Characterization of 4-Coumarate-CoA Ligase (4CL) Genes in Wheat Uncovers Ta4CL91’s Role in Drought and Salt Stress Adaptation
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Identification and Phylogenetic Analysis of 4CL Genes in Wheat
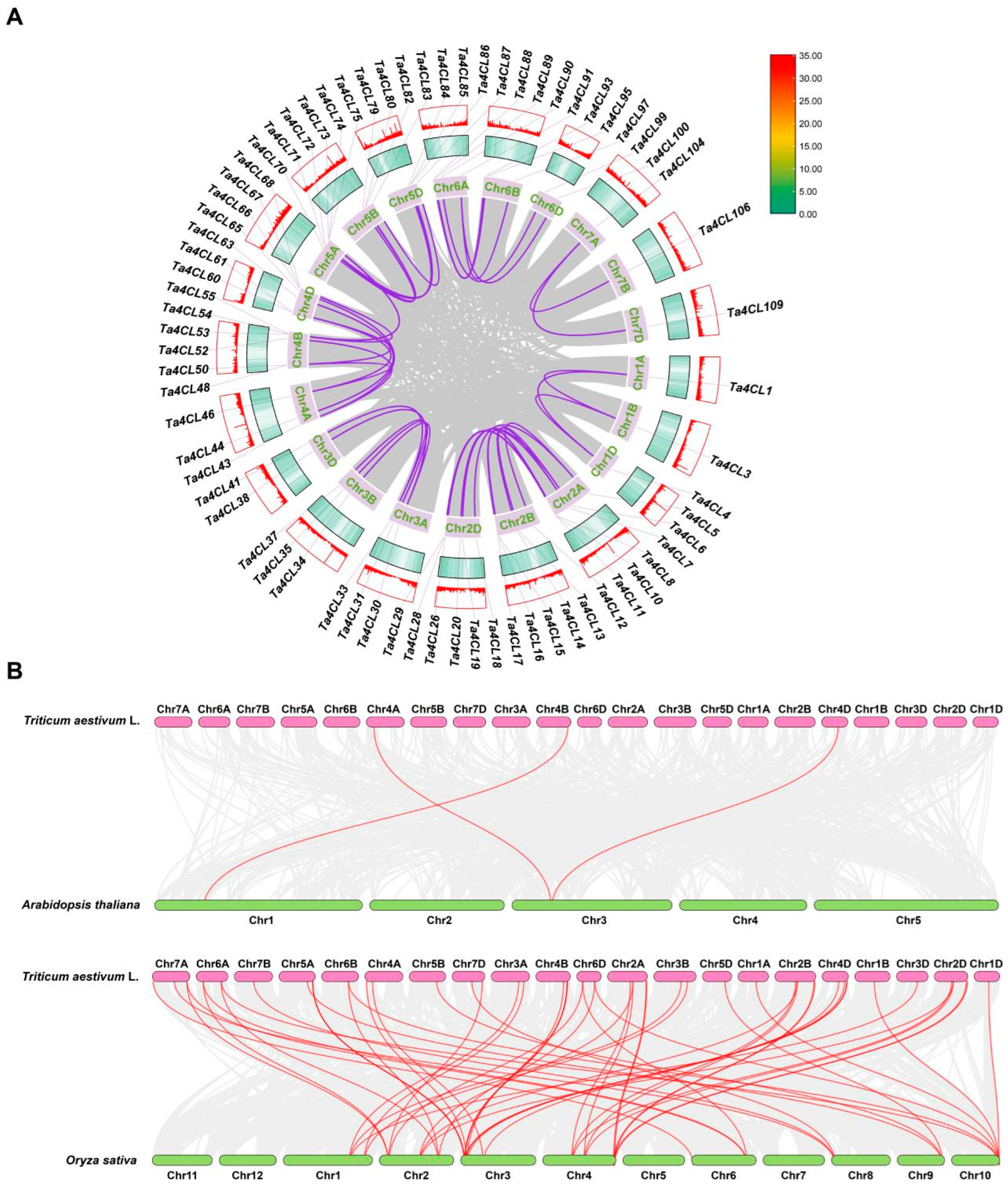
2.2. Chromosomal Distribution and Collinearity Analysis of Ta4CL Genes
2.3. Analysis of Conserved Motifs, Domains, and Gene Structures Within the 4CL Gene Family in Wheat
2.4. Analysis of Cis-Acting Elements for Ta4CLs
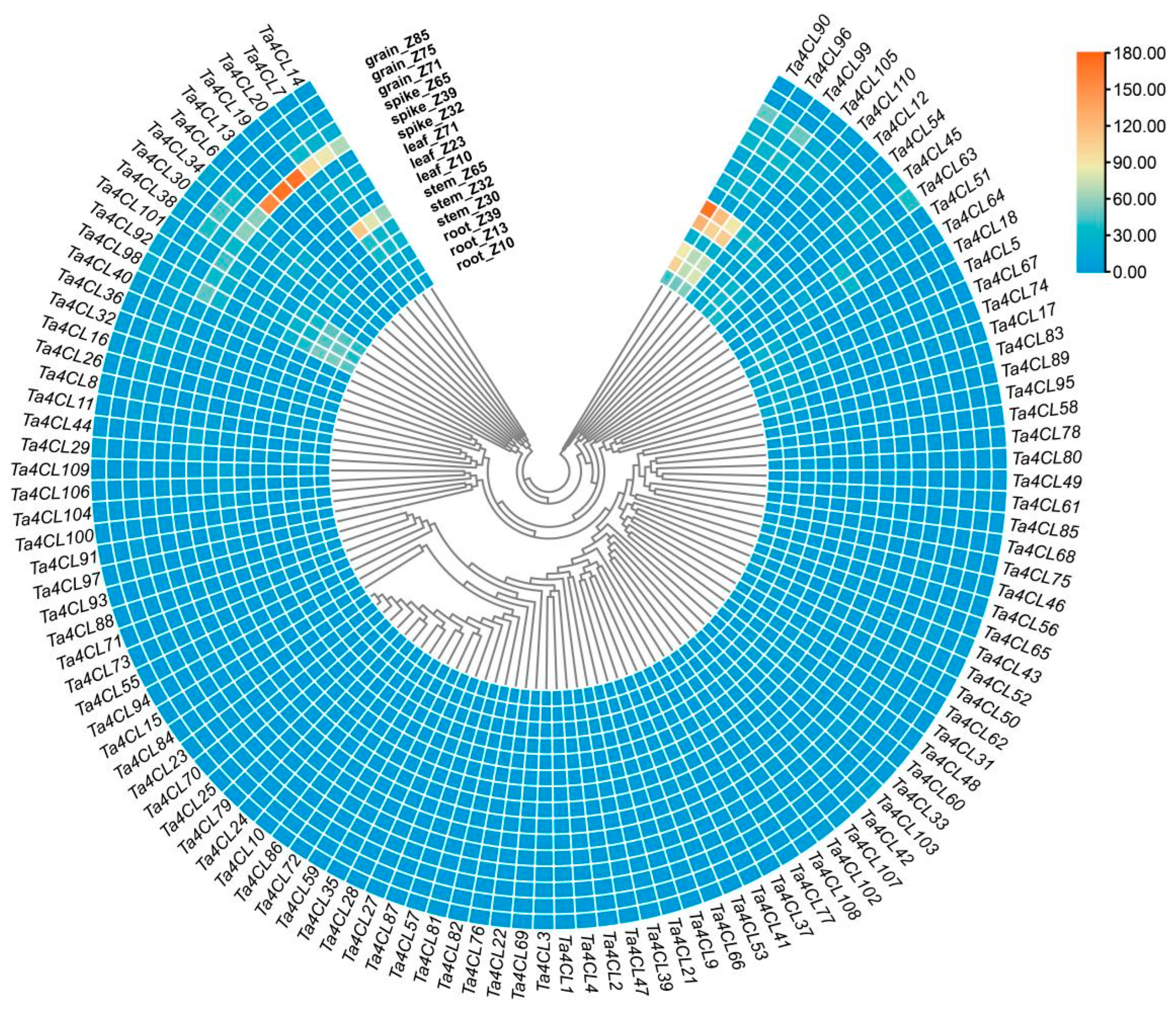
2.5. Analysis of the Tissue Expression Patterns of Ta4CL Genes
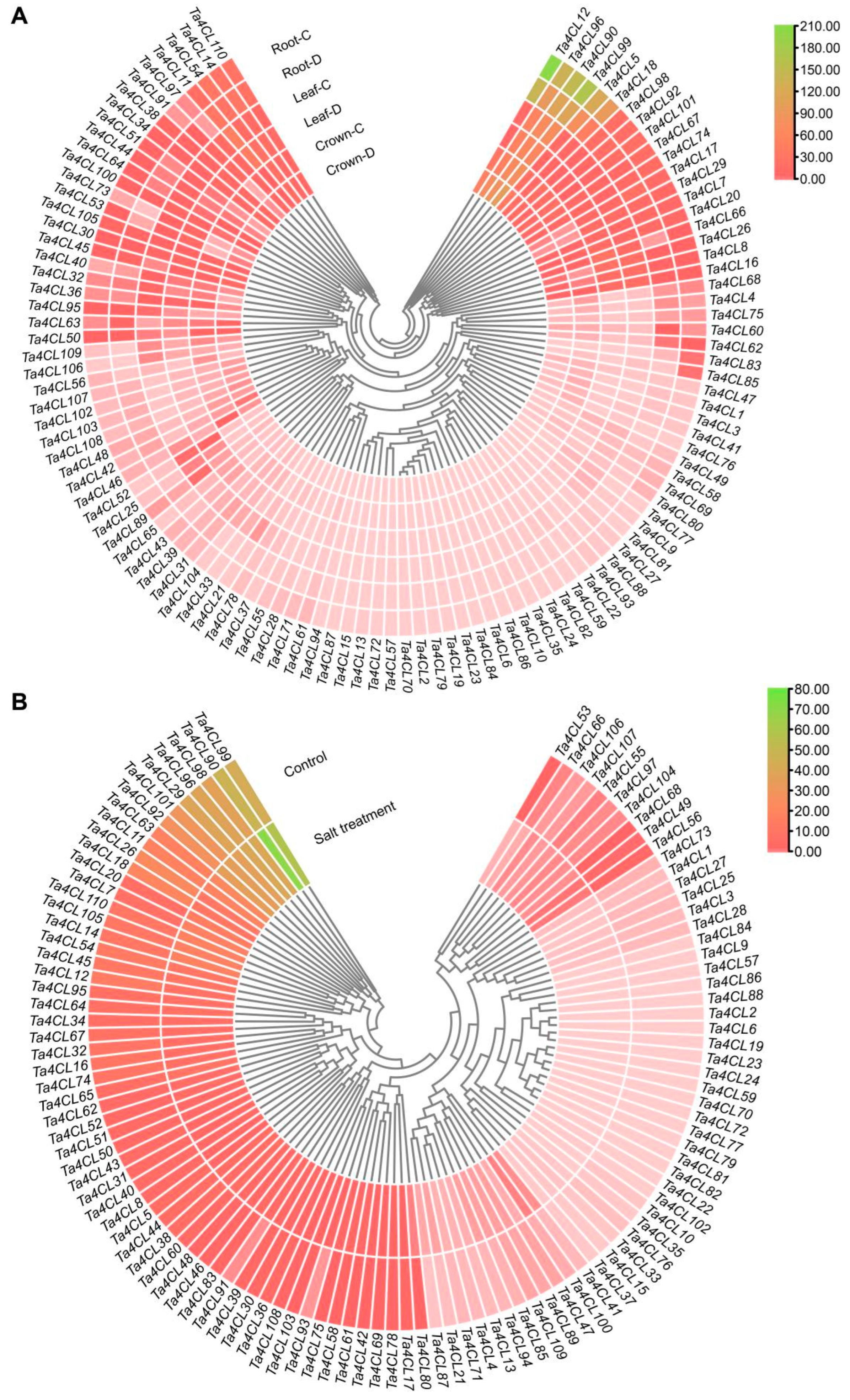
2.6. Expression Analysis of Ta4CLs Under Drought and Salt Stress
2.7. Ta4CL91 Silencing Compromises Drought and Salt Stress Tolerance in Wheat Seedlings
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification of Members of the 4CL Gene Family in Wheat
4.2. Phylogenetic Analysis and Physicochemical Property Assessment
4.3. Analysis of Chromosomal Distribution and Gene Collinearity
4.4. Analysis of Conserved Motifs, Domains, and Gene Structure
4.5. Analysis of Cis-Acting Elements in Promoter Regions
4.6. Expression Analysis Involving Varying Tissues Under Drought and Salinity Stress
4.7. Analysis of Drought and Salt Stress Responses in Selected Ta4CL Genes
4.8. RNA Isolation, Reverse Transcription, and qPCR Analysis
4.9. VIGS Vector Construction and Infection Methodology
4.10. Tolerance Analysis Under Drought and Salt Stress Conditions
4.11. Measurement of Relative Water Content and Water Loss Rate in Detached Leaves
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shiferaw, B.; Smale, M.; Braun, H.-J.; Duveiller, E.; Reynolds, M.; Muricho, G. Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur. 2013, 5, 291–317. [Google Scholar] [CrossRef]
- Dutta, D.; Karmakar, S.; Hossain, A.; Sadhukhan, R.; Atta, K.; Pramanick, S. Chapter 1—Wheat and abiotic stress challenges: An overview. In Abiotic Stresses in Wheat; Khan, M.K., Pandey, A., Hamurcu, M., Gupta, O.P., Gezgin, S., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 1–13. [Google Scholar]
- Alam, M.N.; Islam, M.Z.; Farukh, M.A.; Chan, Z.; Akhter, M.M.; Abedin, M.T.; Hossain, M.M. Detrimental effects of abiotic stress on wheat and its management techniques. In Cereal Research Communications; Springer: Berlin/Heidelberg, Germany, 2024. [Google Scholar] [CrossRef]
- Furtak, K.; Wolińska, A. The impact of extreme weather events as a consequence of climate change on the soil moisture and on the quality of the soil environment and agriculture—A review. CATENA 2023, 231, 107378. [Google Scholar] [CrossRef]
- Lavhale, S.G.; Kalunke, R.M.; Giri, A.P. Structural, functional and evolutionary diversity of 4-coumarate-CoA ligase in plants. Planta 2018, 248, 1063–1078. [Google Scholar] [CrossRef] [PubMed]
- Hamberger, B.; Hahlbrock, K. The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc. Natl. Acad. Sci. USA 2004, 101, 2209–2214. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.; Hövel, K.; Witzel, K.; Hamberger, B.; Schomburg, D.; Kombrink, E.; Stuible, H.P. The substrate specificity-determining amino acid code of 4-coumarate:CoA ligase. Proc. Natl. Acad. Sci. USA 2003, 100, 8601–8606. [Google Scholar] [CrossRef]
- Stuible, H.; Büttner, D.; Ehlting, J.; Hahlbrock, K.; Kombrink, E. Mutational analysis of 4-coumarate:CoA ligase identifies functionally important amino acids and verifies its close relationship to other adenylate-forming enzymes. FEBS Lett. 2000, 467, 117–122. [Google Scholar] [CrossRef]
- Stuible, H.P.; Kombrink, E. Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase allows the rational design of mutant enzymes with new catalytic properties. J. Biol. Chem. 2001, 276, 26893–26897. [Google Scholar] [CrossRef]
- De Azevedo Souza, C.; Barbazuk, B.; Ralph, S.G.; Bohlmann, J.; Hamberger, B.; Douglas, C.J. Genome-wide analysis of a land plant-specific acyl:coenzyme A synthetase (ACS) gene family in Arabidopsis, poplar, rice and Physcomitrella. New Phytol. 2008, 179, 987–1003. [Google Scholar] [CrossRef]
- Ehlting, J.; Büttner, D.; Wang, Q.; Douglas, C.J.; Somssich, I.E.; Kombrink, E. Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999, 19, 9–20. [Google Scholar] [CrossRef]
- Li, Y.; Kim, J.I.; Pysh, L.; Chapple, C. Four Isoforms of Arabidopsis 4-Coumarate:CoA Ligase Have Overlapping yet Distinct Roles in Phenylpropanoid Metabolism. Plant Physiol. 2015, 169, 2409–2421. [Google Scholar] [CrossRef]
- Sun, H.; Li, Y.; Feng, S.; Zou, W.; Guo, K.; Fan, C.; Si, S.; Peng, L. Analysis of five rice 4-coumarate:coenzyme A ligase enzyme activity and stress response for potential roles in lignin and flavonoid biosynthesis in rice. Biochem. Biophys. Res. Commun. 2013, 430, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.E.K.; Choi, B.; Cho, B.-K.; Kim, J.B.; Park, S.U.; Natarajan, S.; Lim, H.-S.; Bae, H. Regulation of 4CL, encoding 4-coumarate: Coenzyme A ligase, expression in kenaf under diverse stress conditions. Plant Omics 2013, 6, 254–262. [Google Scholar]
- Moura, J.C.; Bonine, C.A.; de Oliveira Fernandes Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef] [PubMed]
- Cesarino, I. Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 2019, 56, 209–214. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, Y.; Zhu, C.; Yao, X.; Zheng, Z.; Tian, Z.; Cai, X. EkFLS overexpression promotes flavonoid accumulation and abiotic stress tolerance in plant. Physiol. Plant. 2021, 172, 1966–1982. [Google Scholar] [CrossRef]
- Shomali, A.; Das, S.; Arif, N.; Sarraf, M.; Zahra, N.; Yadav, V.; Aliniaeifard, S.; Chauhan, D.K.; Hasanuzzaman, M. Diverse Physiological Roles of Flavonoids in Plant Environmental Stress Responses and Tolerance. Plants 2022, 11, 3158. [Google Scholar] [CrossRef]
- Li, X.-l.; LÜ, X.; Wang, X.-h.; Peng, Q.; Zhang, M.-s.; Ren, M.-j. Biotic and abiotic stress-responsive genes are stimulated to resist drought stress in purple wheat. J. Integr. Agric. 2020, 19, 33–50. [Google Scholar] [CrossRef]
- Gulzar, F.; Yang, H.; Chen, J.; Hassan, B.; Huang, X.; Qiong, F. 6-BA Reduced Yield Loss under Waterlogging Stress by Regulating the Phenylpropanoid Pathway in Wheat. Plants 2024, 13, 1991. [Google Scholar] [CrossRef]
- Chen, X.; Su, W.; Zhang, H.; Zhan, Y.; Zeng, F. Fraxinus mandshurica 4-coumarate-CoA ligase 2 enhances drought and osmotic stress tolerance of tobacco by increasing coniferyl alcohol content. Plant Physiol. Biochem. PPB 2020, 155, 697–708. [Google Scholar] [CrossRef]
- Chen, X.; Wang, H.; Li, X.; Ma, K.; Zhan, Y.; Zeng, F. Molecular cloning and functional analysis of 4-Coumarate:CoA ligase 4(4CL-like 1)from Fraxinus mandshurica and its role in abiotic stress tolerance and cell wall synthesis. BMC Plant Biol. 2019, 19, 231. [Google Scholar] [CrossRef]
- Wang, C.H.; Yu, J.; Cai, Y.X.; Zhu, P.P.; Liu, C.Y.; Zhao, A.C.; Lü, R.H.; Li, M.J.; Xu, F.X.; Yu, M.D. Characterization and Functional Analysis of 4-Coumarate:CoA Ligase Genes in Mul-berry. PLoS ONE 2016, 11, e0155814. [Google Scholar] [CrossRef]
- Sun, S.C.; Xiong, X.P.; Zhang, X.L.; Feng, H.J.; Zhu, Q.H.; Sun, J.; Li, Y.J. Characterization of the Gh4CL gene family reveals a role of Gh4CL7 in drought tolerance. BMC Plant Biol. 2020, 20, 125. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Ellard, M.; Wanner, L.A.; Davis, K.R.; Douglas, C.J. The Arabidopsis thaliana 4-coumarate:CoA ligase (4CL) gene: Stress and developmentally regulated expression and nucleotide sequence of its cDNA. Plant Mol. Biol. 1995, 28, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Douglas, C.J. Phenylpropanoid metabolism and lignin biosynthesis: From weeds to trees. Trends Plant Sci. 1996, 1, 171–178. [Google Scholar] [CrossRef]
- Yang, M.; Chen, S.; Chao, K.; Ji, C.; Shi, Y. Effects of nano silicon fertilizer on the lodging resistance characteristics of wheat basal second stem node. BMC Plant Biol. 2024, 24, 54. [Google Scholar] [CrossRef]
- Hu, Y.; Qin, F.; Wu, Z.; Wang, X.; Ren, X.; Jia, Z.; Wang, Z.; Chen, X.; Cai, T. Heterogeneous population distribution enhances resistance to wheat lodging by optimizing the light environment. J. Integr. Agric. 2024, 23, 2211–2226. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Li, D.; Lin, Y.; Cai, Y. Systematic Analysis of the 4-Coumarate:Coenzyme A Ligase (4CL) Related Genes and Expression Profiling during Fruit Development in the Chinese Pear. Genes 2016, 7, 89. [Google Scholar] [CrossRef]
- Shockey, J.M.; Fulda, M.S.; Browse, J. Arabidopsis contains a large superfamily of acyl-activating enzymes. Phylogenetic and biochemical analysis reveals a new class of acyl-coenzyme a synthetases. Plant Physiol. 2003, 132, 1065–1076. [Google Scholar] [CrossRef]
- ul Haq, S.; Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.-X.; Zhang, H.-X.; Wei, A.-M.; Gong, Z.-H. Heat Shock Proteins: Dynamic Biomolecules to Counter Plant Biotic and Abiotic Stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar] [CrossRef]
- Hu, C.; Yang, J.; Qi, Z.; Wu, H.; Wang, B.; Zou, F.; Mei, H.; Liu, J.; Wang, W.; Liu, Q. Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm 2022, 3, e161. [Google Scholar] [CrossRef]
- Hong-Bo, S.; Zong-Suo, L.; Ming-An, S. LEA proteins in higher plants: Structure, function, gene expression and regulation. Colloids Surf. B Biointerfaces 2005, 45, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Salgotra, R.K.; Chauhan, B.S. Genetic Diversity, Conservation, and Utilization of Plant Genetic Resources. Genes 2023, 14, 174. [Google Scholar] [CrossRef]
- Abhinandan, K.; Skori, L.; Stanic, M.; Hickerson, N.M.N.; Jamshed, M.; Samuel, M.A. Abiotic Stress Signaling in Wheat—An Inclusive Overview of Hormonal Interactions During Abiotic Stress Responses in Wheat. Front. Plant Sci. 2018, 9, 734. [Google Scholar] [CrossRef]
- Hossain, A.; Skalicky, M.; Brestic, M.; Maitra, S.; Ashraful Alam, M.; Syed, M.A.; Hossain, J.; Sarkar, S.; Saha, S.; Bhadra, P.; et al. Consequences and Mitigation Strategies of Abiotic Stresses in Wheat (Triticum aestivum L.) under the Changing Climate. Agronomy 2021, 11, 241. [Google Scholar] [CrossRef]
- Zhang, C.H.; Ma, T.; Luo, W.C.; Xu, J.M.; Liu, J.Q.; Wan, D.S. Identification of 4CL Genes in Desert Poplars and Their Changes in Expression in Response to Salt Stress. Genes 2015, 6, 901–917. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Danquah, A.; de Zelicourt, A.; Colcombet, J.; Hirt, H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 2014, 32, 40–52. [Google Scholar] [CrossRef]
- Zhang, J.; Jia, W.; Yang, J.; Ismail, A.M. Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Zhang, J.; Yu, D.; Zhang, Y.; Liu, K.; Xu, K.; Zhang, F.; Wang, J.; Tan, G.; Nie, X.; Ji, Q.; et al. Vacuum and Co-cultivation Agroinfiltration of (Germinated) Seeds Results in Tobacco Rattle Virus (TRV) Mediated Whole-Plant Virus-Induced Gene Silencing (VIGS) in Wheat and Maize. Front. Plant Sci. 2017, 8, 393. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, F.; Xie, Q. Balancing growth and adaptation to stress: Crosstalk between brassinosteroid and abscisic acid signaling. Plant Cell Environ. 2020, 43, 2325–2335. [Google Scholar] [CrossRef]
- Kudo, M.; Kidokoro, S.; Yoshida, T.; Mizoi, J.; Todaka, D.; Fernie, A.R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Double overexpression of DREB and PIF transcription factors improves drought stress tolerance and cell elongation in transgenic plants. Plant Biotechnol. J. 2017, 15, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef]
- A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Sade, N.; Vinocur, B.J.; Diber, A.; Shatil, A.; Ronen, G.; Nissan, H.; Wallach, R.; Karchi, H.; Moshelion, M. Improving plant stress tolerance and yield production: Is the tonoplast aquaporin SlTIP2;2 a key to isohydric to anisohydric conversion? New Phytol. 2009, 181, 651–661. [Google Scholar] [CrossRef] [PubMed]







| Gene | Gene ID | No. of Amino Acids | Mol. Wt (Da) | Isoelectric Point (pI) | Instability Index (II) | Aliphatic Index | Grand Average of Hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|---|
| Ta4CL1 | TraesCS1A02G196700 | 568 | 60,239.3 | 7.72 | 43 | 96.94 | 0.141 |
| Ta4CL2 | TraesCS1B02G067600 | 564 | 61,444.37 | 6.17 | 35.62 | 89.95 | −0.048 |
| Ta4CL3 | TraesCS1B02G211300 | 587 | 62,541.94 | 8.78 | 48.46 | 96.46 | 0.119 |
| Ta4CL4 | TraesCS1D02G200200 | 568 | 60,358.44 | 8.54 | 47.57 | 96.06 | 0.13 |
| Ta4CL5 | TraesCS2A02G145800 | 542 | 57,762.92 | 5.65 | 33.4 | 101.11 | 0.173 |
| Ta4CL6 | TraesCS2A02G272900 | 620 | 66,508.68 | 6.27 | 40.33 | 96.42 | 0.102 |
| Ta4CL7 | TraesCS2A02G290900 | 690 | 76,141.63 | 5.64 | 23.98 | 80.55 | −0.187 |
| Ta4CL8 | TraesCS2A02G556900 | 558 | 60,323.35 | 6.87 | 40.81 | 86.99 | −0.024 |
| Ta4CL9 | TraesCS2A02G557100 | 557 | 60,187.48 | 8.3 | 35.91 | 86.62 | −0.006 |
| Ta4CL10 | TraesCS2A02G570500 | 545 | 59,224.46 | 8.56 | 29.14 | 96.99 | 0.036 |
| Ta4CL11 | TraesCS2A02G581600 | 529 | 55,281.21 | 6.16 | 27.28 | 95.69 | 0.15 |
| Ta4CL12 | TraesCS2B02G171200 | 540 | 57,381.47 | 5.58 | 34.13 | 101.15 | 0.173 |
| Ta4CL13 | TraesCS2B02G291100 | 619 | 66,374.5 | 6.21 | 40.07 | 96.25 | 0.106 |
| Ta4CL14 | TraesCS2B02G307300 | 720 | 79,208.5 | 6.48 | 32.26 | 81.29 | −0.162 |
| Ta4CL15 | TraesCS2B02G586300 | 565 | 61,427.81 | 8.03 | 37.25 | 86.96 | −0.025 |
| Ta4CL16 | TraesCS2B02G587100 | 558 | 60,337.42 | 7.16 | 39.98 | 87.69 | −0.029 |
| Ta4CL17 | TraesCS2B02G605800 | 536 | 55,673.58 | 6.1 | 25.21 | 96.27 | 0.167 |
| Ta4CL18 | TraesCS2D02G150400 | 540 | 57,178.25 | 5.7 | 33.88 | 100.43 | 0.186 |
| Ta4CL19 | TraesCS2D02G272200 | 619 | 66,428.59 | 6.17 | 40.07 | 96.58 | 0.11 |
| Ta4CL20 | TraesCS2D02G288800 | 731 | 80,522.71 | 6.01 | 29.53 | 79.53 | −0.179 |
| Ta4CL21 | TraesCS2D02G556000 | 563 | 61,094.64 | 8.72 | 34.52 | 87.96 | −0.009 |
| Ta4CL22 | TraesCS2D02G556100 | 501 | 54,550.86 | 9.3 | 36.53 | 79.38 | −0.105 |
| Ta4CL23 | TraesCS2D02G556500 | 578 | 62,772.43 | 7.63 | 38.39 | 87.37 | 0.015 |
| Ta4CL24 | TraesCS2D02G556600 | 564 | 61,297.7 | 8.01 | 36.13 | 83.83 | −0.028 |
| Ta4CL25 | TraesCS2D02G556900 | 478 | 51,616.28 | 6.16 | 40.48 | 87.28 | −0.049 |
| Ta4CL26 | TraesCS2D02G557900 | 601 | 64,808.66 | 8.23 | 45.49 | 87.77 | −0.03 |
| Ta4CL27 | TraesCS2D02G581200 | 548 | 59,725.13 | 8.41 | 28.24 | 98.41 | 0.055 |
| Ta4CL28 | TraesCS2D02G581900 | 548 | 59,739.14 | 8.56 | 30.03 | 97.14 | 0.04 |
| Ta4CL29 | TraesCS2D02G598500 | 531 | 55,517.3 | 5.85 | 27.58 | 94.97 | 0.127 |
| Ta4CL30 | TraesCS3A02G304600 | 631 | 69,199.48 | 7.29 | 36.07 | 89.95 | −0.143 |
| Ta4CL31 | TraesCS3A02G394200 | 558 | 58,793.98 | 8.56 | 48.52 | 96.58 | 0.191 |
| Ta4CL32 | TraesCS3A02G394300 | 560 | 58,750.7 | 7.18 | 44.01 | 97.29 | 0.18 |
| Ta4CL33 | TraesCS3A02G472200 | 550 | 58,403.73 | 8.77 | 44.6 | 97.42 | 0.177 |
| Ta4CL34 | TraesCS3B02G331900 | 634 | 69,537.89 | 7.03 | 38.38 | 89.35 | −0.13 |
| Ta4CL35 | TraesCS3B02G426200 | 560 | 58,750.76 | 6.44 | 49.26 | 94.11 | 0.213 |
| Ta4CL36 | TraesCS3B02G426300 | 562 | 58,863.77 | 6.85 | 43.96 | 97.63 | 0.188 |
| Ta4CL37 | TraesCS3B02G515400 | 550 | 58,190.55 | 8.93 | 46.19 | 98.13 | 0.218 |
| Ta4CL38 | TraesCS3D02G297300 | 626 | 68,744.91 | 7.63 | 37.9 | 88.93 | −0.174 |
| Ta4CL39 | TraesCS3D02G388000 | 558 | 58,856.93 | 7.71 | 47.81 | 97.1 | 0.155 |
| Ta4CL40 | TraesCS3D02G388100 | 562 | 58,981.88 | 6.85 | 46.12 | 96.42 | 0.164 |
| Ta4CL41 | TraesCS3D02G468000 | 554 | 58,775.37 | 9.2 | 45.59 | 98.12 | 0.204 |
| Ta4CL42 | TraesCS3D02G491300 | 1100 | 120,520.26 | 5.82 | 44.49 | 86.14 | −0.098 |
| Ta4CL43 | TraesCS4A02G036200 | 549 | 58,531.44 | 6.39 | 37.99 | 98.11 | 0.154 |
| Ta4CL44 | TraesCS4A02G119100 | 567 | 61,510.76 | 7.59 | 27.34 | 89.52 | −0.025 |
| Ta4CL45 | TraesCS4A02G119200 | 579 | 63,406.05 | 7.57 | 30.9 | 84.23 | −0.092 |
| Ta4CL46 | TraesCS4A02G239000 | 551 | 58,574.87 | 8.7 | 43.53 | 94.92 | 0.151 |
| Ta4CL47 | TraesCS4A02G239100 | 543 | 57,096.28 | 8.71 | 45.96 | 102.45 | 0.309 |
| Ta4CL48 | TraesCS4B02G075900 | 543 | 57,570.91 | 8.16 | 44.71 | 104.94 | 0.308 |
| Ta4CL49 | TraesCS4B02G076000 | 551 | 58,594.8 | 8.79 | 43.77 | 95.81 | 0.173 |
| Ta4CL50 | TraesCS4B02G076100 | 543 | 57,750.89 | 8.91 | 48 | 99.39 | 0.179 |
| Ta4CL51 | TraesCS4B02G185400 | 567 | 61,574.8 | 7.59 | 27.46 | 88.48 | −0.03 |
| Ta4CL52 | TraesCS4B02G269200 | 555 | 59,004.06 | 6.48 | 38.34 | 98.97 | 0.184 |
| Ta4CL53 | TraesCS4B02G323900 | 581 | 63,176.8 | 7.97 | 32.61 | 91.98 | −0.009 |
| Ta4CL54 | TraesCS4B02G326800 | 552 | 59,349.51 | 8.59 | 36.05 | 96.32 | 0.052 |
| Ta4CL55 | TraesCS4B02G329700 | 549 | 58,957.74 | 6.68 | 36.76 | 86.03 | −0.005 |
| Ta4CL56 | TraesCS4B02G329800 | 650 | 70,840.11 | 6.9 | 41.11 | 85.22 | −0.083 |
| Ta4CL57 | TraesCS4B02G329900 | 606 | 66,458.34 | 7.31 | 45.75 | 87.61 | −0.081 |
| Ta4CL58 | TraesCS4B02G330100 | 682 | 74,136.93 | 7.65 | 38.61 | 84.28 | −0.073 |
| Ta4CL59 | TraesCS4B02G330200 | 558 | 60,755.62 | 6.42 | 33.99 | 84.61 | −0.039 |
| Ta4CL60 | TraesCS4D02G074500 | 543 | 57,442.68 | 8.49 | 46.4 | 103.54 | 0.291 |
| Ta4CL61 | TraesCS4D02G074600 | 551 | 58,505.67 | 8.56 | 43.25 | 95.48 | 0.156 |
| Ta4CL62 | TraesCS4D02G074700 | 546 | 57,945.91 | 9.03 | 48.4 | 96.56 | 0.109 |
| Ta4CL63 | TraesCS4D02G186700 | 579 | 63,262.76 | 7.58 | 30.1 | 82.56 | −0.107 |
| Ta4CL64 | TraesCS4D02G186800 | 567 | 61,535.77 | 7.24 | 27.2 | 89.35 | −0.024 |
| Ta4CL65 | TraesCS4D02G268400 | 547 | 58,328.24 | 6.53 | 39.12 | 98.99 | 0.181 |
| Ta4CL66 | TraesCS4D02G320900 | 583 | 63,280.96 | 7.97 | 33.35 | 91.85 | −0.006 |
| Ta4CL67 | TraesCS4D02G323500 | 551 | 59,036.2 | 8.86 | 35.98 | 96.52 | 0.062 |
| Ta4CL68 | TraesCS4D02G326700 | 585 | 63,149.89 | 6.3 | 38.49 | 90.43 | 0.03 |
| Ta4CL69 | TraesCS4D02G327100 | 656 | 70,903.94 | 7.32 | 40.32 | 80.49 | −0.127 |
| Ta4CL70 | TraesCS5A02G307500 | 528 | 57,725.39 | 8.79 | 40.82 | 86.95 | −0.098 |
| Ta4CL71 | TraesCS5A02G356800 | 565 | 60,791.66 | 7 | 27.92 | 89.63 | −0.025 |
| Ta4CL72 | TraesCS5A02G368300 | 430 | 46,523.43 | 7.66 | 34.52 | 93.65 | −0.006 |
| Ta4CL73 | TraesCS5A02G496400 | 583 | 63,340.09 | 7.97 | 33.31 | 91.36 | −0.005 |
| Ta4CL74 | TraesCS5A02G498800 | 552 | 59,198.41 | 8.74 | 37.86 | 96.34 | 0.061 |
| Ta4CL75 | TraesCS5A02G501200 | 549 | 58,993.9 | 6.48 | 37.54 | 88.52 | 0.016 |
| Ta4CL76 | TraesCS5A02G501300 | 558 | 60,574.43 | 6.66 | 37.31 | 84.46 | −0.06 |
| Ta4CL77 | TraesCS5A02G501400 | 369 | 40,277.61 | 7.74 | 38.7 | 92.55 | 0.067 |
| Ta4CL78 | TraesCS5A02G501700 | 654 | 70,995.27 | 8.03 | 39.95 | 84.31 | −0.077 |
| Ta4CL79 | TraesCS5B02G307900 | 325 | 35,103.07 | 7.62 | 33.36 | 83.05 | −0.141 |
| Ta4CL80 | TraesCS5B02G359300 | 565 | 60,867.83 | 7.32 | 27.07 | 88.41 | −0.014 |
| Ta4CL81 | TraesCS5B02G365000 | 562 | 60,813.73 | 6.61 | 35.77 | 92.38 | −0.015 |
| Ta4CL82 | TraesCS5B02G370600 | 475 | 51,264.79 | 5.84 | 26.99 | 95.45 | 0.033 |
| Ta4CL83 | TraesCS5B02G570100 | 554 | 58,737.12 | 8.54 | 45.32 | 100 | 0.195 |
| Ta4CL84 | TraesCS5D02G314500 | 479 | 52,202.71 | 5.94 | 35.01 | 84.86 | −0.128 |
| Ta4CL85 | TraesCS5D02G365800 | 565 | 60,803.71 | 7.02 | 27.93 | 89.45 | −0.018 |
| Ta4CL86 | TraesCS5D02G377800 | 551 | 59,564.56 | 6.74 | 26.78 | 98.02 | 0.068 |
| Ta4CL87 | TraesCS5D02G561500 | 555 | 58,863.23 | 8.82 | 47.68 | 99.08 | 0.184 |
| Ta4CL88 | TraesCS6A02G029900 | 558 | 60,265.29 | 5.36 | 41.18 | 90.34 | 0.077 |
| Ta4CL89 | TraesCS6A02G059400 | 569 | 60,521.64 | 6.81 | 37.22 | 86.33 | 0.078 |
| Ta4CL90 | TraesCS6A02G151700 | 546 | 58,531.58 | 5.23 | 35.96 | 97.73 | 0.093 |
| Ta4CL91 | TraesCS6A02G266700 | 573 | 60,626.98 | 5.44 | 33.6 | 100.28 | 0.234 |
| Ta4CL92 | TraesCS6A02G390600 | 705 | 77,615.2 | 5.62 | 26.85 | 79.09 | −0.201 |
| Ta4CL93 | TraesCS6B02G042400 | 556 | 60,106.14 | 5.37 | 40.23 | 91.35 | 0.088 |
| Ta4CL94 | TraesCS6B02G042900 | 555 | 59,711.82 | 5.76 | 36.82 | 92.92 | 0.125 |
| Ta4CL95 | TraesCS6B02G079800 | 592 | 62,860.4 | 7.94 | 38.07 | 87.4 | 0.057 |
| Ta4CL96 | TraesCS6B02G179900 | 337 | 36,543.7 | 5.84 | 30.85 | 104.18 | 0.116 |
| Ta4CL97 | TraesCS6B02G294100 | 573 | 60,767.1 | 5.46 | 33.47 | 99.42 | 0.224 |
| Ta4CL98 | TraesCS6B02G431000 | 572 | 63,611.64 | 6.01 | 24.87 | 81.43 | −0.232 |
| Ta4CL99 | TraesCS6D02G141700 | 549 | 58,829.93 | 5.29 | 39.96 | 97.19 | 0.087 |
| Ta4CL100 | TraesCS6D02G248000 | 568 | 60,154.38 | 5.45 | 32.41 | 99.98 | 0.229 |
| Ta4CL101 | TraesCS6D02G376900 | 667 | 73,820 | 5.74 | 25.81 | 78.19 | −0.215 |
| Ta4CL102 | TraesCS7A02G011700 | 1198 | 130,912.24 | 5.76 | 41.49 | 89.67 | −0.022 |
| Ta4CL103 | TraesCS7A02G033700 | 1143 | 124,812.3 | 5.68 | 39.87 | 91.69 | 0.002 |
| Ta4CL104 | TraesCS7A02G310100 | 605 | 65,750.56 | 8.65 | 40.39 | 91.69 | −0.023 |
| Ta4CL105 | TraesCS7A02G496200 | 557 | 59,429.4 | 5.31 | 36.56 | 97.38 | 0.116 |
| Ta4CL106 | TraesCS7B02G210000 | 591 | 63,758.29 | 8.25 | 42.28 | 90.9 | 0.069 |
| Ta4CL107 | TraesCS7B02G448600 | 1145 | 125,202.44 | 5.83 | 41.35 | 88.72 | −0.05 |
| Ta4CL108 | TraesCS7D02G030200 | 891 | 97,629.47 | 5.68 | 39.26 | 91.14 | −0.041 |
| Ta4CL109 | TraesCS7D02G306700 | 551 | 59,312.91 | 6.94 | 40.35 | 92.4 | 0.011 |
| Ta4CL110 | TraesCS7D02G483400 | 560 | 59,574.6 | 5.25 | 36.77 | 97.91 | 0.147 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Yang, X.; Ning, D.; Li, R. Characterization of 4-Coumarate-CoA Ligase (4CL) Genes in Wheat Uncovers Ta4CL91’s Role in Drought and Salt Stress Adaptation. Plants 2025, 14, 1301. https://doi.org/10.3390/plants14091301
Zhang Z, Yang X, Ning D, Li R. Characterization of 4-Coumarate-CoA Ligase (4CL) Genes in Wheat Uncovers Ta4CL91’s Role in Drought and Salt Stress Adaptation. Plants. 2025; 14(9):1301. https://doi.org/10.3390/plants14091301
Chicago/Turabian StyleZhang, Ze, Xiuli Yang, Dongxian Ning, and Rong Li. 2025. "Characterization of 4-Coumarate-CoA Ligase (4CL) Genes in Wheat Uncovers Ta4CL91’s Role in Drought and Salt Stress Adaptation" Plants 14, no. 9: 1301. https://doi.org/10.3390/plants14091301
APA StyleZhang, Z., Yang, X., Ning, D., & Li, R. (2025). Characterization of 4-Coumarate-CoA Ligase (4CL) Genes in Wheat Uncovers Ta4CL91’s Role in Drought and Salt Stress Adaptation. Plants, 14(9), 1301. https://doi.org/10.3390/plants14091301




