- Review
Diabetic Retinopathy Therapeutics: Bridging Conventional Approaches and Gene Therapy with Focus on TXNIP-Targeted Interventions
- Riddhi Tiwari,
- Archana Tiwari and
- Lalit P. Singh
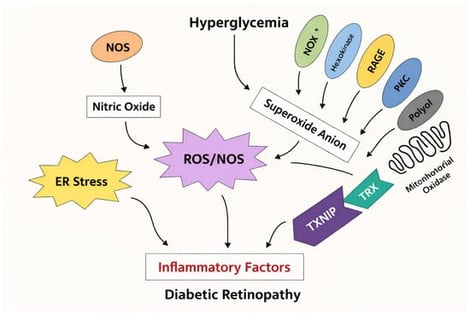
Diabetic retinopathy (DR) is a progressive retinal disorder and a leading cause of vision impairment worldwide affecting the livelihood of millions. Its pathogenesis is driven by chronic hyperglycemia-induced neuronal and microvascular injury, leading to capillary occlusion, increased vascular permeability, and the eventual formation of fragile neo vessels. These changes mark the progression from non-proliferative diabetic retinopathy (NPDR) to proliferative diabetic retinopathy (PDR). Diabetic macular edema (DME), characterized by blood–retinal barrier disruption and macular fluid accumulation, further contributes to vision loss. This review provides an integrative perspective on the cellular and molecular mechanisms of DR, highlighting both vascular and neuroglial contributions to retinal pathology. Current therapeutic approaches, including anti-VEGF agents and corticosteroids, offer symptomatic relief but are limited by the need for repeated administration and variability in patient response. Emerging evidence implicates the role of thioredoxin-interacting protein (TXNIP) as one of mediators of the disease progression. Strongly upregulated under hyperglycaemic stress, TXNIP induces oxidative damage, inflammation, and neuronal apoptosis, exacerbating neurovascular dysfunction. We explore potential therapeutic strategies such as gene therapy, TXNIP-targeted molecular interventions, and stem cell-based approaches aimed at achieving long-term modulation of disease mechanisms. This article thus attempts to address a comprehensive understanding of DR pathophysiology and innovative new strategies to improve long-term visual outcomes.
6 February 2026