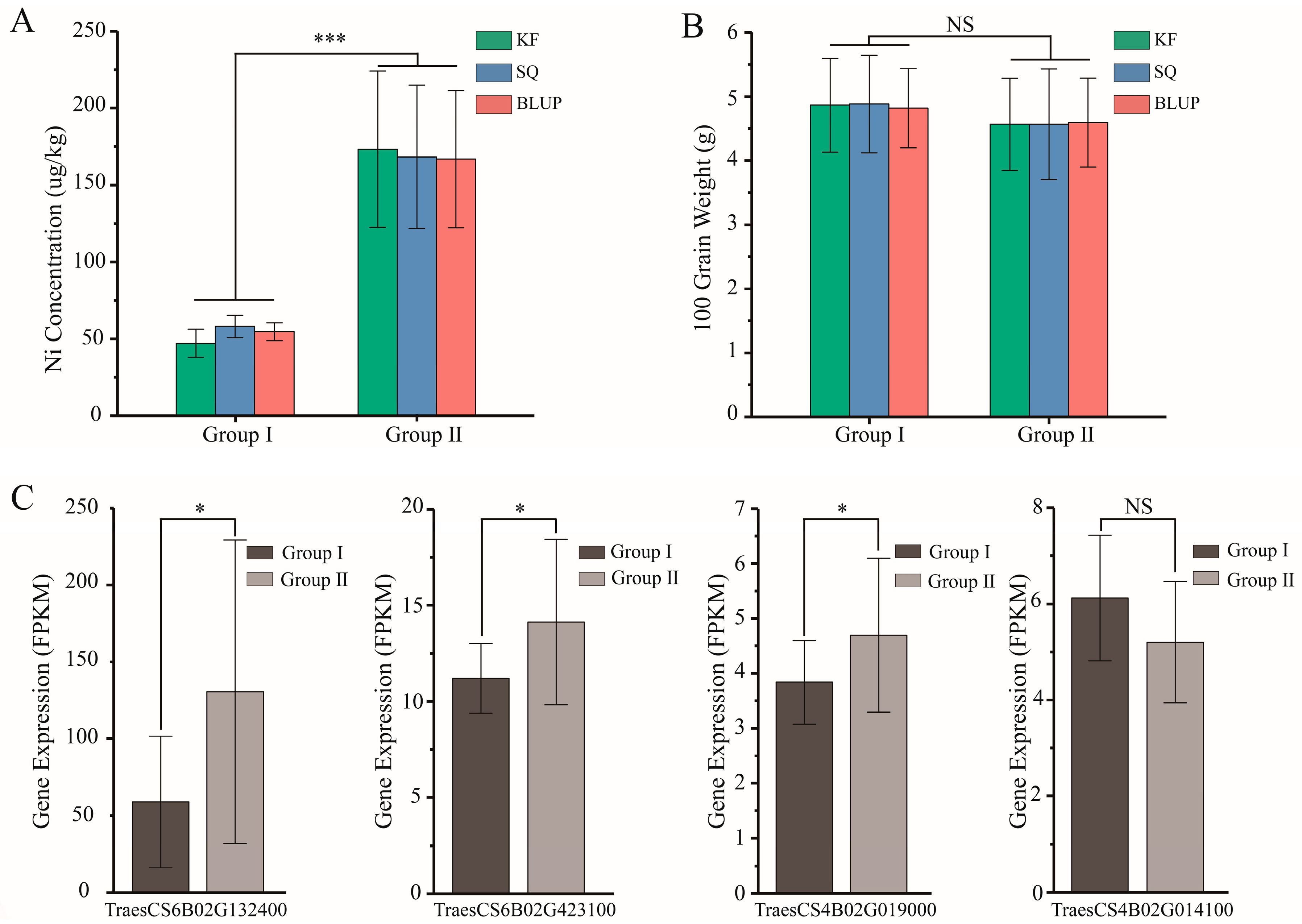
2.4. Phenotypic Effect Evaluation of the Three Stable Genetic Loci
3. Discussion
3.1. High-Density Molecular Markers and Genetic Diversity
3.2. Determining High-Confidence Genetic Loci Through Multi-Model GWAS
3.3. Putative Candidate Genes for Ni Accumulation
3.4. Molecular Pyramiding Breeding for Ni Accumulation in Wheat
4. Materials and Methods
4.1. Plant Materials
4.2. Determination of Ni Content and 100-Grain Weight in Wheat Grains
4.3. Genotyping and Quality Control
4.4. Statistical Analysis and GWAS Mapping
4.5. Putative Candidate Gene Predictions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ANOVA | analysis of variance |
| BLUP | best linear unbiased predictors |
| CAT | catalase |
| CIMMYT | International Maize And Wheat Improvement Center |
| Co | cobalt |
| CTAB | Hexadecyl trimethyl ammonium bromide |
| DNA | Deoxyribonucleic Acid |
| EFSA | European Food Safety Authority |
| FarmCPU | fixed and random model circulating probability unification |
| Fe | ferrous |
| GLM | general linear model |
| GR | glutathione reductase |
| GRPs | glycine-rich proteins |
| GSH | glutathione |
| GWAS | genome-wide association studies |
| HAAS | Henan Academy Of Agricultural Sciences |
| IWGSC | International Wheat Genome Sequence Consortium |
| KASP | Kompetitive Allele-Specific PCR |
| KF | Kaifeng |
| MAF | minor allele frequency |
| MAS | marker-assisted selection |
| MLM | mixed linear model |
| Mn | manganous |
| MTA | marker–trait association |
| MTPs | metal tolerance proteins |
| MTs | metallothioneins |
| Ni | nickel |
| NRAMP | natural resistance-associated macrophage protein |
| PCA | principal component analysis |
| PCs | phytochelatins |
| POD | peroxidase |
| PVE | phenotypic variation explained |
| quantile–quantile | |
| QTL | quantitative trait locus |
| QTN | Quantitative trait nucleotides |
| RNA | Ribonucleic acid |
| SAT | serine acetyltransferase |
| SNP | single nucleotide polymorphism |
| SOD | superoxide dismutase |
| SQ | Shangqiu |
| TMP56 | transmembrane protein 56 |
| ZIP | Zn-regulated, iron-regulated transporter-like protein |
| Zn | zinc |
References
- Rizwan, M.; Usman, K.; Alsafran, M. Ecological impacts and potential hazards of nickel on soil microbes, plants, and human health. Chemosphere 2024, 357, 142028. [Google Scholar] [CrossRef] [PubMed]
- Atta, N.; Shahbaz, M.; Farhat, F.; Maqsood, M.F.; Zulfiqar, U.; Naz, N.; Ahmed, M.M.; Hassan, N.U.; Mujahid, N.; Mustafa, A.E.-Z.M.A.; et al. Proline-mediated redox regulation in wheat for mitigating nickel-induced stress and soil decontamination. Sci. Rep. 2024, 14, 456. [Google Scholar] [CrossRef] [PubMed]
- Cangul, H.; Broday, L.; Salnikow, K.; Sutherland, J.; Peng, W.; Zhang, Q.; Poltaratsky, V.; Yee, H.; Zoroddu, M.A.; Costa, M. Molecular mechanisms of nickel carcinogenesis. Toxicol. Lett. 2002, 127, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Hambidge, M. Human zinc deficiency. J. Nutr. 2000, 130, 1344S–1349S. [Google Scholar] [CrossRef]
- Alfano, M.; Cavazza, C. Structure, function, and biosynthesis of nickel-dependent enzymes. Protein Sci. 2020, 29, 1071–1089. [Google Scholar] [CrossRef]
- Dixon, N.E.; Gazzola, C.; Blakeley, R.L.; Zerner, B. Jack bean urease (EC 3.5.1.5). Metalloenzyme. Simple biological role for nickel. J. Am. Chem. Soc. 1975, 97, 4131–4133. [Google Scholar] [CrossRef]
- Li, Y.; Zamble, D.B. Nickel homeostasis and nickel regulation: An overview. Chem. Rev. 2009, 109, 4617–4643. [Google Scholar] [CrossRef]
- Prajapati, D.H.; Ausma, T.; de Boer, J.; Hawkesford, M.J.; de Kok, L.J. Nickel toxicity in Brassica rapa seedlings: Impact on sulfur metabolism and mineral nutrient content. J. Cultiv. Plants 2020, 72, 473–478. [Google Scholar]
- Nagajyoti, P.C.; Lee, K.D.; Sreekanth, T.V.M. Heavy metals, occurrence and toxicity for plants: A review. Environ. Chem. Lett. 2010, 8, 199–216. [Google Scholar] [CrossRef]
- Ren, X. Functional Analysis of a Putative High-Affinity Ni2+/Co2+ Transporter (AtHAN) in Arabidopsis. Master’s Thesis, Northeast Forestry University, Harbin, China, 2013. [Google Scholar]
- Nishida, S.; Tsuzuki, C.; Kato, A.; Aisu, A.; Yoshida, J.; Mizuno, T. AtIRT1, the primary iron uptake transporter in the root, mediates excess nickel accumulation in Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 1433–1442. [Google Scholar] [CrossRef]
- Bozzi, A.T.; Gaudet, R. Molecular mechanism of Nramp-family transition metal transport. J. Mol. Biol. 2021, 433, 166991. [Google Scholar] [CrossRef]
- Van der Pas, L.; Ingle, R.A. Towards an understanding of the molecular basis of nickel hyperaccumulation in plants. Plants 2019, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Usui, K.; Horie, K.; Nosaka, S.; Mizuno, N.; Obata, H. Cloning of three ZIP/Nramp transporter genes from a Ni hyperaccumulator plant Thlaspi japonicum and their Ni-transport abilities. Plant Physiol. Biochem. 2005, 438, 793–801. [Google Scholar] [CrossRef]
- Angon, P.B.; Islam, M.S.; Kc, S.; Das, A.; Anjum, N.; Poudel, A.; Suchi, S.A. Sources, effects and present perspectives of heavy metals contamination: Soil, plants and human food chain. Heliyon 2024, 10, e28357. [Google Scholar] [CrossRef]
- Bellaloui, N.; Knizia, D.; Yuan, J.; Song, Q.; Betts, F.; Register, T.; Williams, E.; Lakhssassi, N.; Mazouz, H.; Nguyen, H.T.; et al. Genetic mapping for QTL associated with seed nickel and molybdenum accumulation in the soybean ‘Forrest’ by ‘Williams 82’ RIL population. Plants 2023, 12, 3709. [Google Scholar] [CrossRef]
- Bhatta, M.; Baenziger, P.S.; Waters, B.M.; Poudel, R.; Belamkar, V.; Poland, J.; Morgounov, A. Genome-wide association study reveals novel genomic regions associated with 10 grain minerals in synthetic hexaploid wheat. Int. J. Mol. Sci. 2018, 19, 3237. [Google Scholar] [CrossRef] [PubMed]
- Safdar, L.B.; Almas, F.; Rehman Au Umer, M.J.; Ali Shah, S.M.; Uddin, S.; Ashfaq, S.; Rahman, H.U.; Quraishi, U.M. Genetic dissection of Ni toxicity in a spring wheat diversity panel by using 90 K SNP array. Curr. Plant Biol. 2020, 24, 100175. [Google Scholar] [CrossRef]
- Shahzad, B.; Tanveer, M.; Rehman, A.; Cheema, S.A.; Fahad, S.; Rehman, S.; Sharma, A. Nickel; whether toxic or essential for plants and environment—A review. Plant Physiol. Biochem. 2018, 132, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.; Dutta, A.; Mohapatra, K.K.; Pradhan, S. Nickel the ultra-micronutrient: Significant for plant growth and metabolism. Food Chem. Toxicol. 2020, 1, 35–37. [Google Scholar]
- Kan, M.; Fujiwara, T.; Kamiya, T. Golgi-Localized OsFPN1 is involved in Co and Ni transport and their detoxification in rice. Rice 2022, 15, 36. [Google Scholar] [CrossRef]
- Mirete, S.; de Figueras, C.G.; González-Pastor, J.E. Novel nickel resistance genes from the rhizosphere metagenome of plants adapted to acid mine drainage. Appl. Env. Microbiol. 2007, 73, 6001–6011. [Google Scholar] [CrossRef] [PubMed]
- Allen, A.M.; Winfield, M.O.; Burridge, A.J.; Downie, R.C.; Benbow, H.R.; Barker, G.L.A.; Wilkinson, P.A.; Coghill, J.; Waterfall, C.; Davassi, A.; et al. Characterization of a wheat breeders’ array suitable for high-throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum L.). Plant Biotechnol. J. 2017, 15, 390–401. [Google Scholar] [CrossRef]
- Sun, C.; Dong, Z.; Zhao, L.; Ren, Y.; Zhang, N.; Chen, F. The Wheat 660K SNP array demonstrates great potential for marker-assisted selection in polyploid wheat. Plant Biotechnol. J. 2020, 18, 1354–1360. [Google Scholar] [CrossRef]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef]
- Winfield, M.O.; Allen, A.M.; Burridge, A.J.; Barker, G.L.A.; Benbow, H.R.; Wilkinson, P.A.; Coghill, J.; Waterfall, C.; Davassi, A.; Scopes, G.; et al. High-density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol. J. 2016, 14, 1195–1206. [Google Scholar] [CrossRef]
- Long, A.D.; Langley, C.H. The power of association studies to detect the contribution of candidate genetic loci to variation in complex traits. Genome Res. 1999, 9, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and prospects of association mapping in plants. Plant Genome 2008, 1, 5–20. [Google Scholar] [CrossRef]
- Yang, X.; Pan, Y.; Singh, P.K.; He, X.; Ren, Y.; Zhao, L.; Zhang, N.; Cheng, S.; Chen, F. Investigation and genome-wide association study for Fusarium crown rot resistance in Chinese common wheat. BMC Plant Biol. 2019, 19, 153. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, F.; Zhao, C.; Lv, G.; Sun, C.; Pan, Y.; Guo, X.; Chen, F. Genome-wide association study of six quality traits reveals the association of the TaRPP13L1 gene with flour colour in Chinese bread wheat. Plant Biotechnol. J. 2019, 17, 2106–2122. [Google Scholar] [CrossRef]
- Krishnappa, G.; Khan, H.; Krishna, H.; Kumar, S.; Mishra, C.N.; Parkash, O.; Devate, N.B.; Nepolean, T.; Rathan, N.D.; Mamrutha, H.M.; et al. Genetic dissection of grain iron and zinc, and thousand kernel weight in wheat (Triticum aestivum L.) using genome-wide association study. Sci. Rep. 2022, 12, 12444. [Google Scholar] [CrossRef]
- Cu, S.T.; Guild, G.; Nicolson, A.; Velu, G.; Singh, R.; Stangoulis, J. Genetic dissection of zinc, iron, copper, manganese and phosphorus in wheat (Triticum aestivum L.) grain and rachis at two developmental stages. Plant Sci. 2020, 291, 110338. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jia, J.; Cai, Z.; Duan, M.; Jiang, Z.; Xia, Q.; Ma, Q.; Lian, T.; Nian, H. Identification of quantitative trait loci (QTLs) and candidate genes of seed Iron and zinc content in soybean [Glycine max (L.) Merr.]. BMC Genom. 2022, 23, 146. [Google Scholar] [CrossRef]
- Alagarasan, G.; Dubey, M.; Aswathy, K.S.; Chandel, G. Genome wide identification of orthologous ZIP genes associated with zinc and iron translocation in Setaria italica. Front. Plant Sci. 2017, 8, 775. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, X.; Shang, Y.; Zhao, S.; Li, Y.; Zhou, X.; Huo, X.; Qiao, P.; Wang, X.; Dai, K.; et al. Genome-wide association study reveals genetic loci for ten trace elements in foxtail millet (Setaria italica). Theor. Appl. Genet. 2024, 137, 186. [Google Scholar] [CrossRef]
- Mountjoy, E.; Schmidt, E.M.; Carmona, M.; Schwartzentruber, J.; Peat, G.; Miranda, A.; Fumis, L.; Hayhurst, J.; Buniello, A.; Karim, M.A.; et al. An open approach to systematically prioritize causal variants and genes at all published human GWAS trait-associated loci. Nat. Genet. 2021, 53, 1527–1533. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Cheng, K.; Li, J.; Deng, Z.; Zhang, C.; Zhu, H. Roles of plant glycine-rich RNA-binding proteins in development and stress responses. Int. J. Mol. Sci. 2021, 22, 5849. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.O.; Safdar, M.; Kang, H.; Kim, J. Glycine-rich RNA-binding protein AtGRP7 functions in nickel and lead tolerance in Arabidopsis. Plants 2024, 13, 187. [Google Scholar] [CrossRef]
- Mangeon, A.; Junqueira, R.M.; Sachetto-Martins, G. Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal. Behav. 2010, 5, 99–104. [Google Scholar] [CrossRef]
- Sharma, S.S.; Dietz, K.-J. The relationship between metal toxicity and cellular redox imbalance. Trends Plant Sci. 2009, 14, 43–50. [Google Scholar] [CrossRef]
- Jaffré, T.; Reeves, R.D.; Baker, A.J.M.; Schat, H.; van der Ent, A. The discovery of nickel hyperaccumulation in the New Caledonian tree Pycnandra acuminata 40 years on: An introduction to a Virtual Issue. New Phytol. 2018, 218, 397–400. [Google Scholar] [CrossRef]
- Pishchik, V.; Mirskaya, G.; Chizhevskaya, E.; Chebotar, V.; Chakrabarty, D. Nickel stress-tolerance in plant-bacterial associations. PeerJ 2021, 9, e12230. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, N.P.; Walsh, K.B.; Baker, A.J.M. Detection and quantification of ligands involved in nickel detoxification in a herbaceous Ni hyperaccumulator Stackhousia tryonii Bailey. J. Exp. Bot. 2005, 56, 1343–1349. [Google Scholar] [CrossRef]
- Merlot, S.; Hannibal, L.; Martins, S.; Martinelli, L.; Amir, H.; Lebrun, M.; Thomine, S. The metal transporter PgIREG1 from the hyperaccumulator Psychotria gabriellae is a candidate gene for nickel tolerance and accumulation. J. Exp. Bot. 2014, 65, 1551–1564. [Google Scholar] [CrossRef] [PubMed]
- Na, G.; Salt, D.E. Differential regulation of serine acetyltransferase Is involved in nickel hyperaccumulation in thlaspi goesingense. J. Biol. Chem. 2011, 286, 40423–40432. [Google Scholar] [CrossRef]
- Freeman, J.L.; Persans, M.W.; Nieman, K.; Albrecht, C.; Peer, W.; Pickering, I.J.; Salt, D.E. Increased glutathione biosynthesis plays a role in nickel tolerance in thlaspi nickel hyperaccumulators. Plant Cell 2004, 16, 2176–2191. [Google Scholar] [CrossRef]
- Breton, G.; Danyluk, J.; Charron, J.B.; Sarhan, F. Expression profiling and bioinformatic analyses of a novel stress-regulated multispanning transmembrane protein family from cereals and Arabidopsis. Plant Physiol. 2003, 132, 64–74. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, G.; Im, W.; Sugita, Y.; Feig, M. Clustering and dynamics of crowded proteins near membranes and their influence on membrane bending. Proc. Natl. Acad. Sci. USA 2019, 116, 24562–24567. [Google Scholar] [CrossRef]
- Zhang, Q.; Shen, L.; Lin, F.; Liao, Q.; Xiao, S.; Zhang, W. Anionic phospholipid-mediated transmembrane transport and intracellular membrane trafficking in plant cells. New Phytol. 2025, 245, 1386–1402. [Google Scholar] [CrossRef]
- Genchi, G.; Carocci, A.; Lauria, G.; Sinicropi, M.S.; Catalano, A. Nickel: Human health and environmental toxicology. Int. J. Environ. Res. Public Health 2020, 17, 679. [Google Scholar] [CrossRef]
- Khan, M.I.R.; Jalil, S.; Chopra, P.; Chhillar, H.; Ferrante, A.; Khan, N.A.; Ansari, M.I. Role of Ni supply in growth, physiology, and urease activity in wheat (Triticum aestivum L.). Plant Physiol. Biochem. 2021, 162, 171–181. [Google Scholar]
- Fernando Gd, M.; Bresolin, J.D.; Santos, E.F.; Furlan, F.; Lopes da Silva, W.T.; Polacco, J.C.; Lavres, J. Nickel availability in soil as influenced by liming and its role in soybean nitrogen metabolism. Front. Plant Sci. 2016, 7, 1358. [Google Scholar]
- Dalir, N.; Tandy, S.; Gramlich, A.; Khoshgoftarmanesh, A.; Schulin, R. Effects of nickel on zinc uptake and translocation in two wheat cultivars differing in zinc efficiency. Environ. Exp. Bot. 2017, 134, 96–101. [Google Scholar] [CrossRef]
- Clemens, S.; Ma, J.F. Toxic heavy metal and metalloid accumulation in crop plants and foods. Annu. Rev. Plant Biol. 2016, 67, 489–512. [Google Scholar] [CrossRef] [PubMed]
- Gajewska, E.; Skłodowska, M.; Słaba, M.; Mazur, J. Effect of nickel on antioxidative enzyme activities, proline and chlorophyll contents in wheat shoots. Biol. Plant. 2006, 50, 653–659. [Google Scholar] [CrossRef]
- Pandey, N.; Sharma, C.P. Effect of heavy metals Co2+, Ni2+ and Cd2+ on growth and metabolism of cabbage. Plant Sci. 2002, 163, 753–758. [Google Scholar] [CrossRef]
- Bhalerao, S.; Sharma, A.; Poojari, A. Toxicity of nickel in plants. Int. J. Pure Appl. Biosci. 2015, 3, 345–355. [Google Scholar]
- Semagn, K.; Babu, R.; Hearne, S.; Olsen, M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol. Breed. 2014, 33, 1–14. [Google Scholar] [CrossRef]
- Bassi, F.M.; Bentley, A.R.; Charmet, G.; Ortiz, R.; Crossa, J. Breeding schemes for the implementation of genomic selection in wheat (Triticum spp.). Plant Sci. 2016, 242, 23–36. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Zhang, Z.; Ersoz, E.; Lai, C.Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef] [PubMed]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
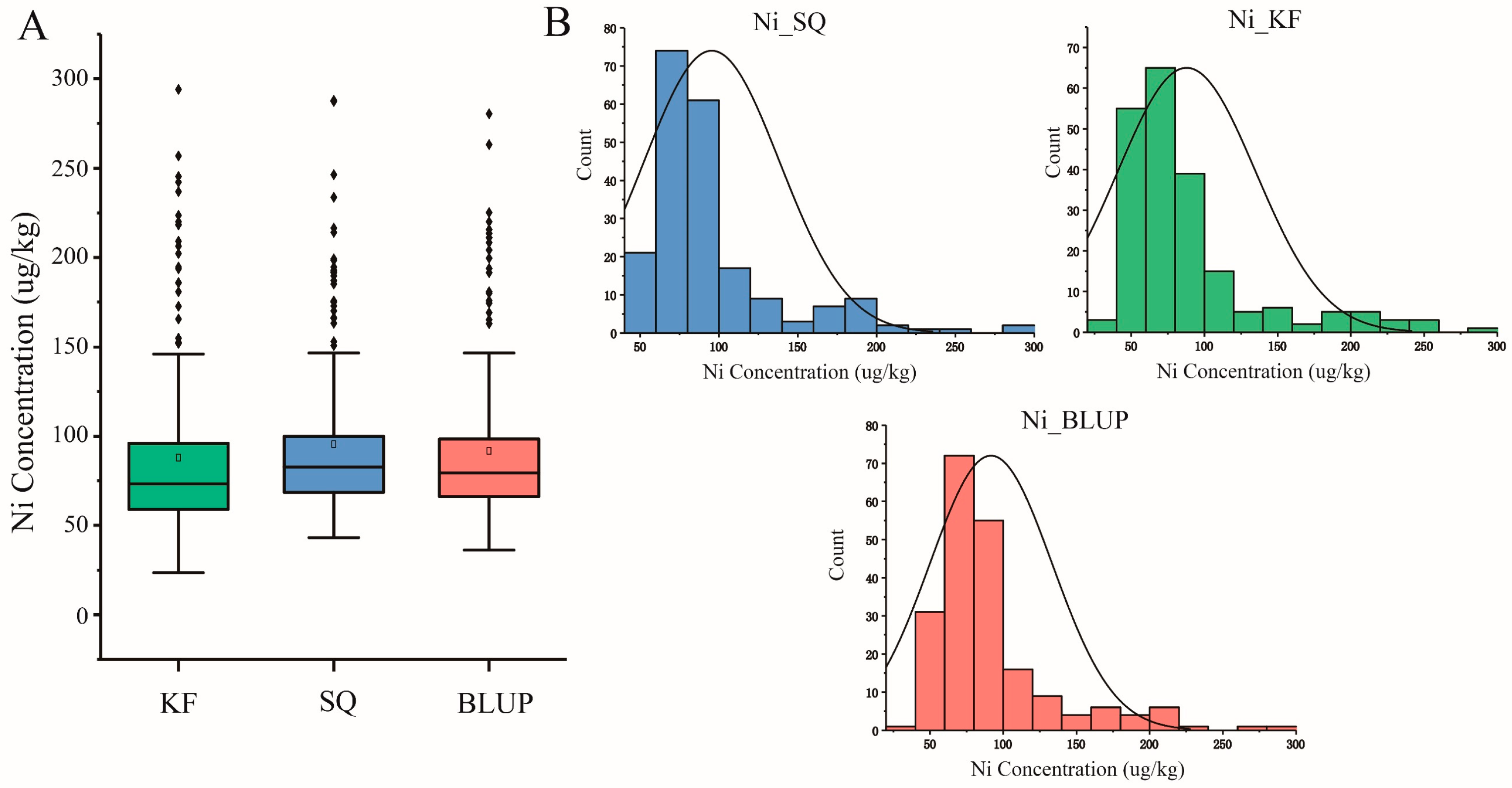

| Location | Year | Trait | Mean ± SD 1 (μg/kg) | Range (μg/kg) | Kurt 2 | Skew 3 |
|---|---|---|---|---|---|---|
| Kaifeng (KF) | 2023 | Ni content | 95.42 ± 43.03 | 43.13–287.93 | 4.33 | 1.99 |
| Shangqiu (SQ) | 2023 | Ni content | 87.85 ± 47.18 | 23.46–294.05 | 3.78 | 1.98 |
| BLUP | 2023 | Ni content | 91.63 ± 41.78 | 36.28–280.47 | 4.18 | 2.02 |
| ID 1 | Chr. | Interval Range (Mb) | No. of SNPs | Loc. | Peak SNP 2 | Position (bp) 3 | p-Value 4 | R2 (%) 5 |
|---|---|---|---|---|---|---|---|---|
| 1 | 3A | 15.42–50.74 | 9 | SQ | AX-110424807 | 15,485,206 | 4.34 × 10−5 | 8.31 |
| 2 | 4B | 6.64–11.66 | 7 | KF | AX-110161827 | 11,593,889 | 4.84 × 10−5 | 9.76 |
| SQ | AX-110161827 | 11,593,889 | 3.18 × 10−5 | 8.63 | ||||
| BLUP | AX-110161827 | 11,593,889 | 3.40 × 10−5 | 9.27 | ||||
| 3 | 4B | 670.43 | 1 | BLUP | AX-109482775 | 670,432,861 | 9.70 × 10−5 | 8.20 |
| 4 | 6B | 125.48–126.30 | 11 | KF | AX-111126872 | 125,482,475 | 1.07 × 10−5 | 11.29 |
| SQ | AX-111126872 | 125,482,475 | 1.47 × 10−5 | 9.43 | ||||
| BLUP | AX-111126872 | 125,482,475 | 7.94 × 10−6 | 10.77 | ||||
| 5 | 6B | 693.58 | 1 | SQ | AX-108754008 | 693,584,519 | 4.94 × 10−6 | 10.58 |
| BLUP | AX-108754008 | 693,584,519 | 6.69 × 10−5 | 8.58 |
| ID 1 | Chr. | SNP_Id | Position (Mb) 2 | Region (Mb) | Near Locus Previously Reported in the Same Locus 3 | Candidate Genes | Annotation |
|---|---|---|---|---|---|---|---|
| 1 | 4B | AX-110161827 | 11.59 | 6.64–11.66 | qNi14 [18] | TraesCS4B02G019000 | Transmembrane protein 56 |
| 2 | 6B | AX-111126872 | 125.48 | 125.48–126.30 | -- | TraesCS6B02G132400 | Glycine rich protein |
| 3 | 6B | AX-108754008 | 693.58 | 693.58 | -- | TraesCS6B02G423100 | Glutathione reductase |
| SNP_Id | Chromosome | Allele Type | Phenotype Value (BLUP) | Allele Number | Allele Percentage (%) | p-Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Superior | Inferior | Superior | Inferior | Superior | Inferior | Superior | Inferior | Ni_KF | Ni_SQ | Ni_BLUP | ||
| AX-110161827 | 4B | TT | CC | 84.76 | 118.14 | 164 | 41 | 80.00 | 20.00 | 7.17 × 10−6 | 3.08 × 10−6 | 3.00 × 10−6 |
| AX-111126872 | 6B | CC | TT | 87.53 | 130.05 | 187 | 20 | 90.34 | 9.66 | 3.15 × 10−5 | 8.22 × 10−6 | 1.02 × 10−5 |
| AX-108754008 | 6B | AA | GG | 88.79 | 145.69 | 195 | 10 | 95.12 | 4.88 | 8.18 × 10−4 | 4.16 × 10−7 | 2.06 × 10−5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, X.; Geng, S.; Hou, J.; Shi, T.; Qin, M.; Li, W.; Dai, Z.; Zhou, Z.; Zhang, M.; Lei, Z. Unveiling Novel Genetic Loci and Superior Alleles for Nickel Accumulation in Wheat via Genome-Wide Association Study. Plants 2025, 14, 1262. https://doi.org/10.3390/plants14081262
Shi X, Geng S, Hou J, Shi T, Qin M, Li W, Dai Z, Zhou Z, Zhang M, Lei Z. Unveiling Novel Genetic Loci and Superior Alleles for Nickel Accumulation in Wheat via Genome-Wide Association Study. Plants. 2025; 14(8):1262. https://doi.org/10.3390/plants14081262
Chicago/Turabian StyleShi, Xia, Shenghui Geng, Jinna Hou, Taotao Shi, Maomao Qin, Wenxu Li, Ziju Dai, Zhengfu Zhou, Minghui Zhang, and Zhensheng Lei. 2025. "Unveiling Novel Genetic Loci and Superior Alleles for Nickel Accumulation in Wheat via Genome-Wide Association Study" Plants 14, no. 8: 1262. https://doi.org/10.3390/plants14081262
APA StyleShi, X., Geng, S., Hou, J., Shi, T., Qin, M., Li, W., Dai, Z., Zhou, Z., Zhang, M., & Lei, Z. (2025). Unveiling Novel Genetic Loci and Superior Alleles for Nickel Accumulation in Wheat via Genome-Wide Association Study. Plants, 14(8), 1262. https://doi.org/10.3390/plants14081262





