Abstract
Xaa-Pro dipeptidase (XPD, EC 3.4.13.9; also known as prolidase) catalyzes the hydrolysis of the iminopeptide bond in the trans-Xaa-Pro dipeptides (Xaa represents any amino acid except proline), which makes it find wide applications in food, medical and environmental protection fields. In the present study, a novel Xaa-Pro dipeptidase from Aspergillus phoenicis ATCC 14332 (ApXPD) was heterologously expressed and biochemically characterized. Reclassification based on phylogenetic analysis and the version 12.5 MEROPS database showed that this enzyme was the only fungal XPD in the unassigned subfamily that shared the highest sequence identity with Xanthomonas campestris prolidase but not with that from the more related fungal species A. niudulans. As compared with other prolidases, ApXPD also contained a long N-terminal tail (residues 1–63) and an additional region (PAPARLREKL) and used a different arginine residue for dipeptide selectivity. After heterologous expression and partial purification, recombinant ApXPD was highly active and stable over the alkaline range from 8.5 to 10.0, with maximum activity at pH 9.0 and more than 80% activity retained after 1 h incubation at pHs of 8.5–10.0 (55 °C). It also had an apparent optimum temperature of 55 °C and remained stable at 20–30 °C. Moreover, this enzyme was a cobalt-dependent prolidase that only cleaved dipeptides Lys-Pro, Gly-Pro, and Ala-Pro rather than other dipeptides, tripeptides, and tetrapeptides. All these distinct features make A. phoenicis ATCC 14332 XPD unique among currently known prolidases, thus defining a novel Xaa-Pro dipeptidase subfamily.
1. Introduction
Xaa-Pro dipeptidase (XPD, EC 3.4.13.9), also called prolidase, proline dipeptidase, peptidase D (PEPD), imidodipeptidase, or organophosphorus anhydrase (OPAA), is the only enzyme catalyzing the hydrolysis of iminopeptide bond in a trans Xaa-Pro dipeptide (Xaa represents any amino acid except proline) and belongs to subfamily M24B within clan MG of metallopeptidases in the version 12.5 MEROPS database (https://www.ebi.ac.uk/merops/ (accessed on 18 October 2023)) [1]. XPDs are found in all living organisms and have been isolated mainly from mammalian tissues, fungi, archaea, and eubacteria [2,3]. Although the physiological roles of prolidases in bacteria and archaea remain unclear, they are thought to act along with other endo- and/or exopeptidases in proline recycling and antitoxin defense [4,5,6]. In mammals, the physiological roles of XPDs are well established under different pathological conditions. Moreover, human prolidases are involved in collagen turnover, matrix remodeling, and protein metabolism [2,3], and they therefore play vital roles in several physiological processes, including inflammation, carcinogenesis, wound healing, angiogenesis, and cell proliferation, especially a rare hereditary syndrome called prolidase deficiency [5,7].
Xaa-Pro dipeptidases find extensive use in food, environmental protection, and medical fields. Because they can hydrolyze proline-containing peptides responsible for bitter flavor in cheese, these enzymes have been used in the dairy industry to lower cheese bitterness, thus enhancing its flavor [8]. Organophosphorus (OP) cholinesterase-inhibiting compounds are constituents of many chemical nerve agents and pesticides. Some XPDs also exhibit OPAA activity and can detoxify these OP compounds by cleaving the P-F and P-O bonds, and this ability allows them to serve as biosensors or environmentally friendly decontaminating agents to detect/degrade OP pesticides and chemical warfare nerve agents [9,10]. Currently, several approaches, such as immobilization [11] and protein engineering [12,13], have also been adopted to improve the OPAA activities of several XPDs. Directed evolution of PlOPAA led to a 30-fold increase in paraoxon hydrolysis [12] and a 10,000-fold increase in methyl-parathion hydrolysis [13], respectively. In addition, human prolidase can be employed as prodrug therapeutic agents and potential biomarkers for prolidase deficiency [14], cancer, diabetes, brain diseases, liver diseases, respiratory diseases, and many other disorders [2,7].
The fungi are always good producers of many industrially important enzymes [15]. To date, however, only one fungal Xaa-Pro dipeptidase (GenBank accession No. CAC39600.1) has been intracellularly overexpressed and biochemically characterized from Aspergillus nidulans [16]. A. phoenicis is an interesting thermotolerant fungus which grows well at 42–45 °C. Due to its great hydrolytic potential, A. phoenicis synthesizes many enzymes with various applications in the biotechnological industry, including carbohydrases, proteases, lipases, and chitinases [17,18], as well as several oxidoreductases, such as laccases [17], catalases [19], and glucose dehydrogenases [20]. Due to the well-known genetics and physiology, fast growth on inexpensive substrates, the ability to post-translationally modify proteins, and high secretory capacity, microbial systems, such as Pichia pastoris, have been extensively used to heterologously express fungal enzymes [21,22,23], which facilitate the biochemical characterization, large-scale production, and practical applications of these enzymes. In July 2018, the release of the genome sequence of A. phoenicis ATCC 13157 opened up opportunities to overexpress and biochemically characterize its proteolytic enzymes for industrial applications and investigate their exact physiological roles. However, only a few of these proteases have been biochemically and functionally identified based on the latest literature survey.
Examination of the genome sequence of A. phoenicis ATCC 13157 revealed that it encodes at least one putative prolidase. In the present study, a putative Xaa-Pro dipeptidase gene was cloned from A. phoenicis ATCC 14332 and expressed in Pichia pastoris. After partial purification, its biochemical characteristics were systematically investigated. Our results will provide a novel insight into fungal XPDs and may facilitate further studies on the physiological roles of this prolidase.
2. Materials and Methods
2.1. Chemicals and Reagents
Restriction enzymes, T4 DNA ligase, PyrobestTM DNA polymerase, DNA Fragment Purification Kit, Agarose Gel DNA Extraction Kit, and Plasmid Purification Kit were purchased from TaKaRa Bio Inc. (Dalian, China). A High Pure RNA Isolation Kit and a Transcriptor High Fidelity cDNA Synthesis Kit were the products of Roche Molecular Biochemicals (Quebec, CA, USA). Tryptone and yeast extract were bought from OXOID (Basingstoke, UK). Casein and pre-stained protein markers were supplied by Sigma-Aldrich (Shanghai, China) and Thermo Fisher Scientific (Shanghai, China), respectively. Dipeptides, tripeptides, and tetrapeptides were purchased from Ontores Biotechnologies (Hangzhou, China). All other chemicals were commercially available in analytical grade.
2.2. Plasmids, Strains and Growth Conditions
Shuttle vector pPIC9K (Invitrogen; Carlsbad, CA, USA) was used for gene cloning and expression. A. phoenicis ATCC 14332, bought from ATCC (Manassas, VI, USA), served as a template for apxpd gene cloning. Escherichia coli JM109 and P. pastoris GS115 (Invitrogen) were employed as the hosts for plasmid amplification and gene expression, respectively. A. phoenicis ATCC 14332 and E. coli JM109 were, respectively, grown in a PDA medium (20% (w/v) potato, 2% dextrose, 2% agar) containing 1% (w/v) casein and Luria–Bertani (LB) medium (1% (w/v) tryptone, 0.5% yeast extract, and 1% NaCl). To cultivate P. pastoris GS115 and its recombinants, yeast extract peptone dextrose (YPD) medium, minimal dextrose (MD) medium, the buffered minimal glycerol complex (BMGY) medium, and the buffered minimal methanol complex (BMMY) medium were prepared according to the manufacturer’s protocols.
2.3. Gene Cloning and Sequence Analysis
The totalRNA of A. phoenicis ATCC 14332 was extracted and reverse transcribed into cDNA by the kits according to the manufacturer’s instructions. Using the cDNA obtained as a template, the apxpd gene was amplified by PCR with primers ApXPD1 (5′-GTACTGGACAAACAGACTGAGTCTCTCCC-3′) and ApXPD2 (5′-TGCTCTAGACTATGGGTCCCAAGGCCCATAT-3′). The PCR products were then cut by XbaI, purified with a DNA Fragment Purification Kit, and ligated with SnaBI/AvrII-digested pPIC9K, generating the recombinant plasmid pPIC9K-apxpd. After this, the plasmid was transformed into E. coli JM109; positive clones were selected on LB plates supplemented with 100 μg/mL ampicillin. The correctness of the inserted sequence was subsequently confirmed via PstI-digested restriction pattern and DNA sequencing (Sangon Biotech Co., Ltd., Shanghai, China).
Based on the deduced full-length amino acid sequence of ApXPD, its phylogenic relationships with other known Xaa-Pro dipeptidases were investigated by an evolutionary tree which was constructed by program MEGA version X using the Neighbor-joining algorithm [24]. Bootstrap resampling with 1000 pseudoreplicates was conducted to assess support for each individual branch. The Prolidase family was designated by reference to the peptidase deposited in the MEROPS database [1]. The online server SignalP 5.0 (https://services.healthtech.dtu.dk/service.php?SignalP-5.0 (accessed on 16 October 2023)) was used to predict the signal peptide cleavage sites of these XPDs. Multiple sequence alignment was performed with Clustal X2 (European Bioinformatics Institute, Cambridge, UK) and BioEdit 7.0.9 (Borland, Scotts Valley, CA, USA) software packages. The three-dimensional structure of ApXPD was constructed by AlphaFold2. The fine structure of XPD was then generated and visualized by the software Discovery Studio 3.5 (Accelrys, BIOVIA, San Diego, CA, USA).
2.4. Expression of ApXPD in P. pastoris
After the correct recombinant plasmid pPIC9K-apxpd was linearized by SacI, it was electroporated into P. pastoris GS115 usinga Gene Pulser X-cell II electroporation system (Bio-Rad, Hercules, CA, USA) under conditions of 1.5 kV, 200 Ω, and 25 μF. His+ transformants were selected on MD plates containing 0.5, 1.0, 1.5, and 2.0 mg/mL G418 with the replica plating method, and the genomic DNAs of these His+ transformants were then extracted and analyzed by PCR to confirm gene integration.
A single colony of the His+ transformants exhibiting resistance to 2.0 mg/mL G418 was inoculated into 25 mL BMGY medium and cultivated at 30 °C and 200 rpm until the optical density at 600 nm (OD600) reached 4–6. The growing cells were then harvested by centrifugation (10,000× g, 4 °C; 10 min), resuspended in 25 mL BMMY medium to an OD600 of 1.0, and cultured at 30 °C and 220 rpm for 120 h. To maintain induction, the cultures were fed with pure methanol to a final concentration of 0.5% (v/v) at 24 h intervals. The induction of GS115-9K cells (P. pastoris GS115 integrated with pPIC9K) was performed in parallel as a negative control. The supernatants were collected via centrifugation at 10,000× g and 4 °C for 10 min and used for the analysis of XPD activity. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was also performed to confirm the expression of ApXPD, using 12% (w/v) polyacrylamide gels (1 mm thickness).
2.5. Enzyme Purification
Unless otherwise stated, all purification procedures were performed at 4 °C. The cell-free supernatant was fractionated with solid ammonium sulfate at 30–70% saturation. The pellet collected by centrifugation (10,000× g, 4 °C; 20 min) was resuspended in a minimal volume of 20 mM Tris-HCl buffer (pH 8.0)/1 mM cobalt chloride and dialyzed extensively against the same buffer. The dialyzed sample was then concentrated and desalted using a VivaFlow 50 tangential flow filtration module (Viva Science, Sartorius group, Göttingen, Germany) with a polyethersulfone membrane (MWCO, 10 kD). The resulting sample was used for enzyme assay and SDS-PAGE analysis.
2.6. Enzyme and Protein Assays
The ninhydrin method [25] was slightly modified to determine the amount of proline liberated from the peptide substrates. Briefly, the reaction mixture consisted (480 μL) of 2 mM Lys-Pro or other dipeptides, 20 mM Tris-HCl buffer (pH 8.0), and 1 mM cobalt chloride. After the addition of the 20 μL enzyme solution in proper dilution, the reaction mixture was incubated at 55 °C for 10 min. An aliquot (200 μL) was withdrawn and mixed with 500 μL of glacial acetic acid and 500 μL of ninhydrin reagent (3% (w/v) ninhydrin, 60% (v/v) glacial acetic acid, and 40% (v/v) phosphoric acid). After boiling for 10 min, the solution was cooled on ice. The resulting chromophore was quantified using 515 nm absorption. All measurements were performed in triplicate. One unit of Xaa-Pro dipeptidase activity is defined as the amount of enzyme liberating 1 μmol proline per minute under assay conditions. Protein concentrations were measured by the Bradford Protein Assay Kit (Beyotime Biotechnology, Nantong, China), using bovine serum albumin as a standard.
2.7. Biochemical Characterization of Recombinant ApXPD
The optimal temperature of the purified ApXPD was determined in reactions using fresh enzymes at various temperatures (20, 30, 35, 40, 45, 50, 55, 60, 65, and 70 °C). To determine its thermostability, recombinant ApXPD was pre-incubated in 20 mM Tris-HCl buffer (pH 8.0) containing 1 mM cobalt chloride at 20, 30, 40, 50, 60, or 70 °C for 1 h, prior to standard enzyme assay. All determinations were carried out in triplicate, and the results were expressed in percentage activity by assuming the maximum activity as 100%.
The optimum pH of recombinant ApXPD was examined using the following buffer solutions in place of the Tris-HCl buffer in the method described above: 20 mM sodium citrate buffer (pH 6.0–7.5), 20 mM Tris-HCl buffer (pH 7.5–9.0), and 20 mM glycine-NaOH buffer (pH 9.0–10.5). The pH stability was determined via pre-incubation of the purified enzyme in the abovementioned buffers at its optimum temperature for 1 h, followed by measurement of the residual activity under standard assay conditions.
To investigate the effects of various metal ions on the activity of recombinant ApXPD, they were added as the salts of chloride to the standard reaction mixtures at a final concentration of 0.1 mM or 1 mM to replace 1 mM cobalt chloride. The procedure used to determine the effect of EDTA was the same as described above. The activity of recombinant ApXPD without any additions was set as 100%.
Relative substrate specificity of the recombinant ApXPD was determined in the presence of 1 mM Co2+, using 2 mM various dipeptides (Lys-Pro, Gly-Pro, Ala-Pro, Pro-Pro, Cys-Gly, Lys-Ala, Lys-Ser, or Lys-Gly), tripeptides (Leu-Ala-Pro, Lys-Pro-Ala, or Gly-Pro-Ala), or tetrapeptides (Lys-Trp-Ala-Pro or Lys-Pro-Ala-Ala) as the substrates. The previously described thin-layer chromatography on Silica Gel 60 F254 [26] wasused to determine the activities of recombinant ApXPD toward dipeptides, tripeptides, or tetrapeptides that do not contain C-terminus prolines. All of the determinations were repeated three times.
Kinetic parameters of enzymatic activity for various substrates and substrate concentrations were studied in the presence of 1 mM Co2+ vianon-linear regression using the software Origin 2021 (OriginLab Corporation, Northampton, MA, USA). The Michaelis constant (Km) and maximum velocity (Vmax) were determined by directly fitting the Michaelis–Menten kinetic equation V= VmaxS/(Km + S) with the experimental data. Here, V and S denote the reaction velocity and substrate concentration, respectively. Using the Vmax obtained, the preliminary value of the Hill’s constant (h) is calculated from the slope of the ln(V/(Vmax − S)) vs. ln(S) plot.
3. Results
3.1. Molecular Cloning and Sequence Analysis of the xpd Gene from A. phoenicis ATCC 14332
DNA sequencing showed that the xpd gene from A. phoenicis ATCC 14332 consisted of 1467 bp and encoded a 488-amino acid polypeptide, with the predicted molecular weight and theoretical pI of 54.9 kD and 5.09, respectively. The DNA sequence of xpd gene from A. phoenicis ATCC 14332 matched well with that of A. phoenicis ATCC 13157 xpd gene (GenBank accession No. KZ851879.1) except for 24 nucleotide differences. Five amino acids (Asn96, Phe125, Met360, Ala362, and Lys402) in the deduced amino acid sequence of xpd gene from A. phoenicis ATCC 14332 (Figure S1) were different from those in the amino acid sequence of A. phoenicis ATCC 13157 xpd gene, which were Asp96, Tyr125, Ile360, Thr362, and Glu402, respectively. As analyzed by the SignalP 5.0 server, this enzyme did not contain a secretory signal peptide.
3.2. Reclassification of XPDs Based on Phylogenetic Analysis and MEROPS Database
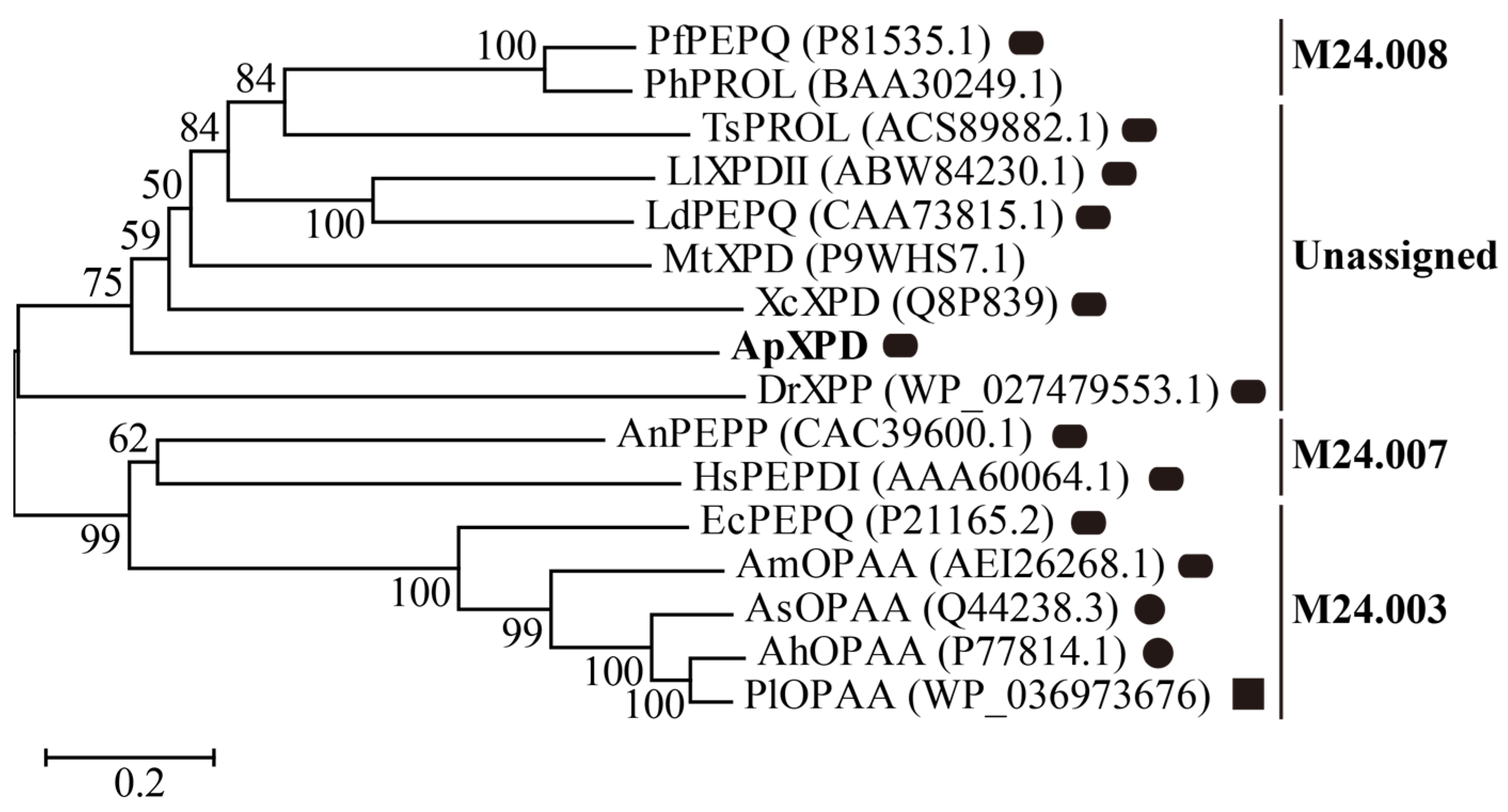
Based on the amino acid sequences of 16 available XPDs that were cloned and biochemically characterized, a phylogenetic tree was constructed by MEGA X. Based on the MEROPS database (version 12.5) [1], these XPDs can be grouped into three subfamilies according to their similarities to EcPEPQ (M24.003), HsPEPDI (M24.007), and PfPEPQ (M24.008), respectively, with one unassigned subfamily containing XPD from A. phoenicis ATCC 14332 (Figure 1). Subfamily M24.003 contains only bacterial XPDs, and M24.007 prolidases are found in eukaryotes (filamentous fungi and humans), while those from subfamilies M24.008 are hyperthermophile prolidases from archaea. In the unassigned subfamily, ApXPD is the only fungal Xaa-Pro dipeptidase ever reported, whereas all other enzymes are bacterial prolidases. It has also been observed that XPDs from subfamilies M24.003, M24.007, and M24.008 exhibit hydrolytic activities toward both Xaa-Pro dipeptides and organophosphate compounds, while members of the unknown subfamily hydrolyze only Xaa-Pro dipeptides (Table S1).
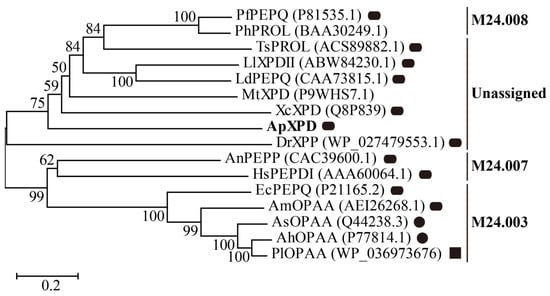
Figure 1.
Evolutionary tree describing the phylogenic relationships among XPD from A. phoenicis ATCC 14332 (ApXPD) and all the currently characterized prolidases from various organisms. Bootstrap resampling with 1000 pseudoreplicates was conducted to assess support for each individual branch. The branch lengths are proportional to the sequence divergence, and the scale of 0.2 amino acid changes per site is indicated at the bottom left of the figure. Filled circles, rounded rectangles, and filled squares represent monomers, dimmers, and tetramers, respectively. PfPEPQ, PhPROL, TsPROL, LlXPDII, LdPEPQ, MtXPD, XcXPD, DrXPP, AnPEPP, HsPEPDI, EcPEPQ, AmOPAA, AsOPAA, AhOPAA, and PlOPAAPlOPAA are prolidases from Pyrococcus furiosus DSM 3638, P. horikoshii OT3, Thermococcus sibiricus MM 739, Lactococcus lactis NRRL B-1821, L. delbrueckii subsp. bulgaricus CNRZ 397, Mycobacterium tuberculosis H37Rv, X. campestris pv. campestris ATCC 33913, Deinococcus radiodurans R1, A. nidulans WG312, Human sapiens, E. coli BL21 (DE3), Alteromonas macleodii NCIMN1963, Alteromonas sp. JD6.5, A. haloplanktis ATCC 23851, and Pseudoalteromonaslipolytica SCSIO04301, respectively. GenBank accession numbers of these polidases are given in parentheses.
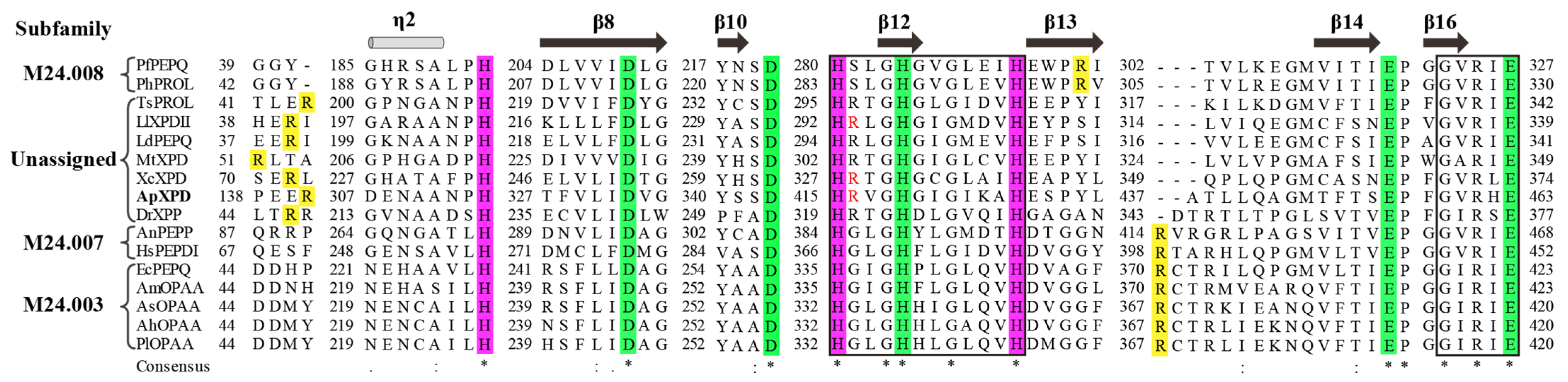
As predicted by SignalP 5.0, all of these XPDs were intracellular enzymes. Multiple sequence alignment showed that ApXPD shared the highest identity (29.30%) with XcXPD but not with AnPEPP from the more related fungal species (Figure 2), while the lowest identity (19.01%) is observed between ApXPD and DrXPP. Despite this divergence, the catalytic triads (His-His-His) and metal-binding residues (Asp-Asp-His-Glu-Glu) are highly conserved (Figure 2). The catalytic triads of ApXPD are His314, His415, and His426, and its putative metal-binding residues are Asp332, Asp343, His419, Glu449, and Glu463. Multiple sequence alignment also showed that these subfamily M24B metallopeptidases contained the conserved motifs HXXGHXXGXXXH and GXRXE (X represents any amino acid, Figure 2), where the N- and C-terminal histidines in the former motif are the nucleophiles, while the middle histidine acts as the proton receptor in the catalysis and participates in metal ion binding [27,28]. The specific arginine residues that mainly determine the length of the ligand and form the allosteric site are supposed to be Arg141 and Arg416, respectively.
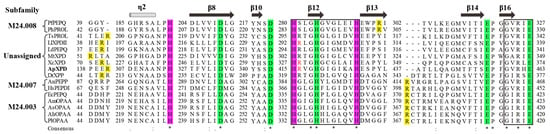
Figure 2.
Multiple sequence alignment of ApXPD with all the characterized XPDs from various organisms. Prolidases from subfamilies M24.003, M24.007, and M24.008 are indicated, while other enzymes are from the unassigned subfamily. (*), the conserved amino acids; (:), the conservative replacement; (.), semiconservative replacement. Xaa-Pro dipeptidase from A. phoenicis ATCC 14332 is in bold. The secondary structural elements of XcXPD are shown above the sequence. The strictly conserved motifs HXXGHXXGXXXH and GXRXE (X represents any amino acid) are boxed. The putative catalytic triads, metal ion binding residues, and the specific arginine residues involved in dipeptide selection are highlighted inpurple, green, and yellow, respectively. The critical arginine residues involved in the formation of allosteric site are shown in red.
3.3. Three-Dimensional Structure Analysis of ApXPD
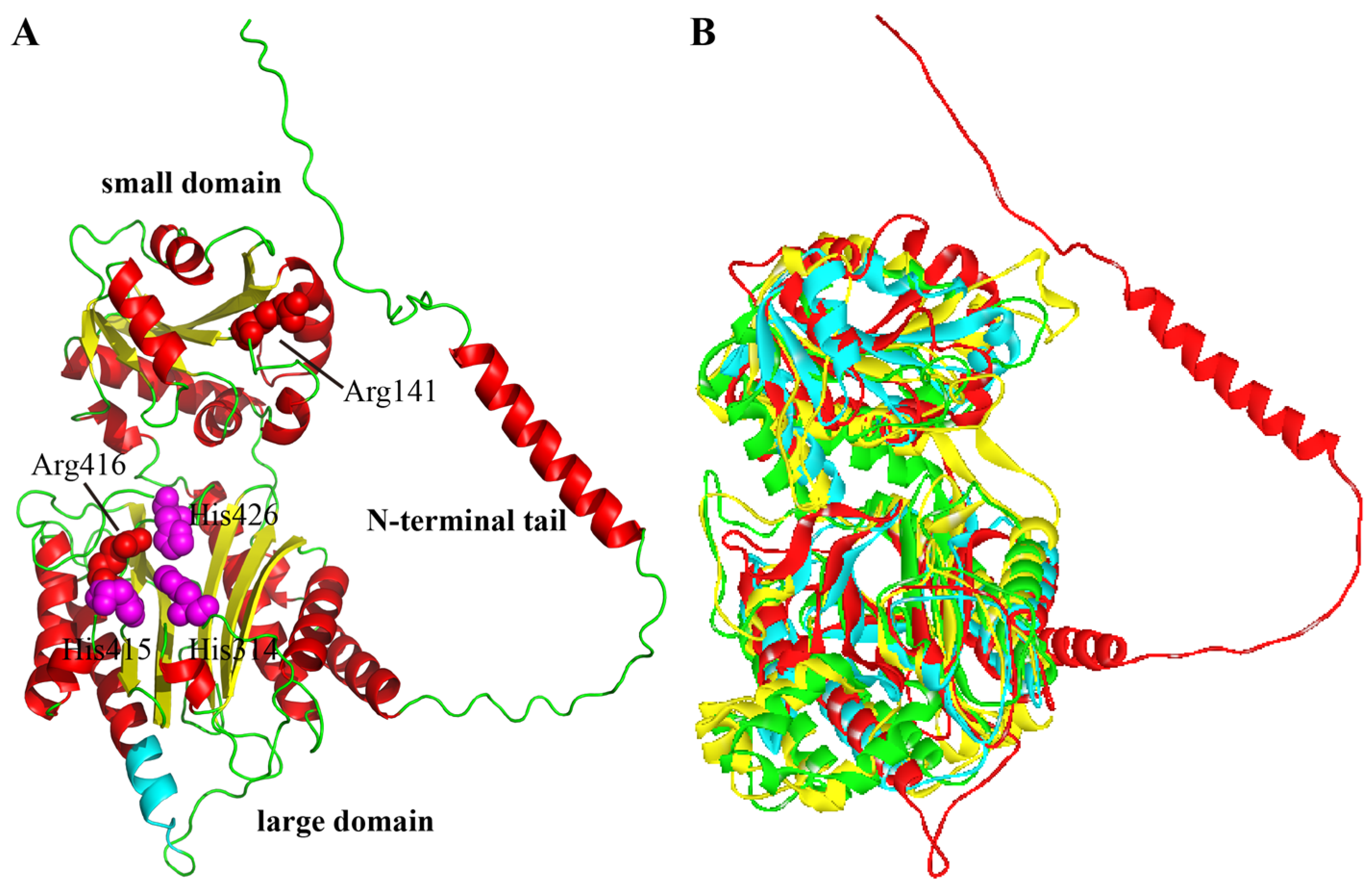
The three-dimensional structure of XPD from A. phoenicis ATCC 14332 was built by AlphaFold2 and subsequently optimized by Discovery Studio. The Ramachandran plot of the modeled structure showed that 98.6% of the residues were in favored and allowed regions, indicating the rationality and credibility of the ApXPD structure. As a member of the ‘pita-bread’ family of metallopeptidases, ApXPD consists of two major domains: a large domain (Figure 3A, residues 64–87 and 251–488) and a small domain (residues 92–245), which are connected by two short loops (residues 88–91 and 246–250). These two domains correspond to the large C-terminal catalytic domain (peptidase-M24 domain) and the small N-domain (creatinase/prolidase-N domain) in other known XPDs, respectively [4,29,30]. But unlike those in other prolidases, the large domain is formed by the pita-bread fold and contains an eight-stranded mixed β-sheet flanked by eight α-helices, while the small domain has a mixed six-stranded β-sheet with seven α-helices on the outer surface. In addition, ApXPD also possesses an N-terminal tail (residues 1 to 63) containing an α-helix, which may make both polar and hydrophobic interactions with the large domain as observed in XcXPD [4]. The putative catalytic triads of ApXPD (His314, His415, and His426) are located in an oval depression created by the pita bread fold at the center of the large domain (Figure 3A). A superimposition of the structures of ApXPD, HsPEPDI, EcPEPQ, and PfPEPQ demonstrates that they vary greatly, especially in the loop area (Figure 3B). Interestingly, ApXPD contains a long N-terminal tail (resides 1–63), but its precise structure and function need to be further elucidated.
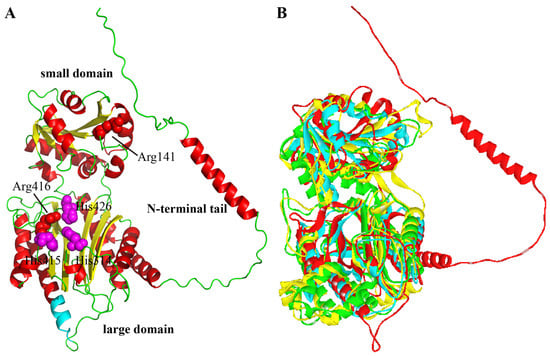
Figure 3.
Three-dimensional structures of ApXPD and other prolidases. (A) Modeled structure of ApXPD. Regions of N-terminal tail and large and small domains are shown. Putative catalytic triads (His314, His415, and H426) and the two critical arginine residues (Arg141 and Arg416) are represented by purple and red spheres, respectively. The ten additional resides (residues 361–370, PAPARLREKL) are colored cyan. (B) Comparison of the 3D structure of ApXPD (red) with those of prolidases from other subfamilies. EcPEPQ (PDB entry: 4qr8; subfamily M24.003), HsPEPDI (PDB entry: 2iw2; subfamily M24.007), and PfPEPQ (PDB entry: 1pv9; subfamily M24.008) are, respectively shown in yellow, green and cyan.
3.4. Expression and Purification of Recombinant ApXPD
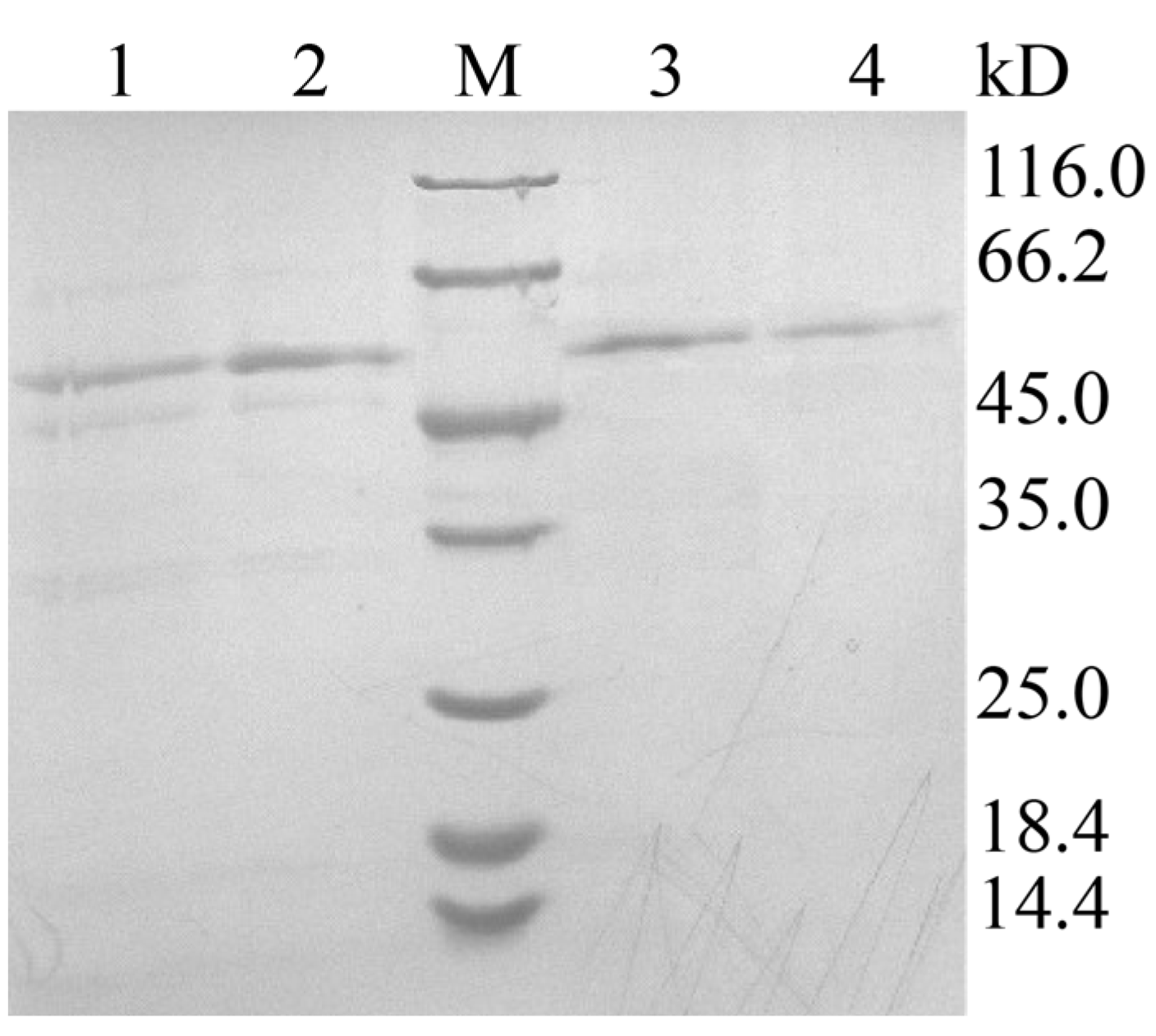
The recombinant plasmid pPIC9K-xpd was linearized with SacI and electroporated into P. pastoris GS115, generating the recombinant GS115 (pPIC9K-apxpd). After five days of shake flask fermentation, the activity of this recombinant toward Lys-Pro was determined to be 35.5 ± 4.1 U/mL, whereas no XPD activity could be detected in the strain GS115-9K (P. pastoris GS115 integrated with pPIC9K). This enzyme was then partially purified to homogeneity by ammonium sulfate precipitation, dialysis, and ultrafiltration (Figure 4), with a yield of the purified ApXPD of 28.7 mg/L culture. SDS-PAGE analysis indicated that the purified ApXPD migrated as a single band corresponding to a molecular mass of approximately 55.0 kD (Figure 4), while it exhibited a single protein peak at a molecular weight corresponding to ~110 kD on gel-filtration chromatography, suggesting that ApXPD exists as a dimer under the given conditions. Moreover, the specific activity of the purified ApXPD toward Lys-Pro was 51.2 U/mg.
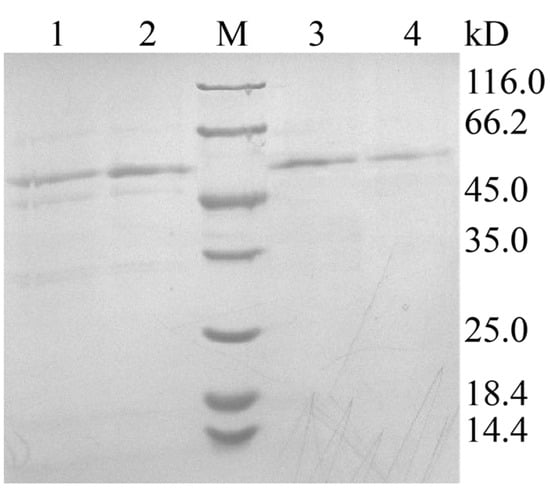
Figure 4.
SDS-PAGE analysis on the expression and purification of ApXPD. Lanes 1 and 2, cell-free supernatants of the recombinant strain; lanes 3 and 4, the purified ApXPD; M, molecular mass markers; the bands at approximately 55.0 kD represent target proteins.
3.5. Effects of Temperature and pH on the Activity and Stability of Recombinant ApXPD
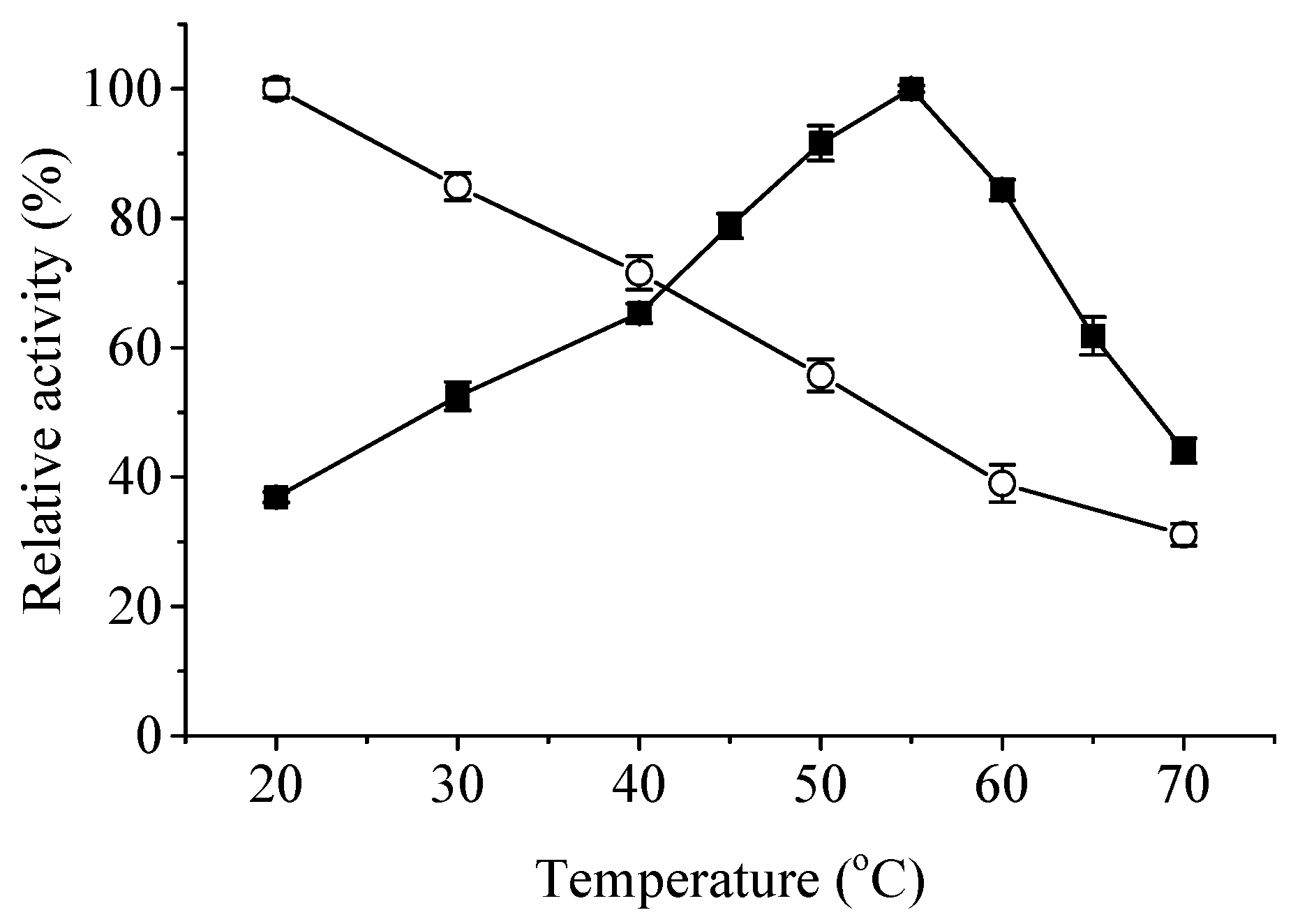
Effects of temperature and pH on the activity and stability of ApXPD were examined. As shown in Figure 5, this enzyme was active at temperatures ranging from 20 to 70 °C. It maintained over 80% of the maximum activity at temperatures of 50–60 °C, and the highest activity was observed at 55 °C. Recombinant ApXPD was stable at 20 and 30 °C because it retained more than 80% of the activity after treatment at these temperatures for 1 h. After 1 h incubation at 40, 50, and 60 °C, its activity was reduced to 71.5%, 55.7%, and 39.0% of the original level, respectively, while 31.1% activity remained after incubation at 70 °C for 1 h.
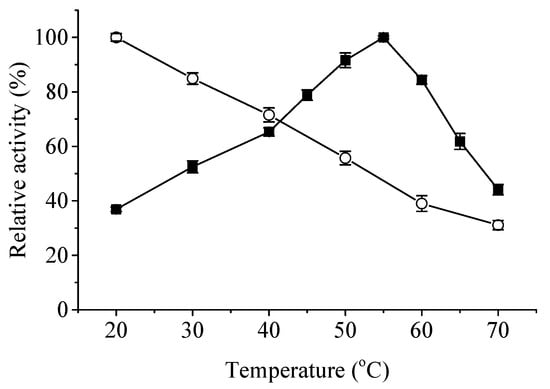
Figure 5.
Effects of temperature on the activity (closed squares) and stability (open circles) of ApXPD. The relative activity is calculated with the highest activity regarded as 100%.
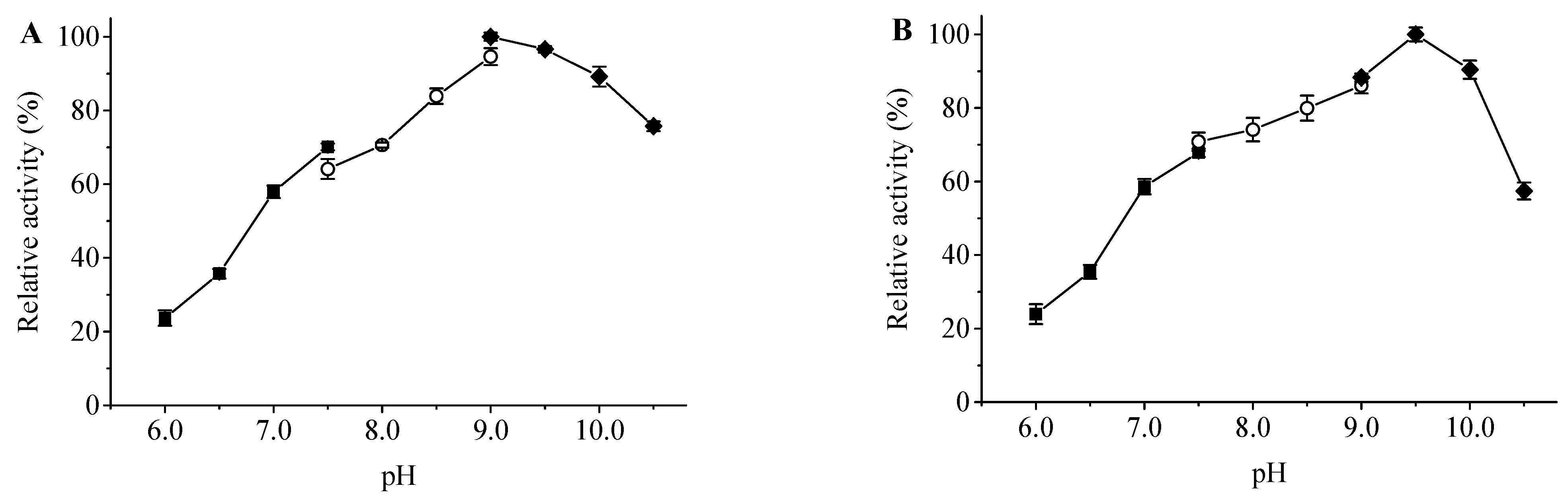
The optimum pH of ApXPD was determined at various pHs using three different buffer solutions at 55 °C. As shown in Figure 6A, recombinant ApXPD was optimally active at pH 9.0, and it retained more than 80% of the maximal activity between pH 8.5–10.5. This enzyme was stable over a narrow pH range of 8.5–10.0, with over 80% residual activities after 1 h incubation at 55 °C (Figure 6B).
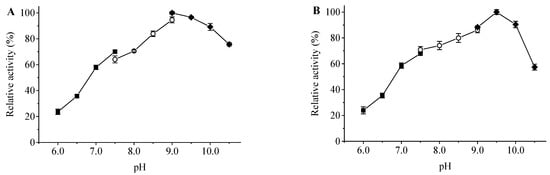
Figure 6.
Effects of pH on the activity (A) and stability (B) of ApXPD. The relative activity is calculated with the maximum activity set as 100%. Filled squares, open circles, and filled diamonds represent 20 mM sodium citrate buffer (pH 6.0–7.5), 20 mM Tris-HCl buffer (pH 7.5–9.0), and 20 mM glycine-NaOH buffer (pH 9.0–10.5), respectively.
3.6. Metal Dependency of Recombinant ApXPD
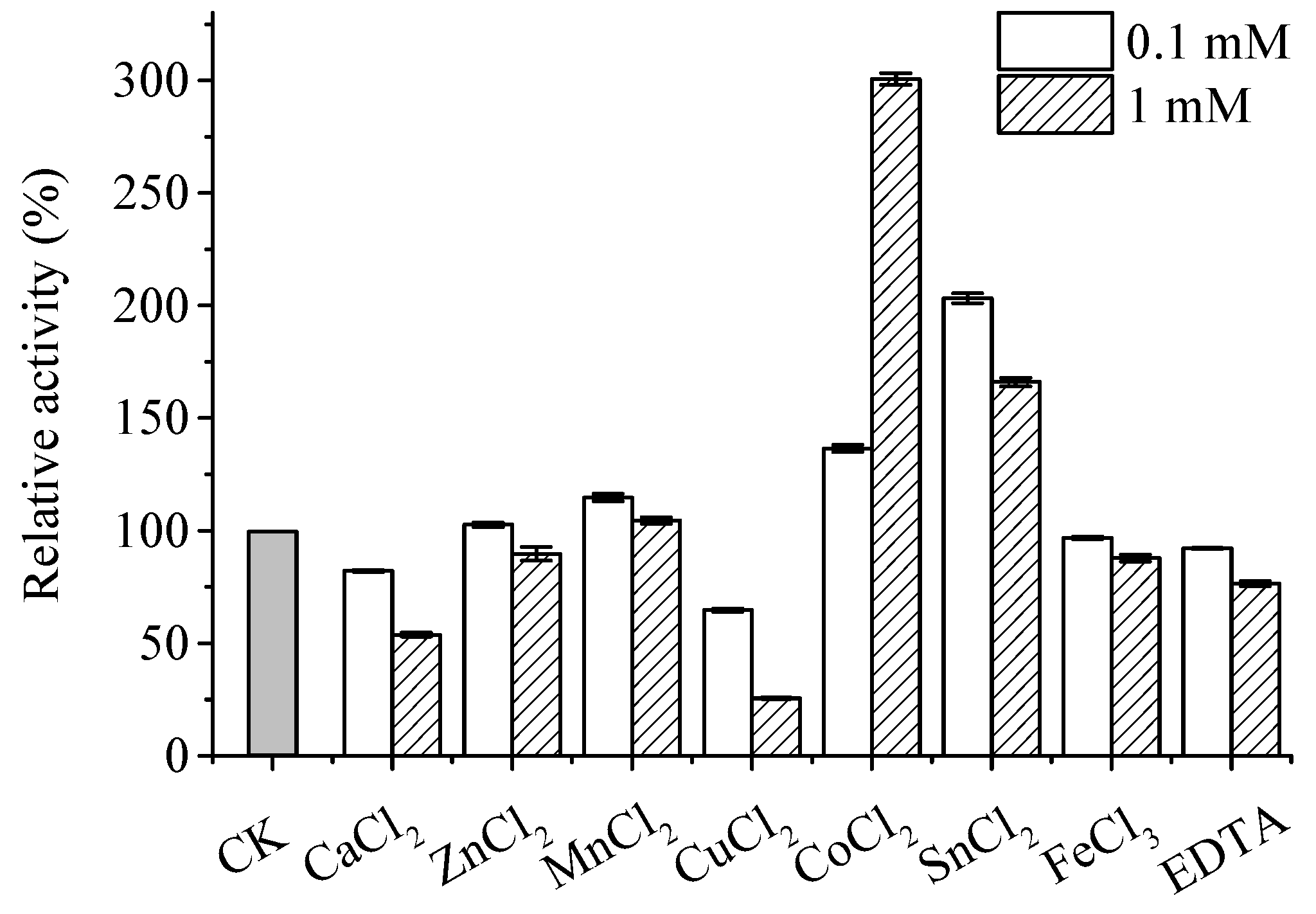
Effects of various metal ions on the activity of recombinant ApXPD areshown in Figure 7. Among the metal ions tested, 0.1 mM and 1 mM Ca2+, Zn2+, Cu2+, and Fe3+ strongly inhibited the activity of ApXPD, and their inhibitory effects increased with the increasing concentrations. EDTA also exerted inhibitory effects on the activity of ApXPD, indicating that this enzyme is metal ion-dependent [4]. On the contrary, the activity of this enzyme was significantly enhanced by 0.1 mM and 1 mM Mn2+, Co2+, and Sn2+. In addition, its activity was increased by twofold by 1 mM Co2+ (Figure 7), which suggests that cobalt is possibly the cofactor of ApXPD.
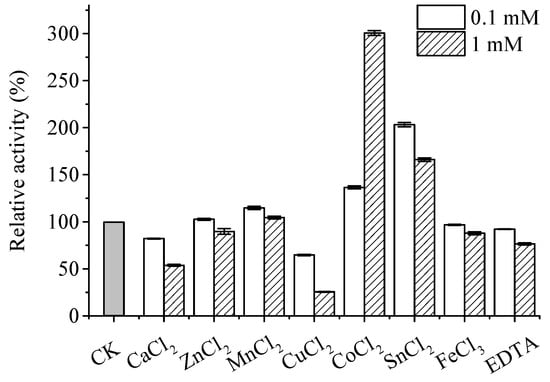
Figure 7.
Metal dependency of recombinant ApXPD. CK represents control (no metal ions addition).
3.7. Substrate Specificity of Recombinant ApXPD
Among the substrates tested, recombinant ApXPD hydrolyzed dipeptides Lys-Pro, Gly-Pro, and Ala-Pro, but it did not cleave other dipeptides, tripeptides, and tetrapeptides tested (Table 1). These results indicated that ApXPD was exclusively a Xaa-Pro dipeptidase degrading only dipeptides with C-terminus pralines rather than Pro-Pro.

Table 1.
Substrate specificity of recombinant ApXPD.
3.8. Enzyme Kinetics
Kinetic parameters were determined for recombinant ApXPD using Lys-Pro and Gly-Pro as substrates (Table 2). As compared with Gly-Pro, recombinant ApXPD showed a high affinity for Lys-Pro as indicated by the low Km value. Additionally, high values of Vmax, kcat and kcat/Km were observed for Lys-Pro, suggesting its high reaction velocity and catalytic efficiency against this substrate.

Table 2.
Kinetic parameters of recombinant ApXPD.
4. Discussion
To date, more than 20 microbial Xaa-Pro dipeptidases have been cloned and/or biochemically characterized. Among them, the prolidase from Aureohacterium esteraromaticum [31] is possibly not a prolidase since it lacks the corresponding active site and metal-binding residues of prolidases as shown by multiple sequence alignment. Previously, prokaryotic prolidases have been classified into two subfamilies (XPDxc-type and XPD48-type) according to their molecular weights and sequence relatedness [4,9,29]. However, this classification system may cause confusion, as observed for prolyl aminopeptidases [32]. In the present study, based on sequence relatedness and structural similarity, the 16 Xaa-Pro dipeptidases that were ever reported were reclassified into four subfamilies by the MEROPS classification system: M24.003, M24.007, M24.008, and one unassigned subfamily (Figure 1). Prolidases from the unassigned subfamily and subfamilies M24.007 and M24.008 are mainly dimers, whereas subfamily M24.003 contains monomers, dimers, and even a tetramer (Figure 1 and Table S1). Multiple sequence alignment demonstrated that four regions containing at least ten residues were present in prolidases from subfamilies M24.003 and M24.007 but absent in members from M24.008 and the unassigned subfamily (Figure S1). In these regions, three residues (EcPEPQ numbering: residues Pro363, Leu369, and Arg370) are strictly conserved. Moreover, an N-terminal loop extension has been found in prolidases from the M24.007 (AnPEPP and HsPEPDI) and unknown (XcXPD, MtXPD and ApXPD) subfamilies (Figure S1). Xaa-Pro dipeptidases from subfamily M24.003 only exist in bacteria, and subfamily M24.007 contains prolidases from eukaryotes (fungi and humans), while enzymes from subfamilies M24.008 proteins are prolidases from hyperthermophilic archaea. In the unassigned subfamily, all prolidases are from bacteria except ApXPD, which is the only reported fungal Xaa-Pro dipeptidase containing a long N-terminal tail (residues 1–63; Figure 3A) and an additional region (PAPARLREKL, residues 361–370; Figure S1). Interestingly, ApXPD also shares the highest sequence identity with XcXPD [4,33] but not with the prolidase from the more related fungal species A. nidulans (AnPEPP, Figure 2). Therefore, XPD from A. phoenicis ATCC 14332 represents a novel Xaa-Pro dipeptidase subfamily.
Xaa-Pro dipeptidases differ greatly in their optimal temperatures, optimal pHs, metal requirements, and substrate specificities. The optimum temperature of ApXPD was 55 °C (Figure 5), which was the same as or higher than those of prolidases from mesophilic sources [2] but much lower than the optimal temperatures (100 °C) of prolidases derived from hyperthermophilic archaea Pyrococcus ssp. [25,34]. Most XPDs are stable at low temperatures (20–50 °C), as is the case of XPD from A. phoenicis ATCC 14332. Unlike most prolidases, which are most active under acidic or neutral pHs (pH 6.0–8.5), ApXPD acts optimally under more alkaline pH (pH 9.0) and is stable over a narrow pH range of 8.5–10.0 (Figure 6).
XPDs require one or two divalent cations for enzyme activity, typically Mn2+, Co2+, or Zn2+ [35,36]. In vitro studies have shown that most prolidases ever reported utilize Mn2+ ion as cofactors (Table S1) [4,28,37,38,39,40], whereas PfPEPQ, PhPROL, and ScXPD require Co2+ in their active sites under aerobic assay conditions [25,34,41], as is the case of XPD from A. phoenicis ATCC 14332 (Figure 7). Furthermore, the enzymatic activities of LdPEPQ, LlXPDII, and TsPROL are dependent on Zn2+ [26,42,43]. In contrast, the activities of AsOPAA, AuOPAA, LcXPD, PfpepQ, PhPROL, and XmXPD were strongly or even completely inactivated by high concentrations of Zn2+ (Table S1), which is due to the fact that Zn binding decreases the volume of active site pocket for accumulating the substrates [30]. Since the in vivo concentrations of Mn2+ and Co2+ are much lower than thoseof Zn2+, the metal ion requirements for XPDs in vivo are quite different, and a preference for Zn2+ might be advantageous [44].Moreover, PfPEPQ and PhPROL require Fe2+ rather than Co2+ for maximal activity under anaerobic conditions, suggesting that their metal preferences in vivo may be changed [34,35].
XPDs are highly specific in the type of substrate that they hydrolyze, and they usually prefer dipeptides to tripeptides and tetrapeptides. Prolidases from subfamilies M24.003, M24.007, and M24.008 hydrolyze both Xaa-Pro dipeptides and OP compounds, whereas those from the unassigned subfamily only cleave Xaa-Pro dipeptides (Table S1). There are also some differences in dipeptide substrate specificities among the prolidases that have been biochemically characterized. Their main activity is Xaa-Pro dipeptides hydrolysis, although several prolidases can cleave dipeptides with proline at the N-terminus or dipeptides that do not contain a proline residue [40,43,45]. Most prolidases show broad substrate specificities and prefer Xaa-Pro dipeptides containing N-terminal hydrophobic amino acids, whereas PfPEPQ [25], PhPROL [34], and EcPEPQ [37] show affinity for negatively charged and/or hydrophilic amino acids at the N-terminus. The recombinant ApXPD characterized in this study, however, shows a preference for positively charged Lys-Pro (Table 1 and Table 2). Additionally, most prolidases do not hydrolyze Pro-Pro and Gly-Pro except MtXPD [4], XcXPD [4], AnPEPP [16], PlOPAA [39], DrXPP [43], and HsPEPDI [46]. As indicated by the kinetic study, ApXPD may also show allosteric behavior (Table 2). This may be because it contains another critical arginine residue (Arg416) responsible for the formation of the allosteric site (Figure 2), as observed in the equivalent positions in allosteric LlXPDII (Arg293) and XcXPD (Arg328). Further kinetic studies using other substrates and crystal structure analyses are highly warranted to delineate the underlying mechanism.
Typically, the substrate length of prolidases is restricted to dipeptides by the guanidinium side chain of an arginine residue that binds to the free carboxylate of dipeptides and prevents the binding of longer peptides [4,9,29,47]. Furthermore, the specific arginine residue can also increase the affinities of XPDs for substrates by introducing allosteric behavior and substrate inhibition [4,27,48]. Prolidases from these four subfamilies vary greatly in their specific arginine residues. As shown by multiple sequence alignment in Figure 2, the specific arginine residues of subfamily M24.003 prolidases are counterparts of those in XPDs from subfamily M24.007, such as Arg370 in AmOPAA [9], Arg370 in EcPEPQ [47] as well as Arg398 in HsPEPDI [49], while the arginine residues in PfPEPQ and PhPROL from subfamily M24.008 are Arg295 and Arg298 [42], respectively, which do not correspond to those in subfamilies M24.003 and M24.007 XPDs (Figure 2). Unlike XPDs in the other three subfamilies, however, prolidases from the unassigned subfamily use a novel arginine-dependent mechanism for dipeptide selectivity. According to the results of structural, mutational, and/or molecular dynamics simulation analyses, the corresponding arginine residues employed by XcXPD [4], DrXPP [43], and LlXPDII [48] have been found to be Arg72, Arg46, and Arg40, respectively, while the equivalent residue in ApXPD is Arg141 (Figure 2). Further investigations are therefore needed to understand the precise mechanism of prolidases from the unassigned subfamily used for dipeptide selectivity. Aside from structural differences in the substrate binding pocket, the variability of prolidase substrate specificity also depends on the catalytic metal cation [4,27,42]. For example, in the presence of Mn2+, LlXPDII shows low activity toward Pro-Pro, whereas it cannot hydrolyze this dipeptide in the presence of Zn2+. Furthermore, its substrate preference changes from Leu-Pro to Arg-Pro in the presence of Mn2+ [42].
5. Conclusions
In summary, a Xaa-Pro dipeptidase from A. phoenicis ATCC 14332 was molecularly and biochemically characterized. Phylogenetic analysis showed that this enzyme was the only fungal XPD in the unassigned subfamily, and it shared the highest sequence identity with XcXPD but not with AnPEPP from the more related species. Multiple sequence alignment and 3D structure modeling revealed that it contained an additional region of PAPARLREKL and an N-terminal tail (residues 81–89) and utilized a different arginine residue for dipeptide selectivity. Taken together with its distinct biochemical properties, including high stability under alkaline conditions, specific metal requirement, and substrate specificity, our results indicate that recombinant ApXPD represents a novel prolidase subfamily. Knowledge gained here not only brings some new knowledge in the area of fungal XPDs but also provides a firm foundation for future research on the industrial application and in vivo physiological roles of this fungal Xaa-Pro dipeptidase.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/fermentation9110978/s1. Table S1: Xaa-Pro dipeptidases that have been characterized by molecular and/or biochemical techniques; Figure S1: Multiple sequence alignment of ApXPD with all the characterized XPDs from various organisms [50,51,52,53,54,55,56,57,58,59,60].
Author Contributions
Conceptualization, Z.D. and L.Y.; methodology, Z.D., S.Y. and K.Z.; software, S.Y. and K.Z.; validation, Z.D. and Y.K.; formal analysis, Z.D. and C.T.; investigation, S.Y. and C.T.; writing—original draft preparation, Z.D. and S.Y.; writing—review and editing, Y.K. and L.Y.; visualization, Z.D. and S.Y.; supervision, Z.D. and L.Y.; funding acquisition, Z.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Specialized Research Fund for the Doctoral Program of Nanyang Normal University (grant No. 2020ZX004), the Training Program for National Natural Science Foundation of China by Nanyang Normal University (grant No. 2023Y005), and the Henan Province Natural Science Foundation (grant No. 222300420251).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rawlings, N.D.; Alan, J.; Thomas, P.D.; Huang, X.D.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef] [PubMed]
- Kitchener, R.; Grunden, A. Prolidase function in proline metabolism and its medical and biotechnological applications. J. Appl. Microbiol. 2012, 113, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Namiduru, E.S. Prolidase. Bratisl. Lek. Listy 2016, 117, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Are, V.N.; Kumar, A.; Kumar, S.; Goyal, V.D.; Ghosh, B.; Bhatnagar, D.; Jamdar, S.N.; Makde, R.D. Crystal structure and biochemical investigations reveal novel mode of substrate selectivity and illuminate substrate inhibition and allostericity in a subfamily of Xaa-Pro dipeptidases. BBA-Proteins Proteom. 2017, 1865, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Wilk, P.; Wator, E.; Weiss, M.S. Prolidase—A protein with many faces. Biochimie 2021, 183, 3–12. [Google Scholar] [CrossRef]
- Misiura, M.; Miltyk, W. Current understanding of the emerging role of prolidase in cellular metabolism. Int. J. Mol. Sci. 2020, 21, 5906. [Google Scholar] [CrossRef]
- Eni-Aganga, I.; Lanaghan, Z.M.; Balasubramaniam, M.; Dash, C.; Pandhare, J. PROLIDASE: A review from discovery to its role in health and disease. Front. Mol. Biosci. 2021, 8, 723003. [Google Scholar] [CrossRef]
- Theriot, C.M.; Tove, S.R.; Grunden, A.M. Chapter 3 Biotechnological applications of recombinant microbial prolidases. In Advances in Applied Microbiology; Academic Press: Cambridge, MA, USA, 2009; Volume 68, pp. 99–132. [Google Scholar]
- Stepankova, A.; Duskova, J.; Skalova, T.; Hasek, J.; Koval, T.; Ostergaard, L.H.; Dohnalek, J. Organophosphorus acid anhydrolase from Alteromonas macleodii: Structural study and functional relationship to prolidases. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 346–354. [Google Scholar] [CrossRef]
- Manco, G.; Porzio, E.; Suzumoto, Y. Enzymatic detoxification: A sustainable means of degrading toxic organophosphate pesticides and chemical warfare nerve agents. J. Chem. Technol. Biot. 2018, 93, 2064–2082. [Google Scholar] [CrossRef]
- Wang, T.-F.; Lo, H.-F.; Chi, M.-C.; Lai, K.-L.; Lin, M.-G.; Lin, L.-L. Affinity immobilization of a bacterial prolidase onto metal-ion-chelated magnetic nanoparticles for the hydrolysis of organophosphorus compounds. Int. J. Mol. Sci. 2019, 20, 3625. [Google Scholar] [CrossRef]
- Yang, J.; Xiao, Y.-Z.; Li, R.; Liu, Y.; Long, L.-J. Repurposing a bacterial prolidase for organophosphorus hydrolysis: Reshaped catalytic cavity switches substrate selectivity. Biotechnol. Bioeng. 2020, 117, 2694–2702. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xiao, Y.; Liu, Y.; Li, R.; Long, L. Structure-based redesign of the bacterial prolidase active-site pocket for efficient enhancement of methyl-parathion hydrolysis. Catal. Sci. Technol. 2021, 11, 5086–5093. [Google Scholar] [CrossRef]
- Spodenkiewicz, M.; Spodenkiewicz, M.; Cleary, M.; Massier, M.; Fitsialos, G.; Cottin, V.; Jouret, G.; Poirsier, C.; Doco-Fenzy, M.; Lèbre, A.S. Clinical genetics of prolidase deficiency: An updated review. Biology 2020, 9, 108. [Google Scholar] [CrossRef] [PubMed]
- Benassi, V.M.; Lucas, R.C.d.; Michelin, M.; Jorge, J.A.; Terenzi, H.F.; Polizeli, M.d.L.T.d.M. Production and action of an Aspergillus phoenicis enzymatic pool using different carbon sources. Braz. J. Food Technol. 2012, 15, 253–260. [Google Scholar] [CrossRef]
- Jalving, R.; Bron, P.; Kester, H.C.; Visser, J.; Schaap, P.J. Cloning of a prolidase gene from Aspergillus nidulans and characterisation of its product. Mol. Genet. Genom. 2002, 267, 218–222. [Google Scholar] [CrossRef]
- Ameen, F.; AlNadhari, S.; Yassin, M.A.; Al-Sabri, A.; Almansob, A.; Alqahtani, N.; Stephenson, S.L. Desert soil fungi isolated from Saudi Arabia: Cultivable fungal community and biochemical production. Saudi J. Biol. Sci. 2022, 29, 2409–2420. [Google Scholar] [CrossRef]
- Li, Q.; Loman, A.A.; Coffman, A.M.; Ju, L. Soybean hull induced production of carbohydrases and protease among Aspergillus and their effectiveness in soy flour carbohydrate and protein separation. J. Biotechnol. 2017, 248, 35–42. [Google Scholar] [CrossRef]
- NKacem, N.C.; Destain, J.; Meraihi, Z.; Dehimat, L.; Haddoum, T.; Wathelet, J.P.; Thonart, P. Optimization of extracellular catalase production from Aspergillus phoenicis K30 by Plackett-Burman design and linear regression using date flour as single carbon source and purification of the enzyme. Afr. J. Biotechnol. 2013, 12, 2646–2653. [Google Scholar]
- Iwasa, H.; Ozawa, K.; Sasaki, N.; Kinoshita, N.; Yokoyama, K.; Hiratsuka, A. Fungal FAD-dependent glucose dehydrogenases concerning high activity, affinity, and thermostability for maltose-insensitive blood glucose sensor. Biochem. Eng. J. 2018, 140, 115–122. [Google Scholar] [CrossRef]
- Lopes, D.C.B.; Carraro, C.B.; Silva, R.N.; de Paula, R.G. Molecular characterization of xyloglucanase cel74a from Trichoderma reesei. Int. J. Mol. Sci. 2021, 22, 4545. [Google Scholar] [CrossRef]
- Campos Antoniêto, A.C.; Maués, D.B.; Nogueira, K.M.V.; de Paula, R.G.; Steindorff, A.S.; Kennedy, J.F.; Pandey, A.; Gupta, V.K.; Silva, R.N. Engineering of holocellulase in biomass-degrading fungi for sustainable biofuel production. J. Clean. Prod. 2022, 371, 133488. [Google Scholar] [CrossRef]
- Alnoch, R.C.; Salgado, J.C.S.; Alves, G.S.; de Andrades, D.; Meleiro, L.P.; Segato, F.; Berto, G.L.; Ward, R.J.; Buckeridge, M.S.; Polizeli, M.d.L.T.M. Biochemical characterization of an endoglucanase GH7 from thermophile Thermothielavioides terrestris expressed on Aspergillus nidulans. Catalysts 2023, 13, 582. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Ghosh, M.; Grunden, A.M.; Dunn, D.M.; Weiss, R.; Adams, M.W. Characterization of native and recombinant forms of an unusual cobalt-dependent proline dipeptidase (prolidase) from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 1998, 180, 4781–4789. [Google Scholar] [CrossRef]
- Morel, F.; Frot-Coutaz, J.; Aubel, D.; Portalier, R.; Atlan, D. Characterization of a prolidase from Lactobacillus delbrueckii subsp. bulgaricus CNRZ 397 with an unusual regulation of biosynthesis. Microbiology 1999, 145 Pt 2, 437. [Google Scholar]
- Kgosisejo, O.; Chen, J.A.; Grochulski, P.; Tanaka, T. Crystallographic structure of recombinant Lactococcus lactis prolidase to support proposed structure-function relationships. BBA-Proteins Proteom. 2017, 1865, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Besio, R.; Alleva, S.; Forlino, A.; Lupi, A.; Meneghini, C.; Minicozzi, V.; Profumo, A.; Stellato, F.; Tenni, R.; Morante, S. Identifying the structure of the active sites of human recombinant prolidase. Eur. Biophys. J. 2010, 39, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Maher, M.J.; Ghosh, M.; Grunden, A.M.; Menon, A.L.; Adams, M.W.W.; Adams, M.W.W.; Freeman, H.C.; Guss, J.M. Structure of the prolidase from Pyrococcus furiosus. Biochemistry 2004, 43, 2771–2783. [Google Scholar] [CrossRef]
- Jeyakanthan, J.; Takada, K.; Sawano, M.; Ogasahara, K.; Mizutani, H.; Kunishima, N.; Yokoyama, S.; Yutani, K. Crystal structural and functional analysis of the putative dipeptidase from Pyrococcus horikoshii OT3. J. Biophys. 2009, 2009, 434038. [Google Scholar] [CrossRef]
- Kabashima, T.; Fujii, M.; Hamasaki, Y.; Ito, K.; Yoshimoto, T. Cloning of a novel prolidase gene from Aureobacterium esteraromaticum. BBA-Mol. Cell Res. 1999, 1429, 516–520. [Google Scholar] [CrossRef]
- Mahon, C.S.; O’Donoghue, A.J.; Goetz, D.H.; Murray, P.G.; Craik, C.S.; Tuohy, M.G. Characterization of a multimeric, eukaryotic prolyl aminopeptidase: An inducible and highly specific intracellular peptidase from the non-pathogenic fungus Talaromyces emersonii. Microbiology 2009, 155, 3673–3682. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Are, V.N.; Ghosh, B.; Agrawal, U.; Jamdar, S.N.; Makde, R.D.; Sharma, S.M. Crystallization and preliminary X-ray diffraction analysis of Xaa-Pro dipeptidase from Xanthomonas campestris. Acta Crystallogr. F Struct. Biol. Commun. 2014, 70, 1268–1271. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.M.; Tove, S.R.; Grunden, A.M. Characterization of two proline dipeptidases (prolidases) from the hyperthermophilic archaeon Pyrococcus horikoshii. Appl. Microbiol. Biotechnol. 2010, 86, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Du, X.L.; Tove, S.; Kast-Hutcheson, K.; Grunden, A.M. Characterization of the dinuclear metal center of Pyrococcus furiosus prolidase by analysis of targeted mutants. FEBS Lett. 2005, 579, 6140–6146. [Google Scholar] [CrossRef]
- Besio, R.; Baratto, M.C.; Gioia, R.; Monzani, E.; Nicolis, S.; Cucca, L.; Profumo, A.; Casella, L.; Basosi, R.; Tenni, R.; et al. A Mn(II)–Mn(II) center in human prolidase. BBA—Proteins Proteom. 2013, 1834, 197–204. [Google Scholar] [CrossRef]
- Park, M.S.; Hill, C.M.; Li, Y.; Hardy, R.K.; Khanna, H.; Khang, Y.H.; Raushel, F.M. Catalytic properties of the PepQ prolidase from Escherichia coli. Arch. Biochem. Biophys. 2004, 429, 224–230. [Google Scholar] [CrossRef]
- Vyas, N.K.; Nickitenko, A.; Rastogi, V.K.; Shah, S.S.; Quiocho, F.A. Structural insights into the dual activities of the nerve agent degrading organophosphate anhydrolase/prolidase. Biochemistry 2010, 49, 547–559. [Google Scholar] [CrossRef]
- Xiao, Y.Z.; Yang, J.; Tian, X.P.; Wang, X.X.; Li, J.; Zhang, S.; Long, L.J. Biochemical basis for hydrolysis of organophosphorus by a marine bacterial prolidase. Process Biochem. 2017, 52, 141–148. [Google Scholar] [CrossRef]
- Fernandez-Espla, M.D.; Martin-Hernandez, M.C.; Fox, P.F. Purification and characterization of a prolidase from Lactobacillus casei subsp. casei IFPL 731. Appl. Environ. Microbiol. 1997, 63, 314–316. [Google Scholar] [CrossRef]
- Kaminogawa, S.; Azuma, N.; Hwang, I.K.; Suzuki, Y.; Yamauchi, K. Isolation and characterization of a prolidase from Streptococcus cremoris H61. J. Agric. Chem. Soc. JPN 1984, 48, 3035–3040. [Google Scholar] [CrossRef]
- Yang, S.I.; Tanaka, T. Characterization of recombinant prolidase from Lactococcus lactis—Changes in substrate specificity by metal cations, and allosteric behavior of the peptidase. FEBS J. 2008, 275, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Are, V.N.; Jamdar, S.N.; Ghosh, B.; Goyal, V.D.; Kumar, A.; Neema, S.; Gadre, R.; Makde, R.D. Crystal structure of a novel prolidase from Deinococcus radiodurans identifies new subfamily of bacterial prolidases. Proteins 2017, 85, 2239–2251. [Google Scholar] [CrossRef] [PubMed]
- Willingham, K.; Maher, M.J.; Grunden, A.M.; Ghosh, M.; Adams, M.W.W.; Freeman, H.C.; Guss, J.M. Crystallization and characterization of the prolidase from Pyrococcus furiosus. Acta Crystallogr. D 2001, 57, 428–430. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Liu, L.; Wang, B.; Wu, J.; DeFrank, J.J.; Anderson, D.M.; Rastogi, V.K.; Hamilton, A.B. Nucleotide sequence of a gene encoding an organophosphorus nerve agent degrading enzyme from Alteromonas haloplanktis. J. Ind. Microbiol. Biotechnol. 1997, 18, 49–55. [Google Scholar] [CrossRef]
- Lupi, A.; Della Torre, S.; Campari, E.; Tenni, R.; Cetta, G.; Rossi, A.; Forlino, A. Human recombinant prolidase from eukaryotic and prokaryotic sources. Expression, purification, characterization and long-term stability studies. FEBS J. 2006, 273, 5466–5478. [Google Scholar] [CrossRef]
- Weaver, J.; Watts, T.; Li, P.; Rye, H.S. Structural basis of substrate selectivity of E. coli prolidase. PLoS ONE 2014, 9, e111531. [Google Scholar] [CrossRef]
- Chen, J.A.; Tanaka, T. Charged residues on a flap-loop structure of Lactococcus lactis prolidase play critical roles in allosteric behavior and substrate inhibition. BBA-Proteins Proteom. 2011, 1814, 1677–1685. [Google Scholar] [CrossRef]
- Wilk, P.; Uehlein, M.; Kalms, J.; Dobbek, H.; Mueller, U.; Weiss, M.S. Substrate specificity and reaction mechanism of human prolidase. FEBS J. 2017, 284, 2870–2885. [Google Scholar] [CrossRef]
- Grunden, A.; Ghosh, M.; Schut, G. Proline dipeptidase from Pyrococcus furiosus. Methods Enzymol. 2001, 330, 433–445. [Google Scholar]
- Theriot, C.M.; Semcer, R.L.; Shah, S.S.; Grunden, A.M. Improving the catalytic activity of hyperthermophilic Pyrococcus horikoshii prolidase for detoxification of organophosphorus nerve agents over a broad range of temperatures. Archaea 2011, 2011, 565127. [Google Scholar] [CrossRef]
- DeFrank, J.J.; Cheng, T.C. Purification and properties of an organophosphorus acid anhydrase from a halophilic bacterial isolate. J. Bacteriol. 1991, 173, 1938–1943. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cheng, T.C.; Harvey, S.P.; Chen, G.L. Cloning and expression of a gene encoding a bacterial enzyme for decontamination of organophosphorus nerve agents and nucleotide sequence of the enzyme. Appl. Environ. Microbiol. 1996, 62, 1636–1641. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.C.; Harvey, S.P.; Stroup, A.N. Purification and properties of a highly active organophosphorus acid anhydrolase from Alteromonas undina. Appl. Environ. Microbiol. 1993, 59, 3138–3140. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.F.; Chi, M.C.; Lai, K.L.; Lin, M.G.; Chen, Y.Y.; Lo, H.F.; Lin, L.L. High-level expression and molecular characterization of a recombinant prolidase from Escherichia coli NovaBlue. PeerJ 2018, 6, e5863. [Google Scholar] [CrossRef]
- Booth, M.; Jennings, V.; Fhaolain, I.N.; O’Cuinn, G. Prolidase activity of Lactococcus lactis subsp. cremoris AM2: Partial purification and characterization. J. Dairy Res. 1990, 57, 245–254. [Google Scholar]
- Timofeev, V.; Slutskaya, E.; Gorbacheva, M.; Boyko, K.; Rakitina, T.; Korzhenevskiy, D.; Lipkin, A.; Popov, V. Structure of recombinant prolidase from Thermococcus sibiricus in space group P21221. Acta Crystallogr. 2015, 71, 951–957. [Google Scholar]
- Trofimov, A.A.; Slutskaya, E.A.; Polyakov, K.M.; Dorovatovskii, P.V.; Gumerov, V.M.; Popov, V.O. Influence of intermolecular contacts on the structure of recombinant prolidase from Thermococcus sibiricus. Acta Crystallogr. 2012, 68, 1275–1278. [Google Scholar]
- Suga, K.; Kabashima, T.; Ito, K.; Tsuru, D.; Okamura, H.; Kataoka, J.; Yoshimoto, T. Prolidase from Xanthomonas maltophilia: Purification and characterization of the enzyme. Biosci. Biotechnol. Biochem. 1995, 59, 2087–2090. [Google Scholar] [CrossRef][Green Version]
- Chandrasekaran, L.; Belinskaya, T.; Saxena, A. In vitro characterization of organophosphorus compound hydrolysis by native and recombinant human prolidase. Toxicol. In Vitro 2013, 27, 499–506. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).