Changes in Physicochemical Characteristics and Microbial Diversity of Traditional Fermented Vinasse Hairtail
Abstract
:1. Introduction
2. Materials and Methods
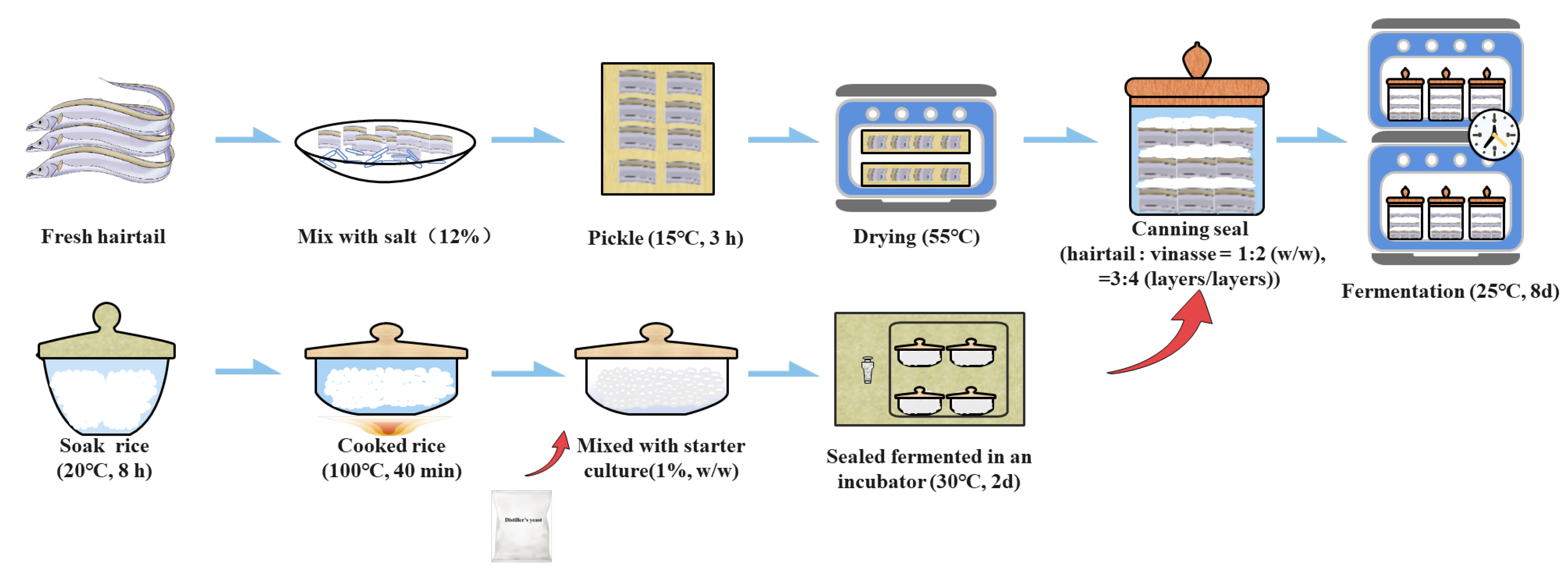
2.1. Samples Collection
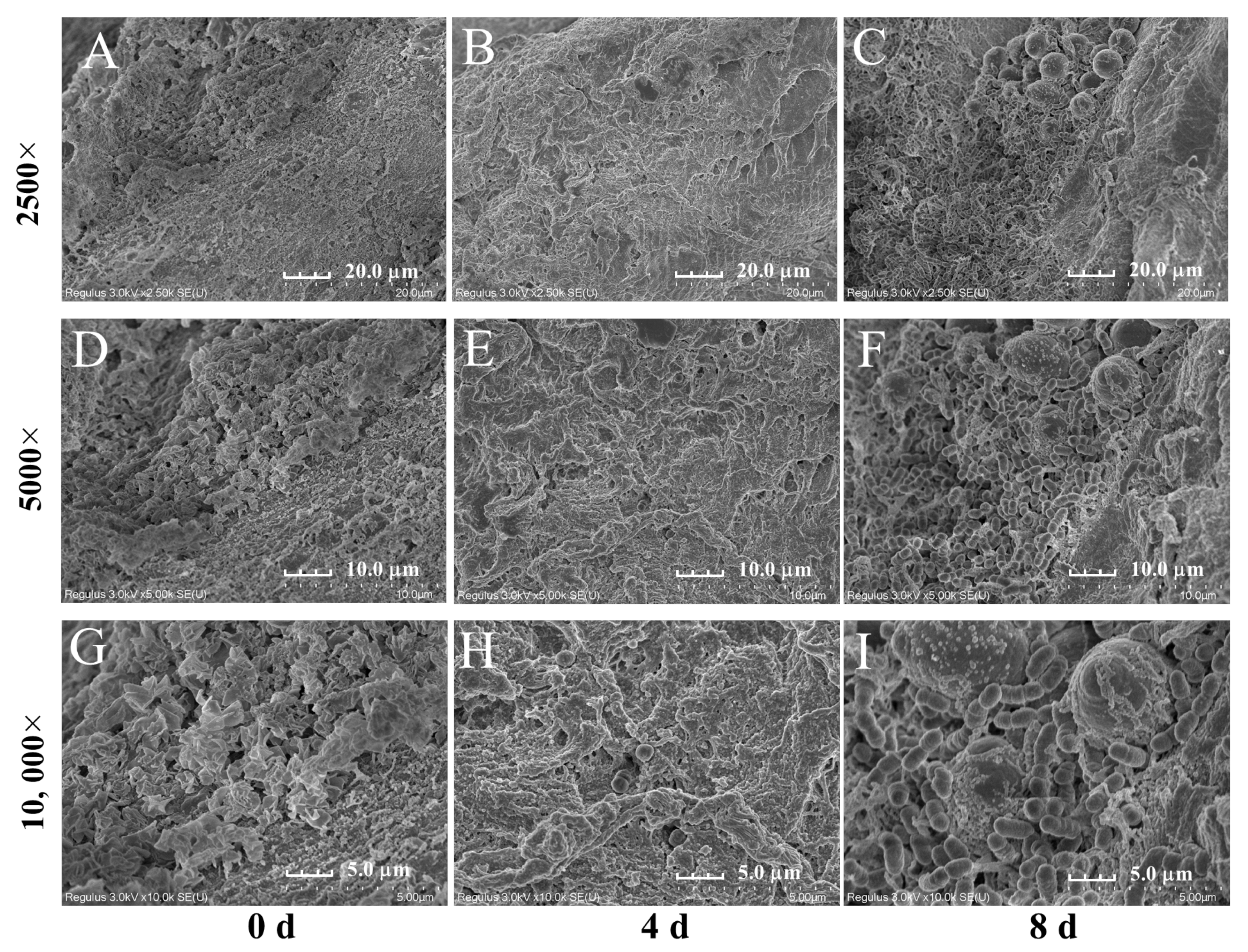
2.2. Observation of Vinasse Hairtails with the Scanning Electron Microscope (SEM)
2.3. Texture Profile Analysis and Color Changes
2.4. Physicochemical Determination
2.5. DNA Extraction, PCR Amplification, and Sequencing
2.6. Data Processing and Statistical Analysis
3. Results and Discussion
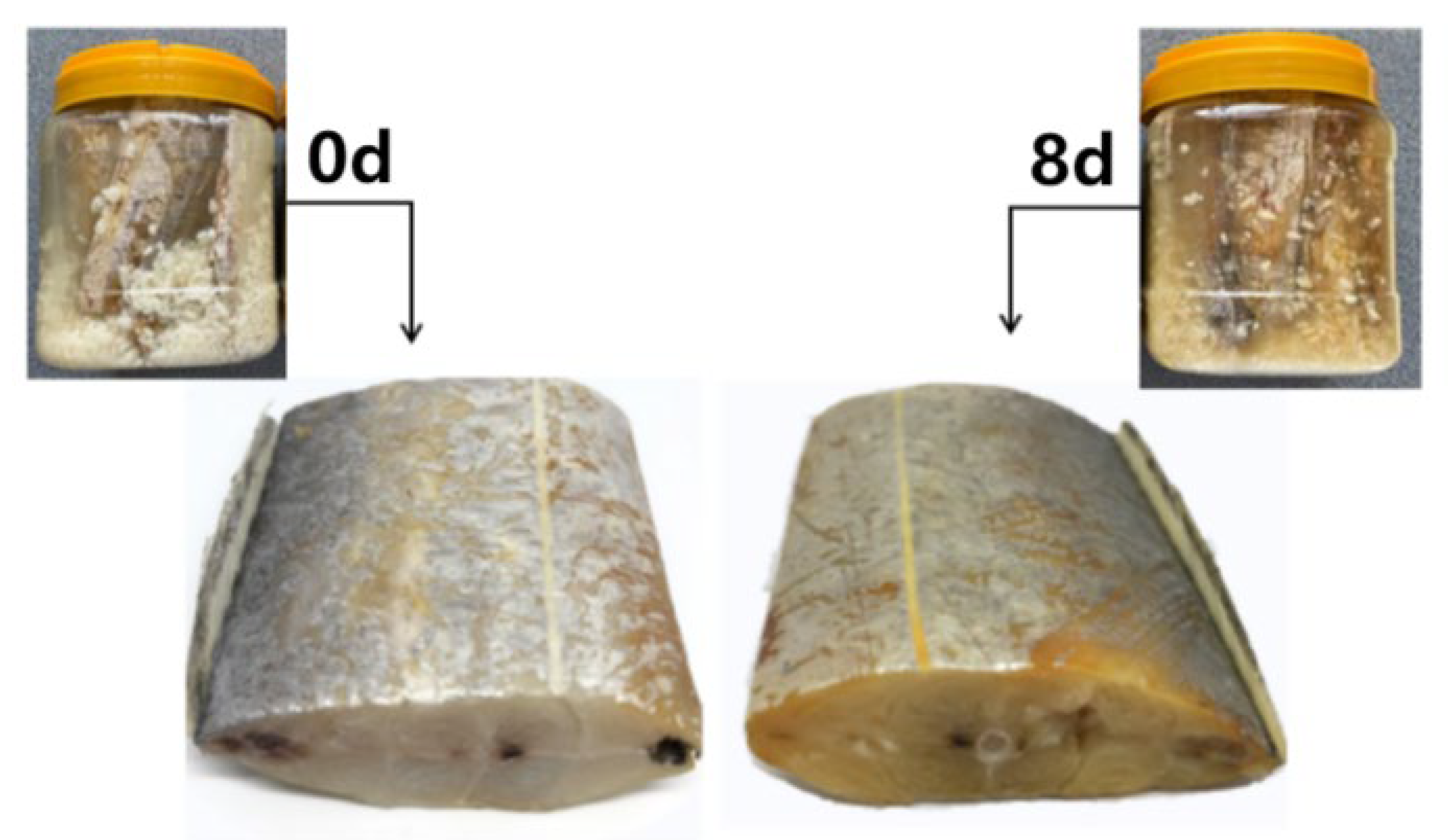
3.1. Macroscopic and Microscopic Properties of Vinasse Hairtail
3.2. Texture Profile Analysis and Color Changes
3.3. Physicochemical Properties of Vinasse Hairtail at Different Stages of Fermentation
3.4. High-Throughput Metagenomic and Alpha Diversity Indexes Analysis of Vinasse Hairtail Microbial Community
3.4.1. Alpha Diversity Indexes
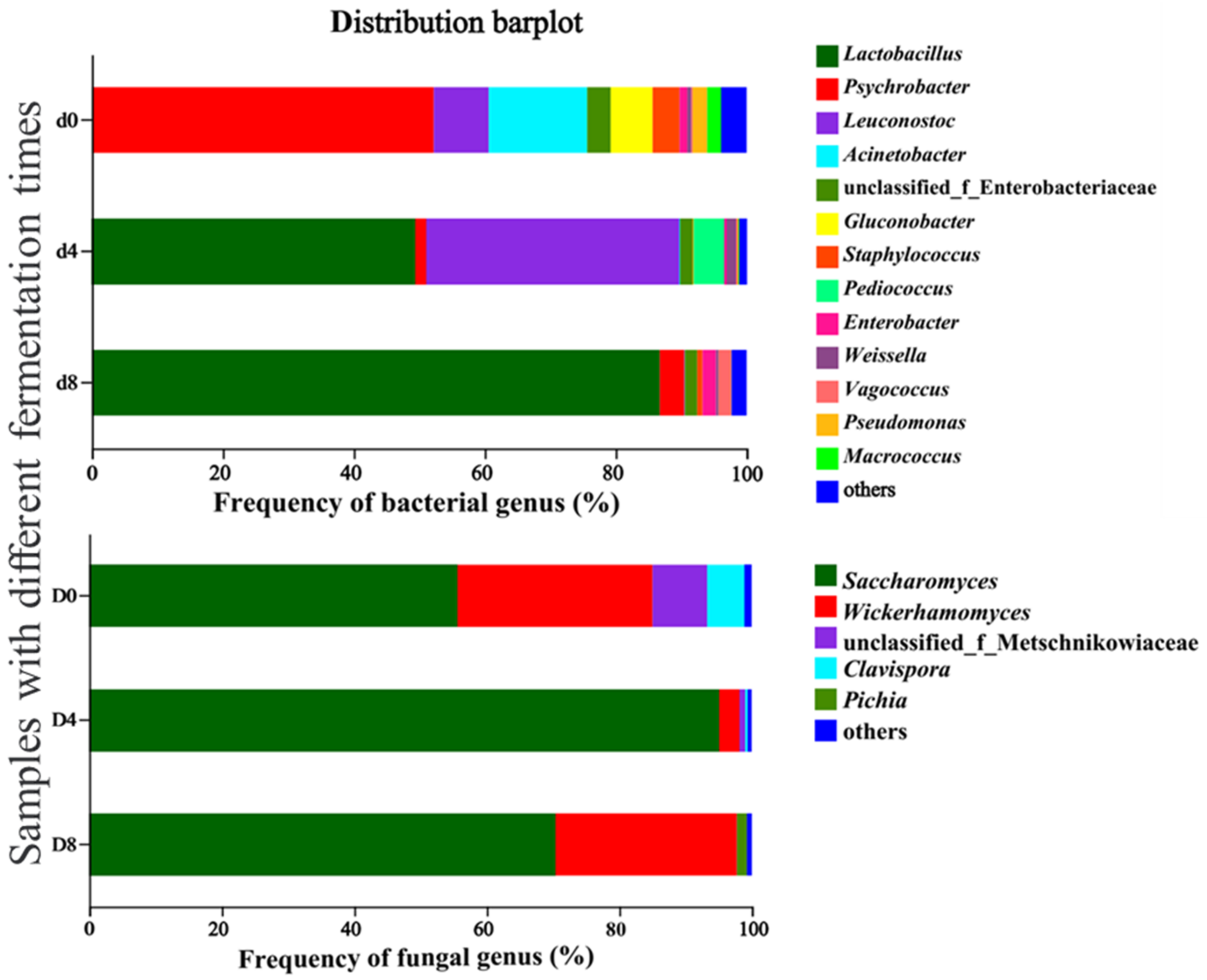
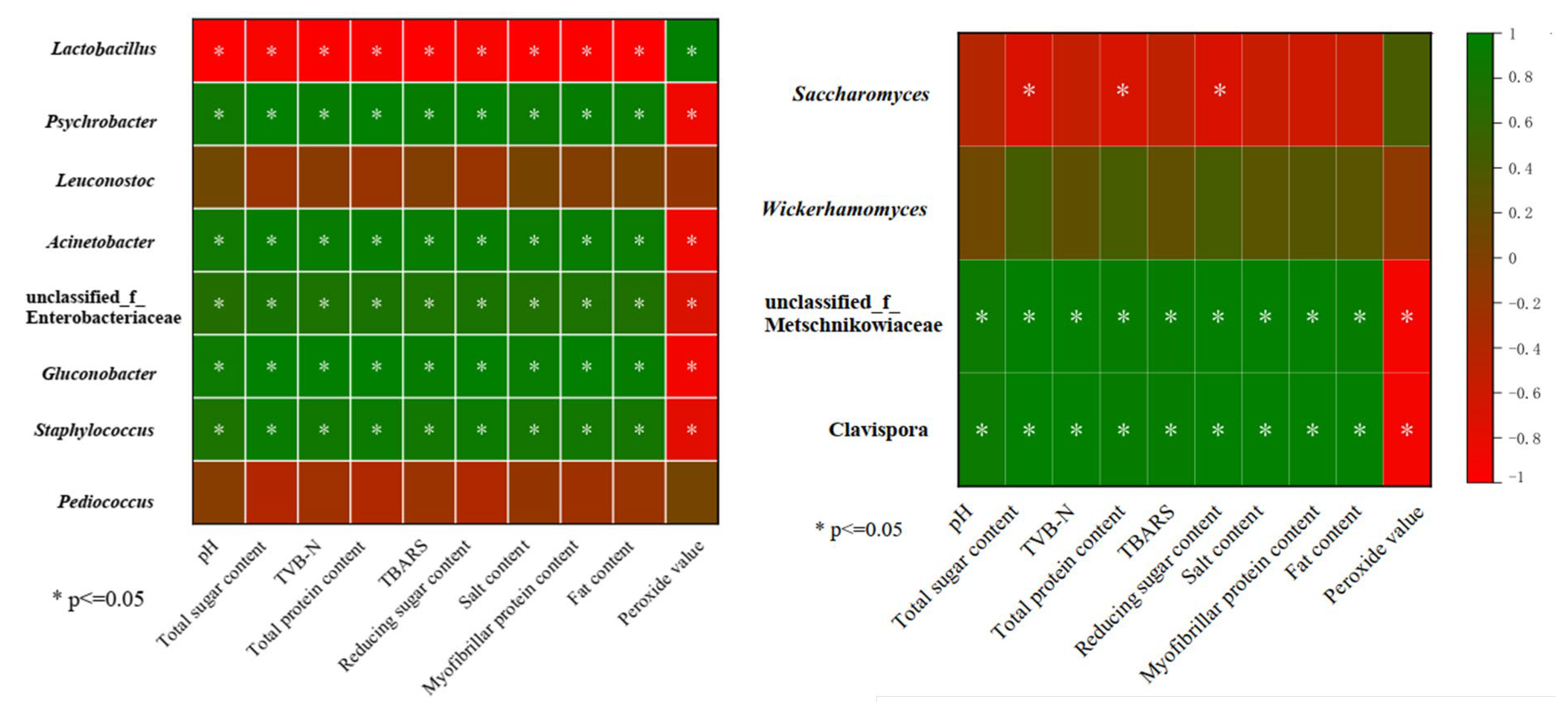
3.4.2. Bacterial and Fungal Community Dynamics during Vinasse Hairtail Fermentation
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligné, B.; Gänzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Marti-Quijal, F.J.; Remize, F.; Meca, G.; Ferrer, E.; Ruiz, M.J.; Barba, F.J. Fermentation in fish and by-products processing: An overview of current research and future prospects. Curr. Opin. Food Sci. 2020, 31, 9–16. [Google Scholar] [CrossRef]
- Wolfe, B.E.; Button, J.E.; Santarelli, M.; Dutton, R.J. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell 2014, 158, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.M.; Lu, Z.M.; Shi, J.S.; Xu, Z.H. Exploring flavour-producing core microbiota in multispecies solid-state fermentation of traditional Chinese vinegar. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Song, Z.; Du, H.; Zhang, Y.; Xu, Y. Unraveling core functional microbiota in traditional solid-state fermentation by high-throughput amplicons and metatranscriptomics sequencing. Front. Microbiol. 2017, 8, 1294. [Google Scholar] [CrossRef]
- Huang, Z.R.; Hong, J.L.; Xu, J.X.; Li, L.; Guo, W.L.; Pan, Y.Y.; Chen, S.J.; Bai, W.D.; Rao, P.F.; Ni, L.; et al. Exploring core functional microbiota responsible for the production of volatile flavour during the traditional brewing of Wuyi Hong Qu glutinous rice wine. Int. J. Syst. Evol. Microbiol. 2018, 76, 487–496. [Google Scholar] [CrossRef]
- Aldrete-Tapia, A.; Escobar-Ramírez, M.C.; Tamplin, M.L.; Hernández-Iturriaga, M. High-throughput sequencing of microbial communities in Poro cheese, an artisanal Mexican cheese. Int. J. Food Microbiol. 2014, 44, 136–141. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, X.; Ding, Y.; Ke, Z.; Zhou, X.; Zhang, J. Diversity and succession of the microbial community and its correlation with lipid oxidation in dry-cured black carp (Mylopharyngodon piceus) during storage. Int. J. Food Microbiol. 2021, 98, 103686. [Google Scholar] [CrossRef]
- Zhao, D.; Hu, J.; Chen, W. Analysis of the relationship between microorganisms and flavour development in dry-cured grass carp by high-throughput sequencing, volatile flavour analysis and metabolomics. Food Chem. 2022, 368, 130889. [Google Scholar] [CrossRef]
- Ding, Y.; Zhou, T.; Liao, Y.; Lin, H.; Deng, S.; Zhang, B. Comparative studies on the physicochemical and volatile flavour properties of traditional deep fried and circulating-air fried hairtail (Trichiurus lepturus). Foods 2022, 11, 2710. [Google Scholar] [CrossRef]
- GB 5009.6-2016; Determination of Fat in Foods. China Standard Press: Beijing, China, 2016.
- GB 5009.228-2016; Determination of Volatile Base Nitrogen in Foods. China Standard Press: Beijing, China, 2016.
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Xiong, Y.L. Role of myofibrillar proteins in water-binding in brine-enhanced meats. Food Res. Int. 2005, 38, 281–287. [Google Scholar] [CrossRef]
- Thorarinsdottir, K.A.; Arason, S.; Sigurgisladottir, S.; Valsdottir, T.; Tornberg, E. Effects of different pre-salting methods on protein aggregation during heavy salting of cod fillets. Food Chem. 2011, 124, 7–14. [Google Scholar] [CrossRef]
- Erickson, M.C. Lipid Oxidation of Muscle Foods. In Food Lipids: Chemistry, Nutrition, and Biotechnology, 2nd ed.; Akoh, C.C., Min, D.B., Eds.; Marcel Dekker: New York, NY, USA, 2002; pp. 365–411. [Google Scholar]
- Murata, M. Browning and pigmentation in food through the Maillard reaction. Glycoconj. J. 2021, 38, 283–292. [Google Scholar] [CrossRef]
- Felisiak, K.; Szymczak, M. Use of rapid capillary zone electrophoresis to determine amino acids indicators of herring ripening during salting. Foods 2021, 10, 2518. [Google Scholar] [CrossRef]
- Lv, X.C.; Huang, X.L.; Zhang, W.; Rao, P.F.; Ni, L. Yeast diversity of traditional alcohol fermentation starters for Hong Qu glutinous rice wine brewing, revealed by culture-dependent and culture-independent methods. Food Control 2013, 34, 183–190. [Google Scholar] [CrossRef]
- Ge, Q.; Pei, H.; Liu, R.; Chen, L.; Gao, X.; Gu, Y.; Hou, Q.; Yin, Y.; Yu, H.; Wu, M.; et al. Effects of Lactobacillus plantarum NJAU-01 from Jinhua ham on the quality of dry-cured fermented sausage. LWT—Food Sci. Technol. 2019, 101, 513–518. [Google Scholar] [CrossRef]
- Xu, Y.; Li, L.; Mac Regenstein, J.; Gao, P.; Zang, J.; Xia, W.; Jiang, Q. The contribution of autochthonous microflora on free fatty acids release and flavor development in low-salt fermented fish. Food Chem. 2018, 256, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Ya-Ting, J.; Jin-Xuan, C.; Yin-Ji, C.; Yang-Ying, S.; Xiao-Qun, Z.; Dao-Dong, P.; Chang-Rong, O.; Ning, G. Study on lipolysis-oxidation and volatile flavour compounds of dry-cured goose with different curing salt content during production. Food Chem. 2016, 190, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Le, T.; Takahashi, K.; Okazaki, E.; Osako, K. Mitigation of lipid oxidation in tuna oil using gelatin pouches derived from horse mackerel (Trachurus japonicus) scales and incorporating phenolic compounds. LWT—Food Sci. Technol. 2020, 128, 109533. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, S.; Lv, J.; Sun, Z.; Xu, W.; Ji, C.; Liang, H.; Li, S.; Yu, C.; Lin, X. Microbial succession and the changes of flavor and aroma in Chouguiyu, a traditional Chinese fermented fish. Food Biosci. 2020, 37, 100725. [Google Scholar] [CrossRef]
- Visessanguan, W.; Benjakul, S.; Riebroy, S.; Yarchai, M.; Tapingkae, W. Changes in lipid composition and fatty acid profile of Nham, a Thai fermented pork sausage, during fermentation. Food Chem. 2006, 94, 580–588. [Google Scholar] [CrossRef]
- Zang, J.; Xu, Y.; Xia, W.; Regenstein, J.M. Quality, functionality, and microbiology of fermented fish: A review. Crit. Rev. Food Sci. Nutr. 2020, 60, 1228–1242. [Google Scholar] [CrossRef]
- Yang, F.; Xia, W.S.; Zhang, X.W.; Xu, Y.S.; Jiang, Q.X. A comparison of endogenous and microbial proteolytic activities during fast fermentation of silver carp inoculated with Lactobacillus plantarum. Food Chem. 2016, 207, 86–92. [Google Scholar] [CrossRef]
- Candogan, K.; Acton, J.C. Proteolytic activity of bacterial starter cultures for meat fermentation. J. Muscle Foods 2004, 15, 23–34. [Google Scholar] [CrossRef]
- Yongsawatdigul, J.; Rodtong, S.; Raksakulthai, N. Acceleration of Thai fish sauce fermentation using proteinases and bacterial starter cultures. J. Food Sci. 2007, 72, M382–M390. [Google Scholar] [CrossRef]
- Kasankala, L.M.; Xiong, Y.L.; Chen, J. Enzymatic activity and flavor compound production in fermented silver carp fish paste inoculated with douchi starter culture. J. Agric. Food Chem. 2012, 60, 226–233. [Google Scholar] [CrossRef]
- Chéret, R.; Delbarre-Ladrat, C.; de Lamballerie-Anton, M.; Verrez-Bagnis, V. Calpain and cathepsin activities in post mortem fish and meat muscles. Food Chem. 2007, 101, 1474–1479. [Google Scholar] [CrossRef]
- Tingman, W.; Jian, Z.; Xiaoshuan, Z. Fish product quality evaluation based on temperature monitoring in cold chain. Afr. J. Biotechnol. 2010, 9, 6146–6151. [Google Scholar]
- Kaale, L.D.; Eikevik, T.M. Changes of proteins during superchilled storage of Atlantic salmon muscle (Salmo salar). J. Food Sci. Technol. 2016, 53, 441–450. [Google Scholar] [CrossRef]
- Bekhit, A.E.D.A.; Holman, B.W.; Giteru, S.G.; Hopkins, D.L. Total volatile basic nitrogen (TVB-N) and its role in meat spoilage: A review. Trends Food Sci. Tech. 2021, 109, 280–302. [Google Scholar] [CrossRef]
- Kritikos, A.; Aska, I.; Ekonomou, S.; Mallouchos, A.; Parlapani, F.F.; Haroutounian, S.A.; Boziaris, I.S. Volatilome of chill-stored european seabass (Dicentrarchus labrax) fillets and atlantic salmon (Salmo salar) slices under modified atmosphere packaging. Molecules 2020, 25, 1981. [Google Scholar] [CrossRef]
- Soto del Rio, M.D.; Dalmasso, A.; Civera, T.; Bottero, M.T. Characterization of bacterial communities of donkey milk by high-throughput sequencing. Int. J. Food Microbiol. 2017, 251, 67–72. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Z.; Ji, L.; Zhang, J.; Zhao, Z.; Zhang, R.; Bai, T.; Hou, B.; Zhang, Y.; Liu, D.; et al. A review: Microbial diversity and function of fermented meat products in China. Front. Microbiol. 2021, 12, 645435. [Google Scholar] [CrossRef]
- Chen, L.; Li, D.; Ren, L.; Song, S.; Ma, X.; Rong, Y. Effects of simultaneous and sequential cofermentation of Wickerhamomyces anomalus and Saccharomyces cerevisiae on physicochemical and flavor properties of rice wine. Food Sci. Nutr. 2021, 9, 71–86. [Google Scholar] [CrossRef]
- Wang, X.H.; Ren, H.Y.; Liu, D.Y.; Zhu, W.Y.; Wang, W. Effects of inoculating Lactobacillus sakei starter cultures on the microbiological quality and nitrite depletion of Chinese fermented sausages. Food Control 2013, 32, 591–596. [Google Scholar] [CrossRef]
- Bianchi, F.; Dall’Asta, M.; Del Rio, D.; Mangia, A.; Musci, M.; Scazzina, F. Development of a headspace solid-phase microextraction gas chromatography–mass spectrometric method for the determination of short-chain fatty acids from intestinal fermentation. Food Chem. 2011, 129, 200–205. [Google Scholar] [CrossRef]
- Liu, S.P.; Yu, J.X.; Wei, X.L.; Ji, Z.W.; Zhou, Z.L.; Meng, X.Y.; Mao, J. Sequencing-based screening of functional microorganism to decrease the formation of biogenic amines in Chinese rice wine. Food Control 2016, 64, 98–104. [Google Scholar] [CrossRef]
- Zhang, X.M.; Dang, X.J.; Wang, Y.B.; Sun, T.; Wang, Y.; Yu, H.; Yang, W.S. Diversity and composition of microbiota during fermentation of traditional Nuodeng ham. J. Microbiol. 2021, 59, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.R.; Liao, H.; Zhao, H.J.; Zhang, Z.D.; Guo, Z. Evaluation of the bacterial diversity in cured fish samples collected from Enshi by PCRDGGE and MiSeq high-throughput sequencing. Mod. Food Sci. Technol. 2018, 34, 208–213. [Google Scholar]
- Mu, Y.; Su, W.; Mu, Y.; Jiang, L. Combined application of high-throughput sequencing and metabolomics reveals metabolically active microorganisms during Panxian ham processing. Front. Microbiol. 2020, 10, 3012. [Google Scholar] [CrossRef]
- Helinck, S.; Perello, M.C.; Deetae, P.; De Revel, G.; Spinnler, H.E. Debaryomyces hansenii, Proteus vulgaris, Psychrobacter sp. and Microbacterium foliorum are able to produce biogenic amines. Dairy Sci. Technol. 2013, 93, 191–200. [Google Scholar] [CrossRef]
- Kim, M.K.; Mah, J.H.; Hwang, H.J. Biogenic amine formation and bacterial contribution in fish, squid and shellfish. Food Chem. 2009, 116, 87–95. [Google Scholar] [CrossRef]
- Park, M.K.; Kim, Y.S. Comparative metabolic expressions of fermented soybeans according to different microbial starters. Food Chem. 2020, 305, 125461. [Google Scholar] [CrossRef]
- Lv, X.C.; Chen, Z.C.; Jia, R.B.; Liu, Z.B.; Zhang, W.; Chen, S.J.; Rao, P.F.; Ni, L. Microbial community structure and dynamics during the traditional brewing of Fuzhou Hong Qu glutinous rice wine as determined by culture-dependent and culture-independent techniques. Food Control 2015, 57, 216–224. [Google Scholar] [CrossRef]

| 0 d | 4 d | 8 d | |
|---|---|---|---|
| Hardness (N) | 8903.77 ± 18.90 c | 5824.86 ± 30.25 b | 5110.09 ± 84.13 a |
| Sprigness (mm) | 0.76 ± 0.09 a | 0.79 ± 0.08 a | 0.82 ± 0.05 a |
| Cohesiveness | 0.88 ± 0.06 a | 0.72 ± 0.08 a | 0.58 ± 0.05 a |
| Chewiness (N) | 4994 ± 98.90 b | 2661.73 ± 112.79 a | 2577.47 ± 81.94 a |
| Shearing force (N) | 21.75 ± 0.91 c | 14.73 ± 1.58 b | 9.11 ± 0.51 a |
| L* | 60.65 ± 0.73 a | 50.58 ± 1.19 b | 48.18 ± 1.37 b |
| a* | −1.82 ± 1.18 b | 2.83 ± 1.05 a | 2.91 ± 0.67 a |
| b* | 10.14 ± 3.43 b | 17.54 ± 1.02 a | 18.89 ± 2.81 a |
| 0 d | 2 d | 4 d | 6 d | 8 d | |
|---|---|---|---|---|---|
| pH | 7.05 ± 0.03 e | 6.22 ± 0.01 d | 5.64 ± 0.05 c | 5.22 ± 0.02 b | 4.54 ± 0.04 a |
| Total sugar content (g/L) | 36.06 ± 1.14 d | 25.81 ± 0.60 c | 16.49 ± 0.63 b | 15.92 ± 0.46 b | 13.81 ± 0.22 a |
| Reducing sugar content (g/L) | 26.06 ± 0.59 e | 18.96 ± 0.83 d | 10.02 ± 0.01 c | 8.66 ± 0.13 b | 7.59 ± 0.12 a |
| Fat content (%) | 9.21 ± 0.18 e | 7.66 ± 0.11 d | 6.17 ± 0.04 c | 5.34 ± 0.14 b | 4.72 ± 0.16 a |
| Salt content (%) | 11.10 ± 0.30 e | 10.03 ± 0.01 d | 9.77 ± 0.02 c | 9.37 ± 0.05 b | 8.95 ± 0.05 a |
| Total protein content (mg/g) | 161.66 ± 0.47 d | 128.11 ± 0.89 c | 86.47 ± 1.02 b | 74.83 ± 0.84 a | 74.65 ± 0.35 a |
| Myofibrillar protein content (mg/g) | 79.52 ± 0.33 d | 67.78 ± 1.36 c | 57.85 ± 0.09 b | 51.58 ± 1.85 a | 49.37 ± 0.91 a |
| TVB-N (mg/100g) | 38.65 ± 0.38 e | 26.96 ± 0.51 d | 20.61 ± 0.50 c | 15.78 ± 0.74 b | 14.40 ± 0.30 a |
| TBARS (nmol/mg) | 7.67 ± 0.03 c | 4.73 ± 0.24 b | 4.64 ± 0.02 b | 3.35 ± 0.09 a | 3.34 ± 0.05 a |
| Peroxide value (g/100 g) | 0.07 ± 0.01 a | 0.09 ± 0.01 b | 0.21 ± 0.01 c | 0.29 ± 0.01 d | 0.33 ± 0.01 e |
| Samples | Sobs | Seq Num | OUT Num | Shannon Index | Simpson | ACE Index | Chao1 Index | Coverage | |
|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA | 0 d | 92 | 33,875 | 111 | 2.63 | 0.11 | 105.13 | 102.3 | 0.99 |
| 4 d | 63 | 33,348 | 79 | 1.53 | 0.33 | 76.64 | 72.81 | 0.99 | |
| 8 d | 103 | 36,890 | 138 | 2.01 | 0.25 | 118.67 | 123.58 | 0.99 | |
| ITS | 0 d | 31 | 56,393 | 36 | 1.18 | 0.41 | 35.24 | 35.28 | 0.99 |
| 4 d | 19 | 38,140 | 28 | 0.24 | 0.91 | 31.55 | 27 | 0.99 | |
| 8 d | 30 | 45,877 | 44 | 0.72 | 0.57 | 32.57 | 32.56 | 0.99 |
| Relative Abundances (%) | ||||
|---|---|---|---|---|
| Bacteria | Phylum | 0 d | 4 d | 8 d |
| Firmicutes | 15.76% | 95.36% | 91.52% | |
| Proteobacteria | 82.81% | 4.55% | 8.19% | |
| Actinobacteria | 1.23% | 0.07% | 0.23% | |
| others | 0.19% | 0.02% | 0.06% | |
| Family | ||||
| Lactobacillaceae | 0.04% | 54.17% | 86.73% | |
| Enterobacteriaceae | 4.99% | 2.30% | 3.94% | |
| Moraxellaceae | 67.32% | 1.94% | 3.77% | |
| Vagococcaceae | 0.22% | 0.26% | 1.94% | |
| Staphylococcaceae | 6.19% | 0.13% | 0.84% | |
| Leuconostocaceae | 9.02% | 40.02% | 0.46% | |
| Halomonadaceae | 1.44% | 0.01% | 0.07% | |
| Pseudomonadaceae | 2.21% | 0.10% | 0.05% | |
| Acetobacteraceae | 6.49% | 0.06% | 0.00% | |
| Others | 2.08% | 1.01% | 2.20% | |
| Fungi | Phylum | 0 d | 4 d | 8 d |
| Ascomycota | 99.90% | 99.97% | 99.78% | |
| others | 0.10% | 0.03% | 0.22% | |
| Family | ||||
| Saccharomycetaceae | 55.85% | 95.08% | 70.64% | |
| Phaffomycetaceae | 29.59% | 3.14% | 27.37% | |
| Pichiaceae | 0.00% | 0.00% | 1.50% | |
| Metschnikowiaceae | 14.18% | 1.73% | 0.04% | |
| Others | 0.38% | 0.05% | 0.45% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Tu, C.; Lin, H.; Hu, Y.; Jia, J.; Shui, S.; Wang, J.; Hu, Y.; Zhang, B. Changes in Physicochemical Characteristics and Microbial Diversity of Traditional Fermented Vinasse Hairtail. Fermentation 2023, 9, 173. https://doi.org/10.3390/fermentation9020173
Zhang Y, Tu C, Lin H, Hu Y, Jia J, Shui S, Wang J, Hu Y, Zhang B. Changes in Physicochemical Characteristics and Microbial Diversity of Traditional Fermented Vinasse Hairtail. Fermentation. 2023; 9(2):173. https://doi.org/10.3390/fermentation9020173
Chicago/Turabian StyleZhang, Yue, Chuanhai Tu, Huimin Lin, Yuwei Hu, Junqi Jia, Shanshan Shui, Jiaxing Wang, Yi Hu, and Bin Zhang. 2023. "Changes in Physicochemical Characteristics and Microbial Diversity of Traditional Fermented Vinasse Hairtail" Fermentation 9, no. 2: 173. https://doi.org/10.3390/fermentation9020173
APA StyleZhang, Y., Tu, C., Lin, H., Hu, Y., Jia, J., Shui, S., Wang, J., Hu, Y., & Zhang, B. (2023). Changes in Physicochemical Characteristics and Microbial Diversity of Traditional Fermented Vinasse Hairtail. Fermentation, 9(2), 173. https://doi.org/10.3390/fermentation9020173






