Machine Learning to Predict Drug-Induced Liver Injury and Its Validation on Failed Drug Candidates in Development †
Abstract
1. Introduction
2. Materials and Methods
2.1. Data and Annotation for DILI Likelihood
2.2. Molecular Descriptors
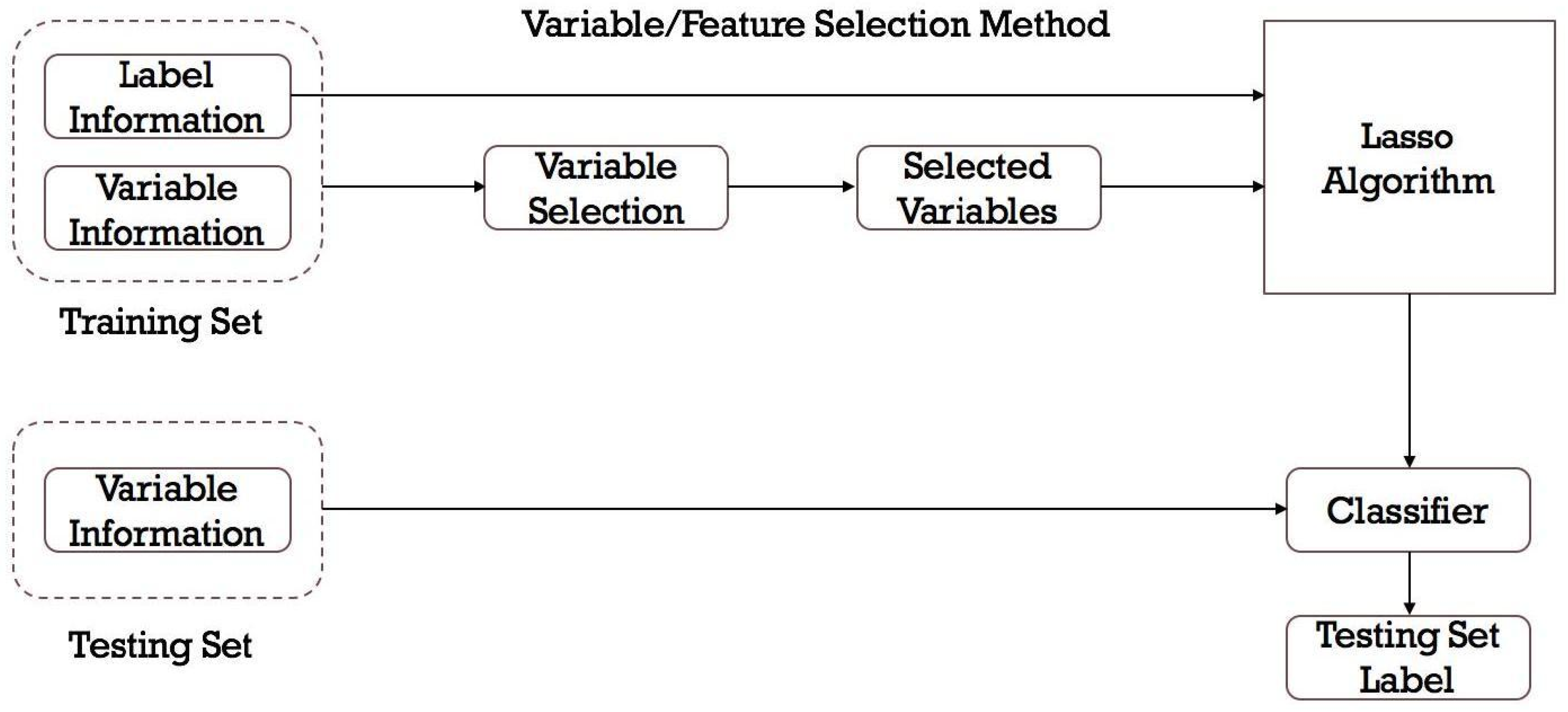
2.3. Model Development
- Feature selection: Not all features may be relevant for predicting DILI. Feature selection techniques can help identify the features that have the most impact on the prediction.
- Model selection: There are various machine learning algorithms that can be used, such as decision trees (DTs), RFs, support vector machines (SVMs), naive Bayes classifier, and neural networks. The choice of the model depends on the characteristics of the data and the specific problem at hand.
- Model training and hyperparameter tuning: The dataset is divided into training and validation sets. The model is developed on the training set using the selected algorithm and the chosen performance metric. Hyperparameters of the machine learning model can significantly affect its performance. Techniques like grid search or random search are used to find the optimal hyperparameters.
- Model evaluation: The model’s performance is assessed on the validation set using appropriate evaluation metrics as detailed below.
- Model test and interpretation: After finding the best model with optimal hyperparameters, it will be tested on an independent test set to get an estimate of its real-world performance. Additionally, interpretability techniques can help understand which features are driving the predictions and provide insights into the underlying mechanisms of DILI.
2.3.1. Feature Selection from Chemical Descriptors
2.3.2. Model Selection and Mathematical Analysis
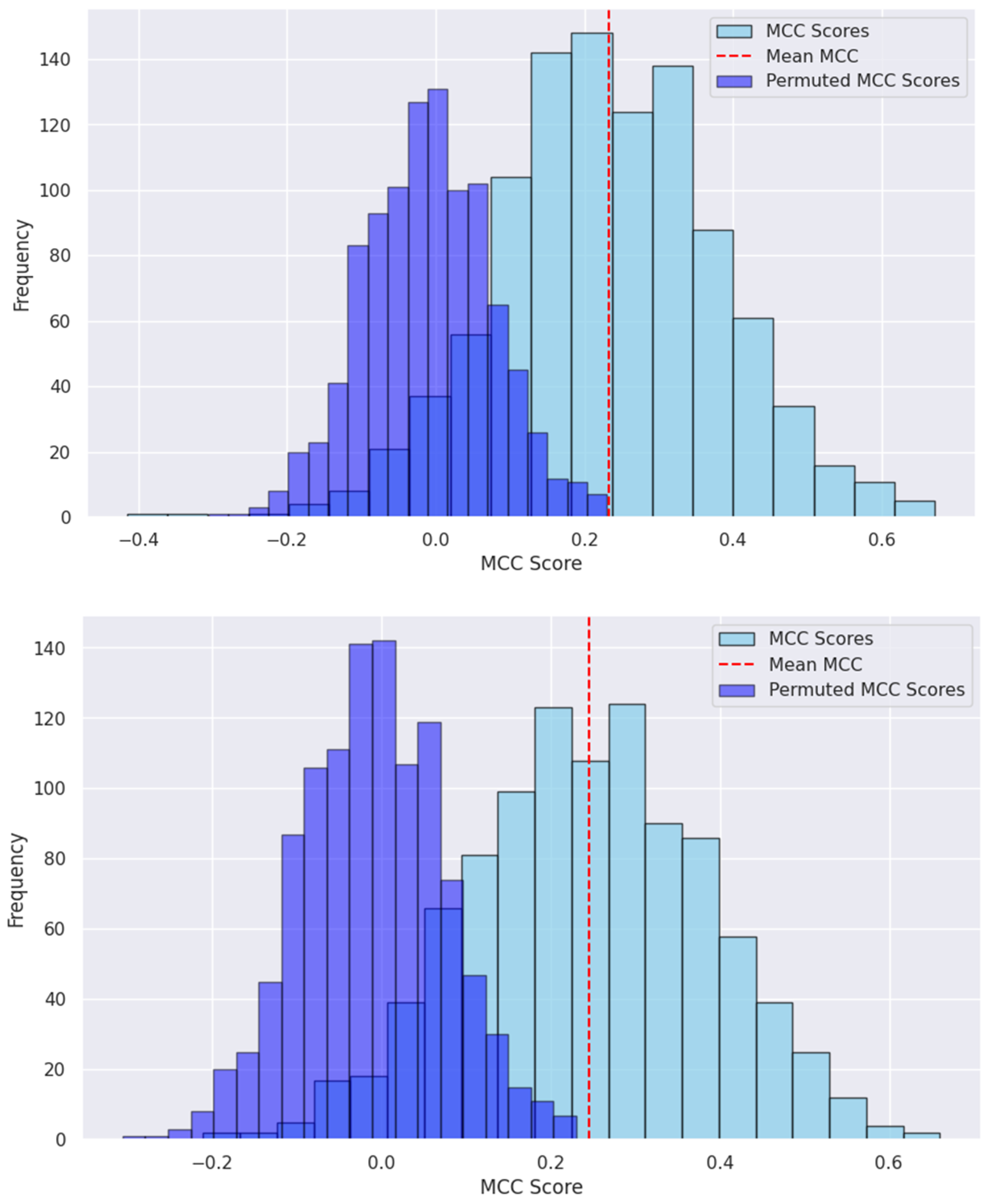
2.3.3. Model Evaluations and Statistical Analysis
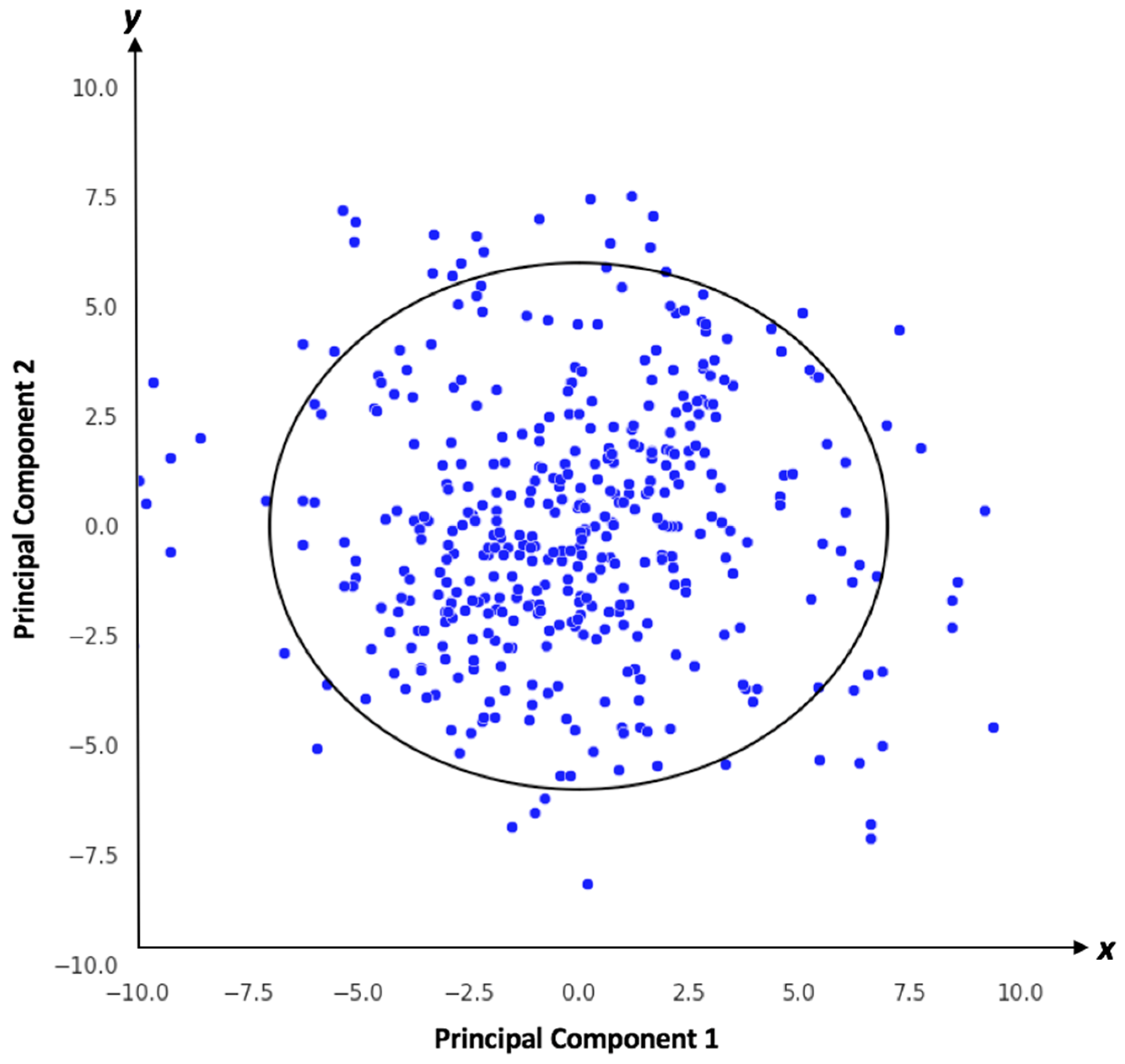
2.3.4. Applicability Domain Analysis
3. Results
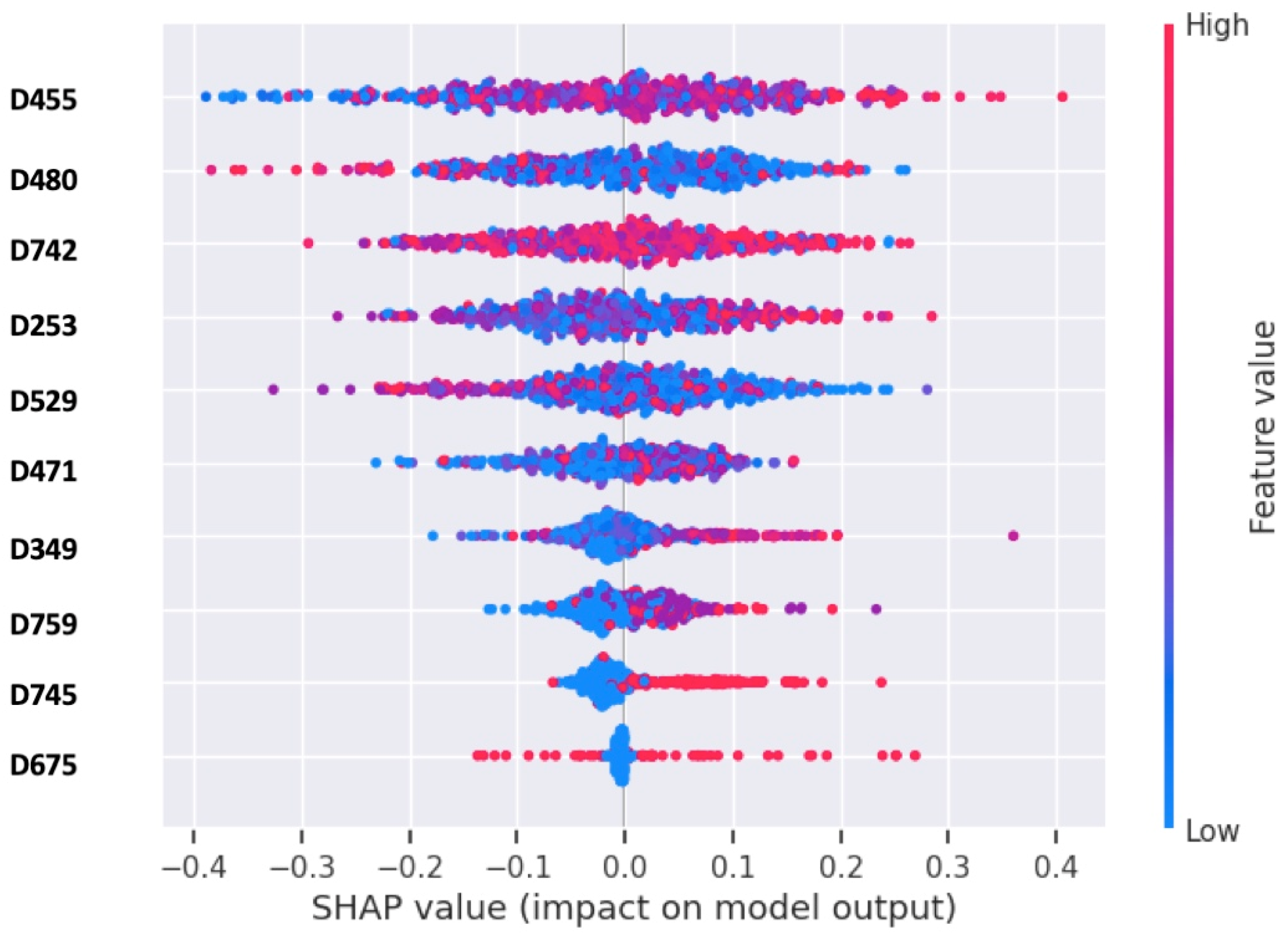
3.1. Assessments of Feature Selection
3.2. Model Development and Performance Assessments
3.3. Model Validation by Drug Candidates That Failed in Development
4. Discussions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weaver, R.J.; Blomme, E.A.; Chadwick, A.E.; Copple, I.M.; Gerets, H.H.J.; Goldring, C.E.; Guillouzo, A.; Hewitt, P.G.; Ingelman-Sundberg, M.; Jensen, K.G.; et al. Managing the challenge of drug-induced liver injury: A roadmap for the development and deployment of preclinical predictive models. Nat. Rev. Drug Discov. 2020, 19, 131–148. [Google Scholar] [CrossRef] [PubMed]
- Raschi, E.; Poluzzi, E.; Koci, A.; Salvo, F.; Pariente, A.; Biselli, M.; Moretti, U.; Moore, N.; De Ponti, F. Liver injury with novel oral anticoagulants: Assessing post-marketing reports in the US Food and Drug Administration adverse event reporting system. Br. J. Clin. Pharmacol. 2015, 80, 285–293. [Google Scholar] [CrossRef]
- Ashby, K.; Zhuang, W.; Gonzalez-Jimenez, A.; Alvarez-Alvarez, I.; Lucena, M.I.; Andrade, R.J.; Aithal, G.P.; Suzuki, A.; Chen, M. Elevated bilirubin, alkaline phosphatase at onset, and drug metabolism are associated with prolonged recovery from DILI. J. Hepatol. 2021, 75, 333–341. [Google Scholar] [CrossRef] [PubMed]
- George, N.; Chen, M.; Yuen, N.; Hunt, C.M.; Suzuki, A. Interplay of gender, age and drug properties on reporting frequency of drug-induced liver injury. Regul. Toxicol. Pharmacol. 2018, 94, 101–107. [Google Scholar] [CrossRef]
- Chen, M.; Bisgin, H.; Tong, L.; Hong, H.; Fang, H.; Borlak, J.; Tong, W. Toward predictive models for drug-induced liver injury in humans: Are we there yet? Biomark. Med. 2014, 8, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Dirven, H.; Vist, G.E.; Bandhakavi, S.; Mehta, J.; Fitch, S.E.; Pound, P.; Ram, R.; Kincaid, B.; Leenaars, C.H.C.; Chen, M.; et al. Performance of preclinical models in predicting drug-induced liver injury in humans: A systematic review. Sci. Rep. 2021, 11, 6403. [Google Scholar] [CrossRef]
- Bassan, A.; Alves, V.M.; Amberg, A.; Anger, L.T.; Auerbach, S.; Beilke, L.; Bender, A.; Cronin, M.T.; Cross, K.P.; Hsieh, J.-H.; et al. In silico approaches in organ toxicity hazard assessment: Current status and future needs in predicting liver toxicity. Comput. Toxicol. 2021, 20, 100187. [Google Scholar] [CrossRef]
- Chen, M.; Hong, H.; Fang, H.; Kelly, R.; Zhou, G.; Borlak, J.; Tong, W. Quantitative Structure-Activity Relationship Models for Predicting Drug-Induced Liver Injury Based on FDA-Approved Drug Labeling Annotation and Using a Large Collection of Drugs. Toxicol. Sci. 2013, 136, 242–249. [Google Scholar] [CrossRef]
- Mahalmani, V.; Sinha, S.; Prakash, A.; Medhi, B. Translational research: Bridging the gap between preclinical and clinical research. Indian J. Pharmacol. 2022, 54, 393–396. [Google Scholar]
- Yu, H.W. Bridging the translational gap: Collaborative drug development and dispelling the stigma of commercialization. Drug Discov. Today 2016, 21, 299–305. [Google Scholar] [CrossRef]
- Björnsson, E.S.; Hoofnagle, J.H. Categorization of drugs implicated in causing liver injury: Critical assessment based on published case reports. Hepatology 2016, 63, 590–603. [Google Scholar] [CrossRef] [PubMed]
- Categorization of the Likelihood of Drug Induced Liver Injury. In LiverTox: Clinical and Research Information on Drug-Induced Liver Injury; Bethesda: Rockville, MD, USA, 2012.
- Leeson, P.D. Impact of Physicochemical Properties on Dose and Hepatotoxicity of Oral Drugs. Chem. Res. Toxicol. 2018, 31, 494–505. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Xie, Q.; Ge, W.; Qian, F.; Fang, H.; Shi, L.; Su, Z.; Perkins, R.; Tong, W. Mold2, Molecular Descriptors from 2D Structures for Chemoinformatics and Toxicoinformatics. J. Chem. Inf. Model. 2008, 48, 1337–1344. [Google Scholar] [CrossRef] [PubMed]
- Fonti, V.; Belitser, E. Feature selection using lasso. VU Amst. Res. Pap. Bus. Anal. 2017, 30, 1–25. [Google Scholar]
- Zhan, X.; Wang, F.; Gevaert, O. Reliably Filter Drug-Induced Liver Injury Literature with Natural Language Processing and Conformal Prediction. IEEE J. Biomed. Health Inform. 2022, 26, 5033–5041. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Zhu, J.; Chen, M.; Gong, P.; Zhang, C.; Tong, W. Quantitative structure–activity relationship models for predicting risk of drug-induced liver injury in humans. Drug-Induc. Liver Toxic. 2018, 136, 77–100. [Google Scholar]
- Aguirre-Plans, J.; Piñero, J.; Souza, T.; Callegaro, G.; Kunnen, S.J.; Sanz, F.; Fernandez-Fuentes, N.; Furlong, L.I.; Guney, E.; Oliva, B. An ensemble learning approach for modeling the systems biology of drug-induced injury. Biol. Direct 2021, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wainberg, M.; Alipanahi, B.; Frey, B.J. Are random forests truly the best classifiers? J. Mach. Learn. Res. 2016, 17, 1–5. [Google Scholar]
- Kawaguchi, K.; Lu, H. Ordered sgd: A new stochastic optimization framework for empirical risk minimization. In Proceedings of the International Conference on Artificial Intelligence and Statistics, PMLR, Atlanta, GA USA, 13–18 July 2020. [Google Scholar]
- Shamir, O.; Zhang, T. Stochastic gradient descent for non-smooth optimization: Convergence results and optimal averaging schemes. In Proceedings of the International Conference on Machine Learning, PMLR, Atlanta, GA USA, 16–21 June 2013. [Google Scholar]
- Chierici, M.; Francescatto, M.; Bussola, N.; Jurman, G.; Furlanello, C. Predictability of drug-induced liver injury by machine learning. Biol. Direct. 2020, 15, 1–10. [Google Scholar] [CrossRef]
- Dalianis, H. Evaluation metrics and evaluation. In Clinical Text Mining: Secondary Use of Electronic Patient Records; Springer Nature: Berlin/Heidelberg, Germany, 2018; pp. 45–53. [Google Scholar]
- Hanser, T.; Barber, C.; Marchaland, J.F.; Werner, S. Applicability domain: Towards a more formal definition. SAR QSAR Environ. Res. 2016, 27, 865–881. [Google Scholar] [CrossRef]
- Tropsha, A.; Gramatica, P.; Gombar, V.K. The Importance of Being Earnest: Validation is the Absolute Essential for Successful Application and Interpretation of QSPR Models. QSAR Comb. Sci. 2003, 22, 69–77. [Google Scholar] [CrossRef]
- McLachlan, G.J. Mahalanobis distance. Resonance 1999, 4, 20–26. [Google Scholar] [CrossRef]
- Wang, H.; Liang, Q.; Hancock, J.T.; Khoshgoftaar, T.M. Feature selection strategies: A comparative analysis of SHAP-value and importance-based methods. J. Big Data 2024, 11, 1–16. [Google Scholar] [CrossRef]
- Shin, H.K.; Chun, H.S.; Lee, S.; Park, S.M.; Park, D.; Kang, M.G.; Hwang, S.; Oh, K.H.; Han, H.Y.; Kim, W.K.; et al. ToxSTAR: Drug-induced liver injury prediction tool for the web environment. Bioinformatics 2022, 38, 4426–4427. [Google Scholar] [CrossRef]
- Chen, M.; Suzuki, A.; Thakkar, S.; Yu, K.; Hu, C.; Tong, W. DILIrank: The largest reference drug list ranked by the risk for developing drug-induced liver injury in humans. Drug Discov. Today 2016, 21, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Dai, Z.; Chen, F.; Gao, S.; Pei, J.; Lai, L. Deep learning for drug-induced liver injury. J. Chem. Inf. Model. 2015, 55, 2085–2093. [Google Scholar] [CrossRef]
- Li, T.; Tong, W.; Roberts, R.; Liu, Z.; Thakkar, S. DeepDILI: Deep Learning-Powered Drug-Induced Liver Injury Prediction Using Model-Level Representation. Chem. Res. Toxicol. 2020, 34, 550–565. [Google Scholar] [CrossRef]
- Wu, L.; Liu, Z.; Auerbach, S.; Huang, R.; Chen, M.; McEuen, K.; Xu, J.; Fang, H.; Tong, W. Integrating Drug’s Mode of Action into Quantitative Structure–Activity Relationships for Improved Prediction of Drug-Induced Liver Injury. J. Chem. Inf. Model. 2017, 57, 1000–1006. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Thakkar, S.; Chen, M.; Tong, W. Development of Decision Forest Models for Prediction of Drug-Induced Liver Injury in Humans Using A Large Set of FDA-approved Drugs. Sci. Rep. 2017, 7, 17311. [Google Scholar] [CrossRef]
- Zhang, H.; Ding, L.; Zou, Y.; Hu, S.-Q.; Huang, H.-G.; Kong, W.-B.; Zhang, J. Predicting drug-induced liver injury in human with Naïve Bayes classifier approach. J. Comput. Mol. Des. 2016, 30, 889–898. [Google Scholar] [CrossRef]
- McPhail, B.; Tie, Y.; Hong, H.; Pearce, B.A.; Schnackenberg, L.K.; Ge, W.; Valerio, L.G.; Fuscoe, J.C.; Tong, W.; Buzatu, D.A.; et al. Modeling Chemical Interaction Profiles: I. Spectral Data-Activity Relationship and Structure-Activity Relationship Models for Inhibitors and Non-inhibitors of Cytochrome P450 CYP3A4 and CYP2D6 Isozymes. Molecules 2012, 17, 3383–3406. [Google Scholar] [CrossRef] [PubMed]
- Nembri, S.; Grisoni, F.; Consonni, V.; Todeschini, R. In Silico Prediction of Cytochrome P450-Drug Interaction: QSARs for CYP3A4 and CYP2C9. Int. J. Mol. Sci. 2016, 17, 914. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Geng, X.; Chen, M.; Zhang, J.; Wang, B.; Ilic, K.; Tong, W. High Daily Dose and Being a Substrate of Cytochrome P450 Enzymes Are Two Important Predictors of Drug-Induced Liver Injury. Drug Metab. Dispos. 2014, 42, 744–750. [Google Scholar] [CrossRef] [PubMed]
| Model | Accuracy | Precision | Recall | MCC | F1 |
|---|---|---|---|---|---|
| RF | 0.631 ± 0.072 | 0.651 ± 0.092 | 0.716 ± 0.096 | 0.226 ± 0.145 | 0.677 ± 0.073 |
| SVM | 0.617 ± 0.071 | 0.663 ± 0.096 | 0.656 ± 0.099 | 0.225 ± 0.143 | 0.654 ± 0.079 |
| DT | 0.571 ± 0.077 | 0.623 ± 0.102 | 0.609 ± 0.102 | 0.132 ± 0.156 | 0.610 ± 0.084 |
| KNN | 0.600 ± 0.070 | 0.640 ± 0.095 | 0.664 ± 0.095 | 0.188 ± 0.146 | 0.648 ± 0.074 |
| MLP | 0.626 ± 0.068 | 0.673 ± 0.097 | 0.658 ± 0.096 | 0.245 ± 0.141 | 0.660 ± 0.077 |
| Failed Drug Candidates | Rule-of-Two Model Prediction a | RF/MLP Model Prediction |
|---|---|---|
| ADX-10059 | Most-DILI-Concern | Most-DILI-Concern |
| Aplaviroc | Most-DILI-Concern | Most-DILI-Concern |
| CP-085958 | Most-DILI-Concern | Most-DILI-Concern |
| CP-368296 | Ambiguous * | Most-DILI-Concern |
| CP-422935 | Most-DILI-Concern | Most-DILI-Concern |
| CP-456773 | Most-DILI-Concern | Most-DILI-Concern |
| CP-457920 | Ambiguous * | Most-DILI-Concern |
| CP-724714 | Most-DILI-Concern | Most-DILI-Concern |
| Darbufelone | Ambiguous * | No-DILI-Concern |
| Falnidamol | Most-DILI-Concern | Most-DILI-Concern |
| Fialuridine | No-DILI-Concern | Most-DILI-Concern |
| Fiduxosin | Most-DILI-Concern | Most-DILI-Concern |
| LY-2409021 | Ambiguous * | Most-DILI-Concern |
| MK-0893 | Most-DILI-Concern | Most-DILI-Concern |
| Pafuramidine | Most-DILI-Concern | Most-DILI-Concern |
| Pralnacasan | Ambiguous * | No-DILI-Concern |
| Sitaxentan | Most-DILI-Concern | Most-DILI-Concern |
| Solithromycin | Most-DILI-Concern | Most-DILI-Concern |
| TAK-875 | Ambiguous * | Most-DILI-Concern |
| Tasosartan | Ambiguous * | Most-DILI-Concern |
| Telcagepant | Most-DILI-Concern | Most-DILI-Concern |
| Zamifenacin | Ambiguous * | Most-DILI-Concern |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mostafa, F.; Howle, V.; Chen, M. Machine Learning to Predict Drug-Induced Liver Injury and Its Validation on Failed Drug Candidates in Development. Toxics 2024, 12, 385. https://doi.org/10.3390/toxics12060385
Mostafa F, Howle V, Chen M. Machine Learning to Predict Drug-Induced Liver Injury and Its Validation on Failed Drug Candidates in Development. Toxics. 2024; 12(6):385. https://doi.org/10.3390/toxics12060385
Chicago/Turabian StyleMostafa, Fahad, Victoria Howle, and Minjun Chen. 2024. "Machine Learning to Predict Drug-Induced Liver Injury and Its Validation on Failed Drug Candidates in Development" Toxics 12, no. 6: 385. https://doi.org/10.3390/toxics12060385
APA StyleMostafa, F., Howle, V., & Chen, M. (2024). Machine Learning to Predict Drug-Induced Liver Injury and Its Validation on Failed Drug Candidates in Development. Toxics, 12(6), 385. https://doi.org/10.3390/toxics12060385







