Comprehensive Identification of HD-Zip Family Genes in Coix lacryma-jobi L. and Their Potential Roles in Response to Abiotic Stress
Abstract
:1. Introduction
2. Results
2.1. Identification and Characteristic Analysis of HD-Zip Genes in Coix
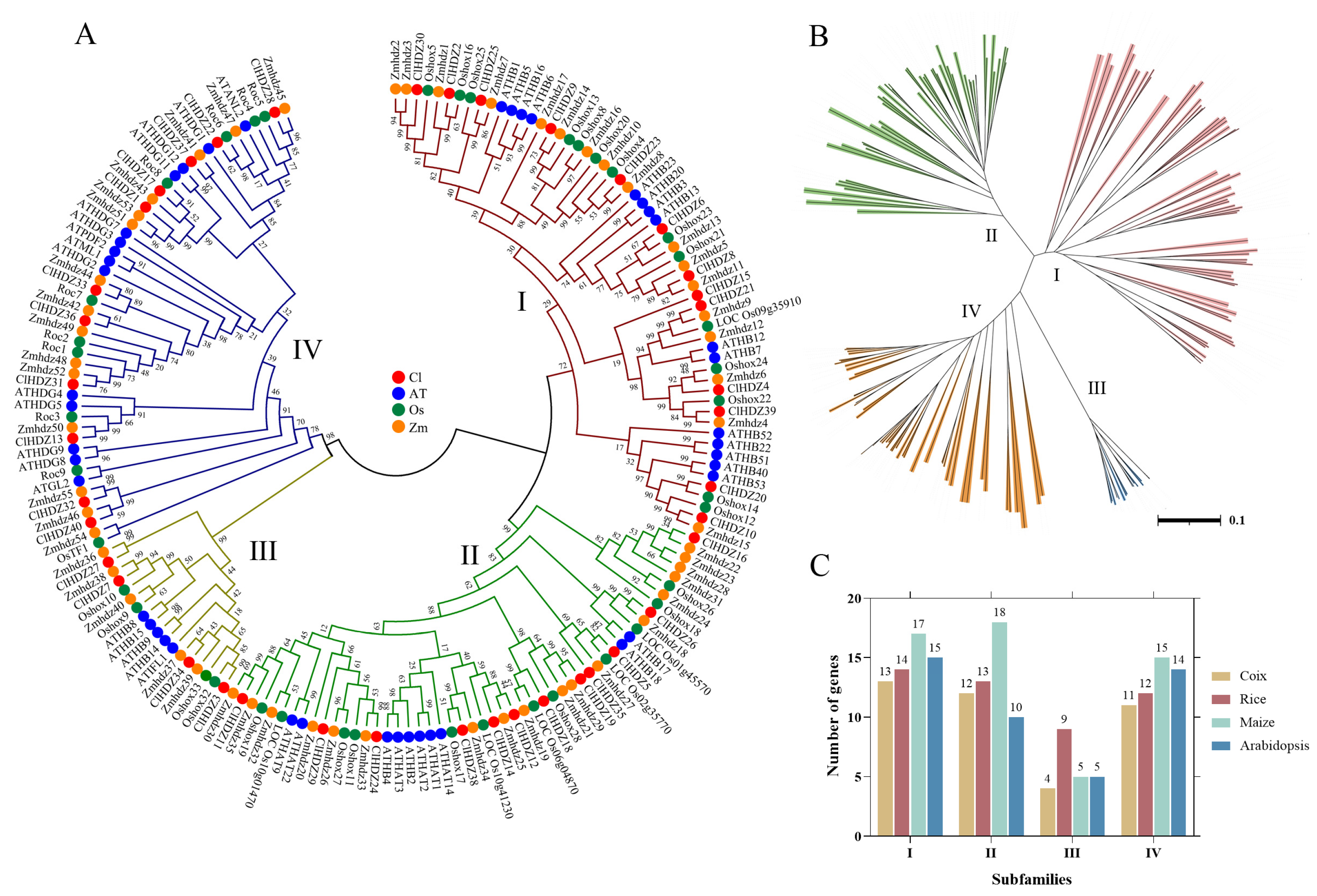
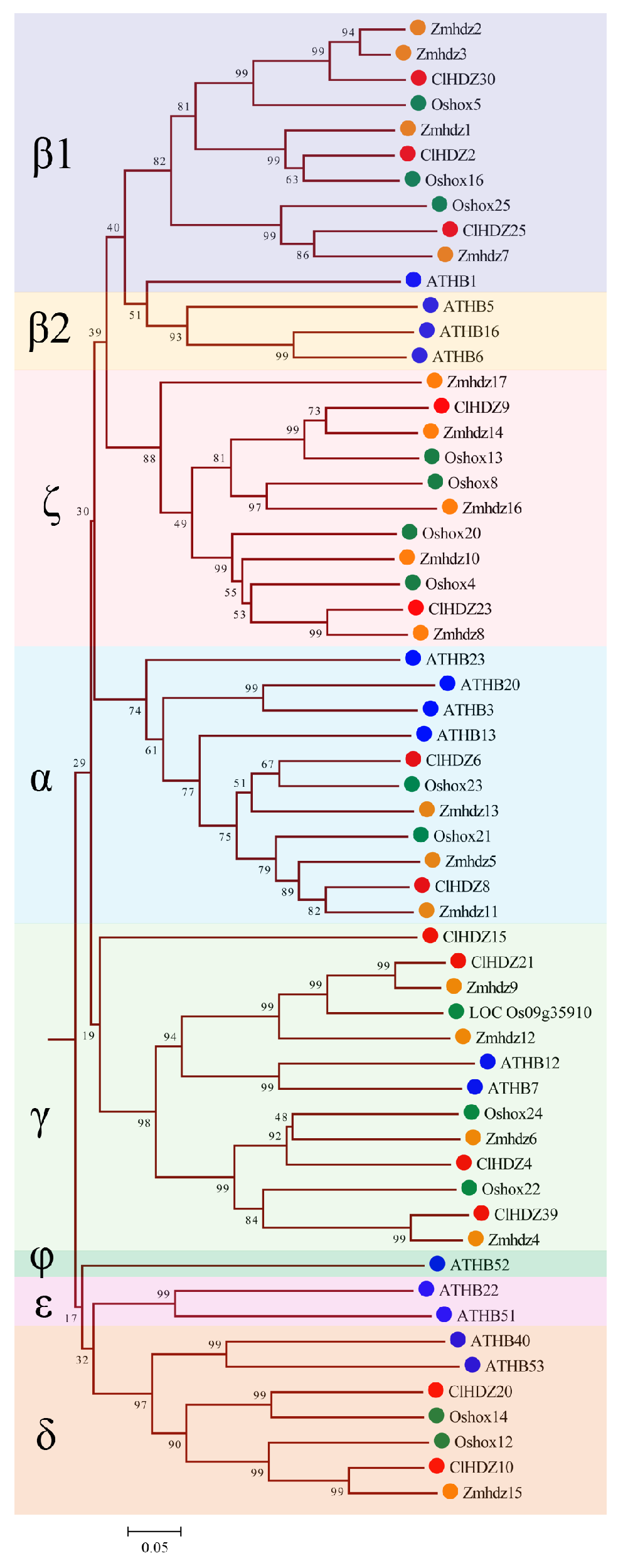
2.2. Phylogenetic Trees of HD-Zip Genes in Arabidopsis, Rice, Maize, and Coix
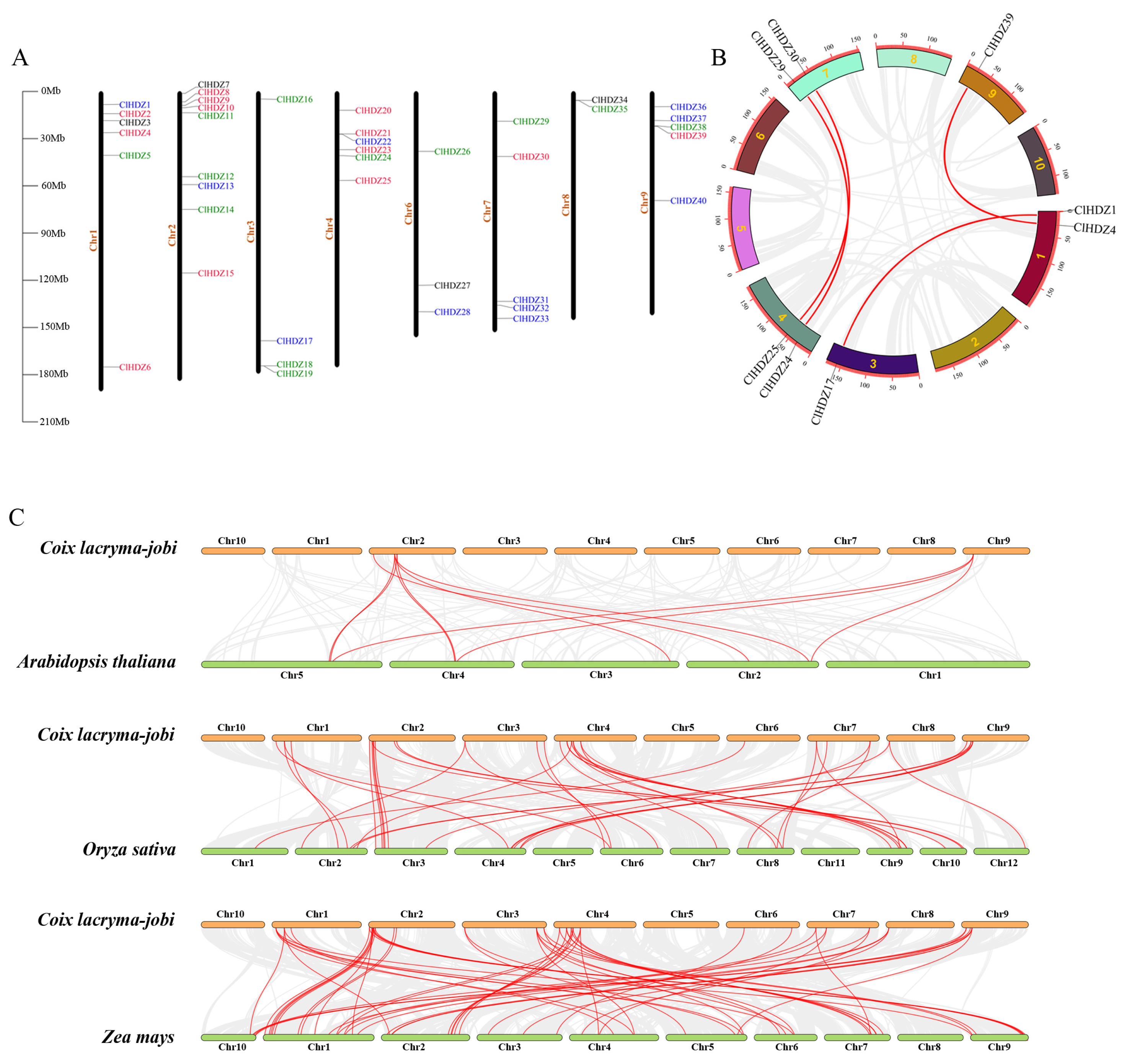
2.3. Chromosomal Distribution and Collinear Analysis
2.4. Structural Feature Analysis of HD-Zip Genes in Coix
2.5. Prediction and Enrichment Analysis of Transcription Factor Binding Sites in Coix HD-Zip
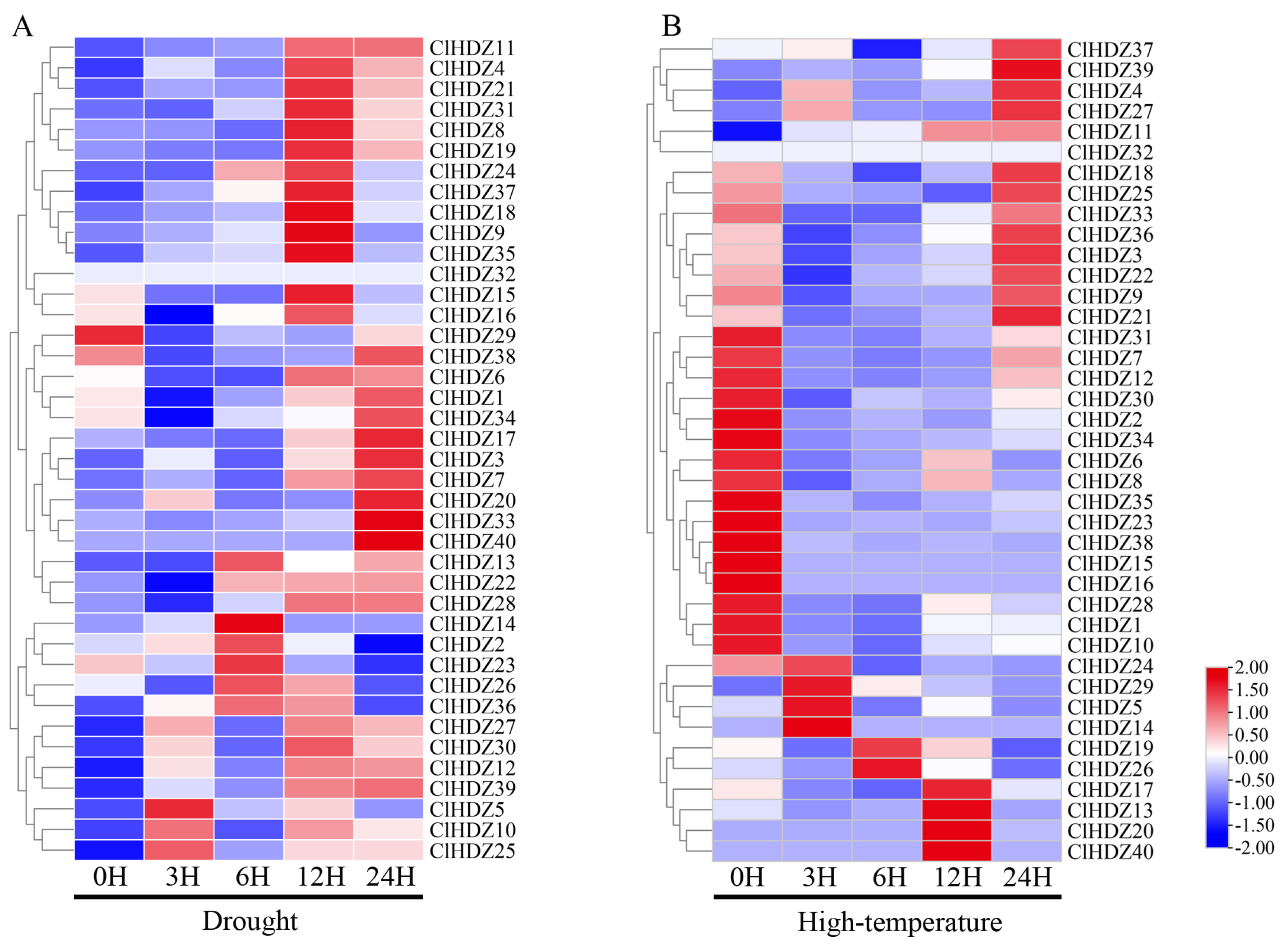
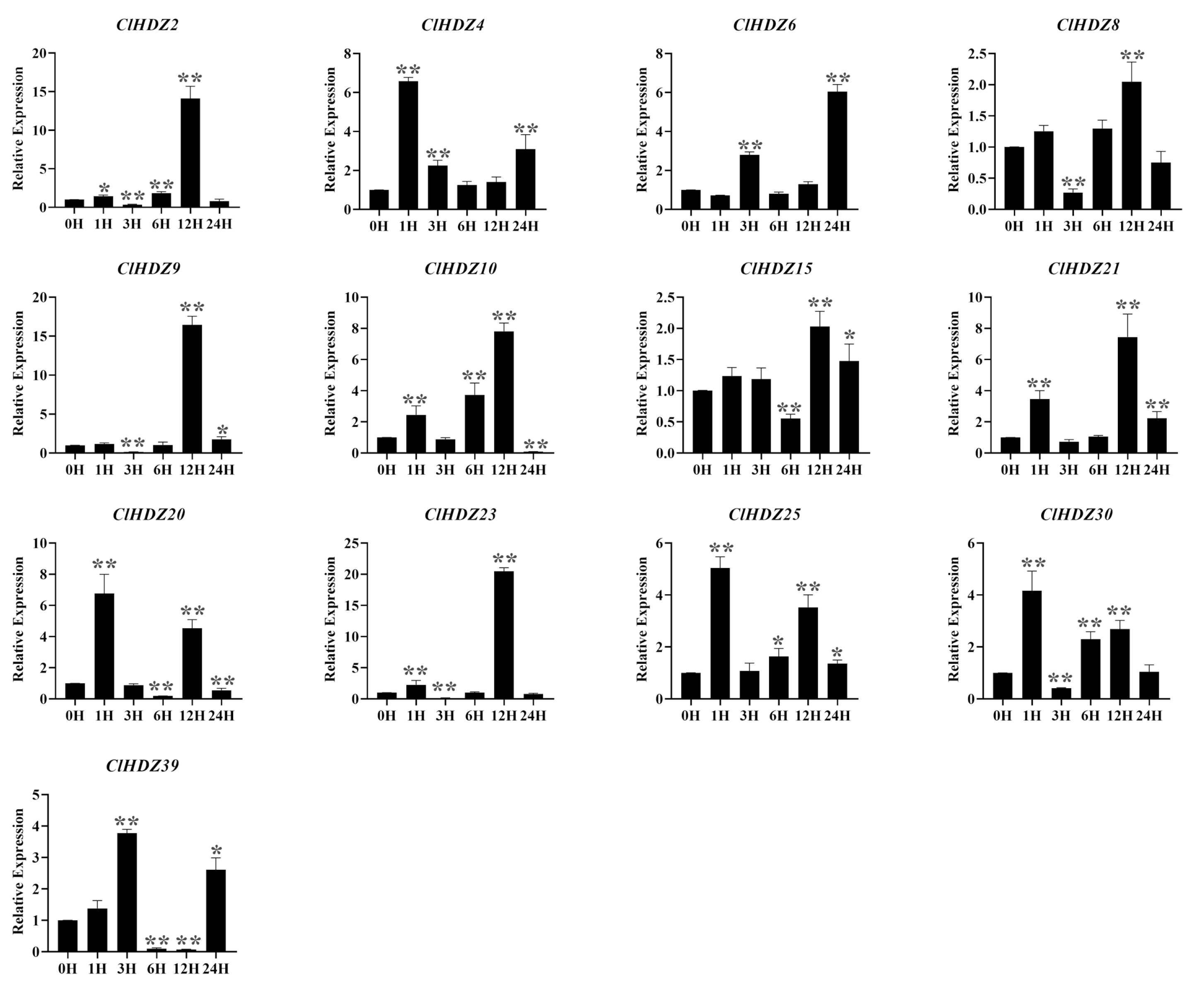
2.6. The Expression Files of Coix HD-Zip Genes in Response to Drought and Heat Stress
2.7. Expression Pattern of the HD-Zip I Genes in Coix Under Salt Stress
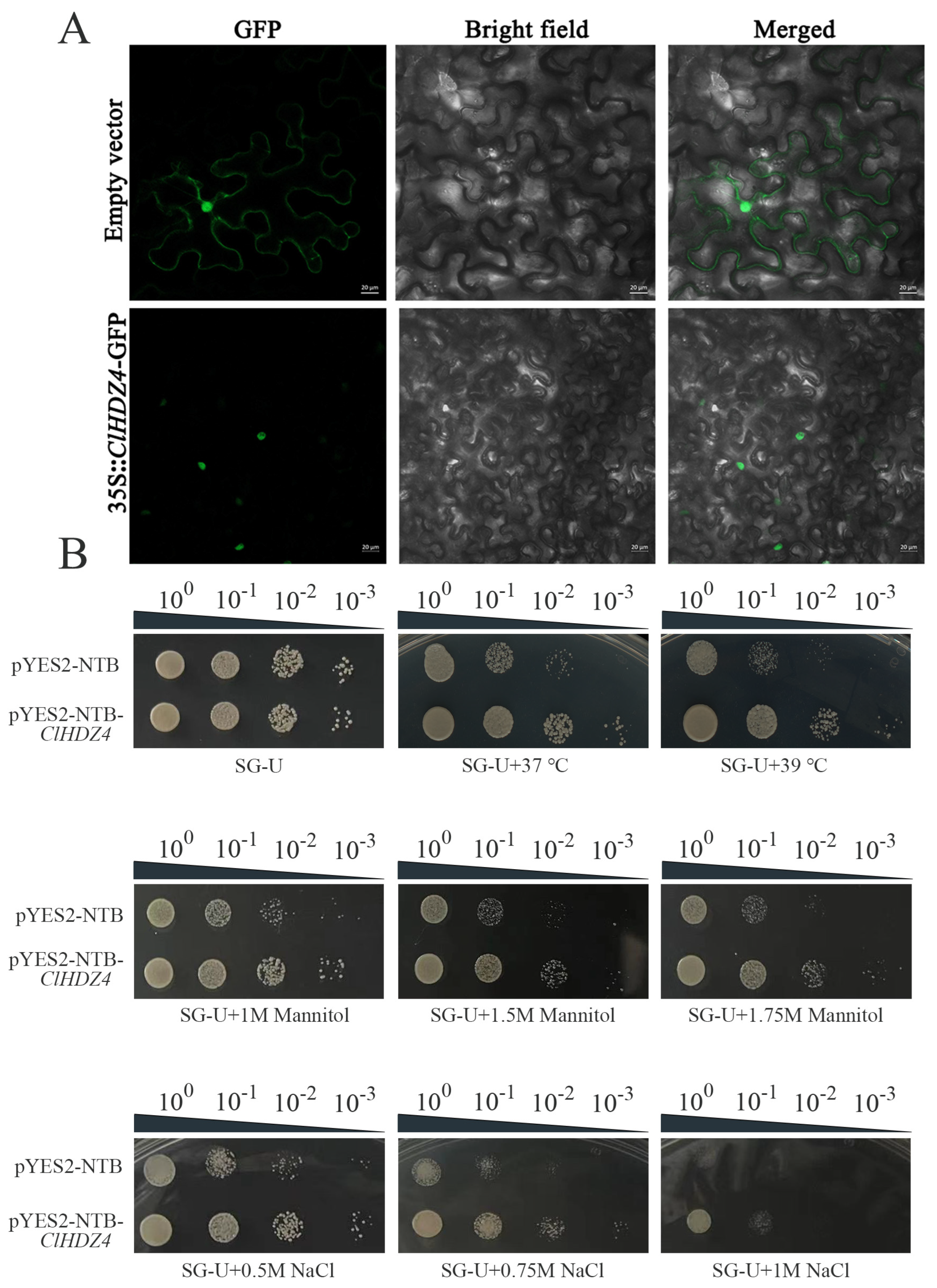
2.8. Subcellular Localization of ClHDZ4 and Its Role in Enhancing Yeast Stress Tolerance Through Ectopic Expression
3. Discussion
4. Materials and Methods
4.1. Retrieving and Identifying HD-Zip Family Genes in Coix
4.2. Phylogenetic Analysis, Chromosome Localization, and Collinear Analysis
4.3. Gene Structure, Conserved Motifs, Transcription Factor Binding Site (TFBS) Analysis, and Enrichment Analysis
4.4. Expression Profiles of HD-Zip Genes in Coix
4.5. Plant Material Acquisition, RNA Extraction, and qRT-PCR
4.6. Subcellular Localization Analysis of ClHDZ4
4.7. The Overexpression of ClHDZ4 in Yeast
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Javed, T.; Shabbir, R.; Ali, A.; Afzal, I.A.-O.; Zaheer, U.; Gao, S.J. Transcription Factors in Plant Stress Responses: Challenges and Potential for Sugarcane Improvement. Plants 2020, 9, 491. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L.; Chan, R.L. The True Story of The HD-Zip Family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Katsumata, H.; Abe, M.; Yabe, N.; Komeda, Y.; Yamamoto, K.T.; Takahashi, T. Characterization of the class IV homeodomain-Leucine Zipper gene family in Arabidopsis. Plant Physiol. 2006, 141, 1363–1375. [Google Scholar] [CrossRef] [PubMed]
- Sessa, G.; Steindler, C.; Morelli, G.; Ruberti, I. The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-Zip proteins. Plant Mol. Biol. 1998, 38, 609–622. [Google Scholar] [CrossRef]
- Côté, C.L.; Boileau, F.; Roy, V.; Ouellet, M.; Levasseur, C.; Morency, M.-J.; Cooke, J.E.; Séguin, A.; MacKay, J.J. Gene family structure, expression and functional analysis of HD-Zip III genes in angiosperm and gymnosperm forest trees. BMC Plant Biol. 2010, 10, 273. [Google Scholar] [CrossRef]
- Viola, I.L.; Gonzalez, D.H. Structure and Evolution of Plant Homeobox Genes. In Plant Transcription Factors; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, X.; Zhang, B.; Xiao, Y.; Guo, J.; Liu, J.; Chen, Q.; Peng, F. Genome-wide identification, bioinformatics and expression analysis of HD-Zip gene family in peach. BMC Plant Biol. 2023, 23, 122. [Google Scholar] [CrossRef]
- Li, R.; Wang, X.; Zhang, S.; Liu, X.; Zhou, Z.; Liu, Z.; Wang, K.; Tian, Y.; Wang, H.; Zhang, Y.; et al. Two zinc-finger proteins control the initiation and elongation of long stalk trichomes in tomato. J. Genet. Genom. 2021, 48, 1057–1069. [Google Scholar] [CrossRef]
- He, G.; Liu, P.; Zhao, H.; Sun, J. The HD-Zip II Transcription Factors Regulate Plant Architecture through the Auxin Pathway. Int. J. Mol. 2020, 21, 3250–3255. [Google Scholar] [CrossRef]
- Xie, Q.; Gao, Y.; Li, J.; Yang, Q.; Qu, X.; Li, H.; Zhang, J.; Wang, T.; Ye, Z.; Yang, C. The HD-Zip IV transcription factor SlHDZIV8 controls multicellular trichome morphology by regulating the expression of Hairless-2. J. Exp. Bot. 2020, 71, 7132–7145. [Google Scholar] [CrossRef]
- Depège-Fargeix, N.; Javelle, M.; Chambrier, P.; Frangne, N.; Gerentes, D.; Perez, P.; Rogowsky, P.M.; Vernoud, V. Functional characterization of the HD-Zip IV transcription factor OCL1 from maize. J. Exp. Bot. 2011, 62, 293–305. [Google Scholar] [CrossRef]
- Liu, X.; Wu, C.; Su, D.; Yang, Y.; Xian, Z.; Yu, C.; Li, Z.; Hao, Y.A.-O.; Chen, R. The SlHB8 Acts as a negative regulator in stem development and lignin biosynthesis. Int. J. Mol. Sci. 2021, 22, 13343. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, M.; Liu, Y.; Li, Z.; Hu, Z.A.-O.; Zhang, M.; Yang, J.A.-O. An HD-Zip III transcription factor, BjPHVa, negatively regulates non-glandular trichome formation in Brassica juncea. Physiol. Plant. 2024, 176, e14553. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.C.; Hrmova, M.; Lopato, S.; Langridge, P. Modulation of plant growth by HD-Zip class I and II transcription factors in response to environmental stimuli. New Phytol. 2011, 190, 823–837. [Google Scholar] [CrossRef]
- Li, S.; Chen, N.; Li, F.; Mei, F.; Wang, Z.; Cheng, X.; Kang, Z.; Mao, H.A.-O. Characterization of wheat homeodomain-leucine zipper family genes and functional analysis of TaHDZ5-6A in drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2020, 20, 50. [Google Scholar] [CrossRef]
- Wang, K.; Xu, L.; Wang, Y.; Ying, J.; Li, J.; Dong, J.; Li, C.; Zhang, X.; Liu, L.A.-O. Genome-wide characterization of homeodomain-leucine zipper genes reveals RsHDZ17 enhances the heat tolerance in radish (Raphanus sativus L.). Physiol. Plant. 2022, 174, e13789. [Google Scholar] [CrossRef]
- Zhao, S.; Wang, H.; Jia, X.; Gao, H.; Mao, K.; Ma, F.A.-O. The HD-Zip I transcription factor MdHB7-like confers tolerance to salinity in transgenic apple (Malus domestica). Physiol. Plant. 2021, 173, 1452–1464. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Yan, X.; Wang, N.; Zenda, T.; Dong, A.; Zhai, X.; Yang, Q.; Duan, H.A.-O. ZmHB53, a maize Homeodomain-Leucine Zipper I transcription factor family gene, contributes to abscisic acid sensitivity and confers seedling drought tolerance by promoting the activity of ZmPYL4. Plant Cell Environ. 2025. [Google Scholar] [CrossRef]
- Jiao, P.; Jiang, Z.; Miao, M.; Wei, X.; Wang, C.; Liu, S.; Guan, S.; Ma, Y. Zmhdz9, an HD-Zip transcription factor, promotes drought stress resistance in maize by modulating ABA and lignin accumulation. Int. J. Biol. Macromol. 2024, 258, 128849. [Google Scholar] [CrossRef]
- Wu, Z.A.-O.; Li, T.; Zhang, Y.; Zhang, D.; Teng, N.A.-O. HD-Zip I protein LlHOX6 antagonizes homeobox protein LlHB16 to attenuate basal thermotolerance in lily. Plant Physiol. 2024, 194, 1870–1888. [Google Scholar] [CrossRef]
- Sharif, R.; Xie, C.; Wang, J.; Cao, Z.; Zhang, H.; Chen, P.; Yuhong, L. Genome wide identification, characterization and expression analysis of HD-Zip gene family in Cucumis sativus L. under biotic and various abiotic stresses. Int. J. Biol. Macromol. 2020, 158, 502–520. [Google Scholar] [CrossRef]
- Wang, J.; Liu, L.; Ball, T.; Yu, L.; Li, Y.; Xing, F. Revealing a 5,000-y-old beer recipe in China. Proc. Natl. Acad. Sci. USA 2016, 113, 6444–6448. [Google Scholar] [CrossRef]
- Guo, C.; Wang, Y.; Yang, A.; He, J.; Xiao, C.; Lv, S.; Han, F.; Yuan, Y.; Yuan, Y.; Dong, X.; et al. The Coix genome provides insights into panicoideae evolution and papery hull domestication. Mol. Plant 2020, 13, 309–320. [Google Scholar] [CrossRef]
- Liu, H.; Shi, J.; Cai, Z.; Huang, Y.; Jin, W. Evolution and domestication footprints uncovered from the genomes of Coix. Mol. Plant 2019, 13, 295–308. [Google Scholar] [CrossRef]
- Chen, X.; Chen, Z.; Zhao, H.; Zhao, Y.; Cheng, B.; Xiang, Y. Genome-wide analysis of soybean HD-Zip gene family and expression profiling under salinity and drought treatments. PLoS ONE 2014, 9, e87156. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Brocchieri, L.; Bürglin, T.R. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 2009, 26, 2775–2794. [Google Scholar] [CrossRef]
- Agalou, A.; Purwantomo, S.; Overnäs, E.; Johannesson, H.; Zhu, X.; Estiati, A.; de Kam, R.J.; Engström, P.; Slamet-Loedin, I.H.; Zhu, Z.; et al. A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 2008, 66, 87–103. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, Y.; Jiang, H.; Li, X.; Gan, D.; Peng, X.; Zhu, S.; Cheng, B. Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-Zip gene family in maize. PLoS ONE 2011, 6, e28488. [Google Scholar] [CrossRef] [PubMed]
- Henriksson, E.; Olsson, A.S.; Johannesson, H.; Hanson, J.; Engström, P.; Söderman, E. Homeodomain Leucine Zipper Class I genes in Arabidopsis. Expression Patterns and Phylogenetic Relationships. Plant Physiol. 2005, 139, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhuang, L.; Zhang, J.; Yu, J.; Yang, Z.; Huang, B. Identification and characterization of novel homeodomain leucine zipper (HD-Zip) transcription factors associated with heat tolerance in Perennial ryegrass. Environ. Exp. Bot. 2019, 160, 1–11. [Google Scholar] [CrossRef]
- Yan, X.; Yue, Z.; Pan, X.; Si, F.; Li, J.; Chen, X.; Li, X.; Luan, F.; Yang, J.; Zhang, X.; et al. The HD-Zip gene family in Watermelon: Genome-wide identification and expression analysis under abiotic stresses. Genes 2022, 13, 2242. [Google Scholar] [CrossRef]
- Ye, K.Y.; Li, J.W.; Wang, F.M.; Gao, J.Y.; Liu, C.X.; Gong, H.J.; Qi, B.B.; Liu, P.P.; Jiang, Q.S.; Tang, J.M.; et al. Genome-wide analysis and expression profiling of the HD-Zip gene family in kiwifruit. BMC Genom. 2024, 25, 354. [Google Scholar] [CrossRef] [PubMed]
- Ré, D.A.; Dezar, C.A.; Chan, R.L.; Baldwin, I.T.; Bonaventure, G. Nicotiana attenuata NaHD20 plays a role in leaf ABA accumulation during water stress, benzylacetone emission from flowers, and the timing of bolting and flower transitions. J. Exp. Bot. 2011, 62, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.Q.; Fan, J.D.; Liu, C.Y.; Zhao, Y.-Q.; Xu, Z.-S.; Lu, X.-J.; Ge, J.; Zhang, B.-W.; Li, M.-Q.; Yang, Y.; et al. Physiological and transcriptome analysis of changes in endogenous hormone contents and related synthesis and signaling genes during the heat stress in garlic (Allium sativum L.). BMC Plant Biol. 2025, 25, 464. [Google Scholar] [CrossRef]
- Balhara, R.; Verma, D.; Kaur, R.; Singh, K. MYB transcription factors, their regulation and interactions with non-coding RNAs during drought stress in Brassica juncea. BMC Plant Biol. 2024, 24, 999. [Google Scholar] [CrossRef]
- Rabeh, K.; Hnini, M.; Oubohssaine, M. A comprehensive review of transcription factor-mediated regulation of secondary metabolites in plants under environmental stress. Stress Biol. 2025, 5, 15. [Google Scholar] [CrossRef]
- Ding, S.; Cai, Z.; Du, H.; Wang, H. Genome-Wide Analysis of TCP Family Genes in Zea mays L. Identified a Role for ZmTCP42 in Drought Tolerance. Int. J. Mol. Sci. 2019, 20, 2762. [Google Scholar] [CrossRef]
- Liu, Y.J.; An, J.P.; Gao, N.; Wang, X.; Chen, X.; Wang, X.; Zhang, S.; You, C. MdTCP46 interacts with MdABI5 to negatively regulate ABA signalling and drought response in apple. Plant Cell Environ. 2022, 45, 3233–3248. [Google Scholar] [CrossRef]
- Ré, D.A.; Capella, M.; Bonaventure, G.; Chan, R.L. Arabidopsis AtHB7 and AtHB12 evolved divergently to fine tune processes associated with growth and responses to water stress. BMC Plant Biol. 2014, 14, 150. [Google Scholar] [CrossRef]
- Harris, J.C.; Sornaraj, P.; Taylor, M.; Bazanova, N.; Baumann, U.; Lovell, B.; Langridge, P.; Lopato, S.; Hrmova, M. Molecular interactions of the γ-clade homeodomain-leucine zipper class I transcription factors during the wheat response to water deficit. Plant Mol. Biol. 2016, 90, 435–452. [Google Scholar] [CrossRef]
- Romani, F.; Ribone, P.A.; Capella, M.; Miguel, V.N.; Chan, R.L. A matter of quantity: Common features in the drought response of transgenic plants overexpressing HD-Zip I transcription factors. Plant Sci. 2016, 251, 139–154. [Google Scholar] [CrossRef]
- Zhao, S.; Gao, H.; Jia, X.; Zhou, K.; Wang, H.; Mao, K.; Ma, F. MdHB7-like confers drought tolerance and improves water-use efficiency through modulating stomatal density in apple (Malus domestica). Sci. Hortic. 2022, 294, 110758. [Google Scholar] [CrossRef]
- Sharif, R.; Raza, A.; Chen, P.; Li, Y.; El-Ballat, E.M.; Rauf, A.; Hano, C.; El-Esawi, M.A. HD-Zip gene family: Potential roles in improving plant growth and regulating stress-responsive mechanisms in plants. Genes 2021, 12, 1256. [Google Scholar] [CrossRef]
- Mou, S.A.-O.; He, W.; Jiang, H.A.-O.; Meng, Q.A.-O.; Zhang, T.A.-O.; Liu, Z.A.-O.X.; Qiu, A.A.-O.; He, S.A.-O. Transcription factor CaHDZ15 promotes pepper basal thermotolerance by activating HEAT SHOCK FACTORA6a. Plant Physiol. 2024, 195, 812–831. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, D.; Yan, F.; Wang, L.; Wang, Y.; Li, J.; Yang, X.; Gao, Z.; Liu, X.; Liu, Y.; et al. Genome-wide analysis of HD-Zip genes in Sophora alopecuroides and their role in salt stress response. Plant Genome 2024, 17, e20504. [Google Scholar] [CrossRef]
- Xie, X.; Ren, Z.; Su, H.; Abou-Elwafa, S.F.; Shao, J.; Ku, L.; Jia, L.; Tian, Z.; Wei, L. Functional study of ZmHDZ4 in maize (Zea mays) seedlings under drought stress. BMC Plant Biol. 2024, 24, 1209. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004, 13, 1402. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Dreos, R.; Ambrosini, G.; Périer, R.C.; Bucher, P. The Eukaryotic Promoter Database: Expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res. 2015, 43, 92–96. [Google Scholar] [CrossRef]
- Rauluseviciute, I.; Riudavets-Puig, R.; Blanc-Mathieu, R.; Castro-Mondragon, J.A.; Ferenc, K.; Kumar, V.; Lemma, R.B.; Lucas, J.; Chèneby, J.; Baranasic, D.; et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2024, 52, D174–D182. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Wang, H.; Lu, X.; Yu, C.; Jiang, B.; Zhu, J.; Wang, Y. Comprehensive Identification of HD-Zip Family Genes in Coix lacryma-jobi L. and Their Potential Roles in Response to Abiotic Stress. Plants 2025, 14, 1318. https://doi.org/10.3390/plants14091318
Wang Y, Wang H, Lu X, Yu C, Jiang B, Zhu J, Wang Y. Comprehensive Identification of HD-Zip Family Genes in Coix lacryma-jobi L. and Their Potential Roles in Response to Abiotic Stress. Plants. 2025; 14(9):1318. https://doi.org/10.3390/plants14091318
Chicago/Turabian StyleWang, Yongle, Hongjuan Wang, Xianyong Lu, Chun Yu, Benli Jiang, Jiabao Zhu, and Yujiao Wang. 2025. "Comprehensive Identification of HD-Zip Family Genes in Coix lacryma-jobi L. and Their Potential Roles in Response to Abiotic Stress" Plants 14, no. 9: 1318. https://doi.org/10.3390/plants14091318
APA StyleWang, Y., Wang, H., Lu, X., Yu, C., Jiang, B., Zhu, J., & Wang, Y. (2025). Comprehensive Identification of HD-Zip Family Genes in Coix lacryma-jobi L. and Their Potential Roles in Response to Abiotic Stress. Plants, 14(9), 1318. https://doi.org/10.3390/plants14091318




