Response of Soil Fungal-Community Structure to Crop-Tree Thinning in Pinus massoniana Plantation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Sampling Design
2.2. Soil Sampling and Survey of Vegetation
2.3. Analysis of Soil Physical and Chemical Properties
2.4. DNA Extraction, PCR Amplification, and ITS Sequencing
2.5. Statistical Analyses
3. Results
3.1. Response of Understory Plant Diversity and Soil Physicochemical Properties to Crop-Tree Thinning
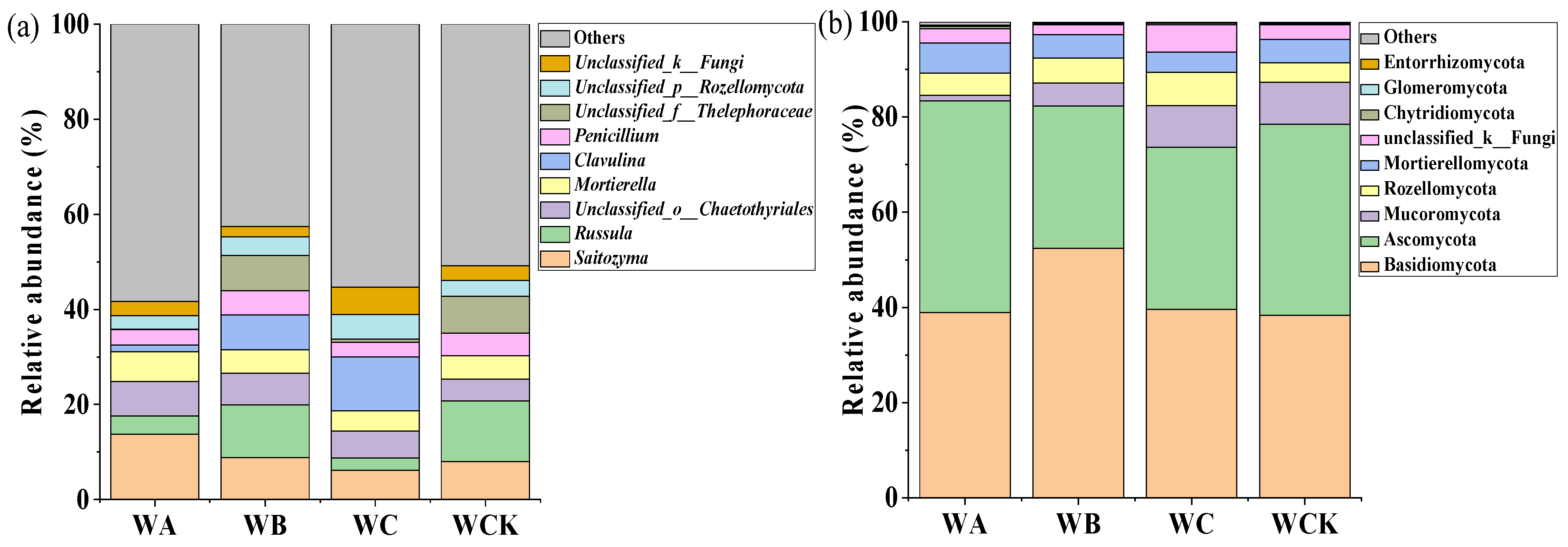
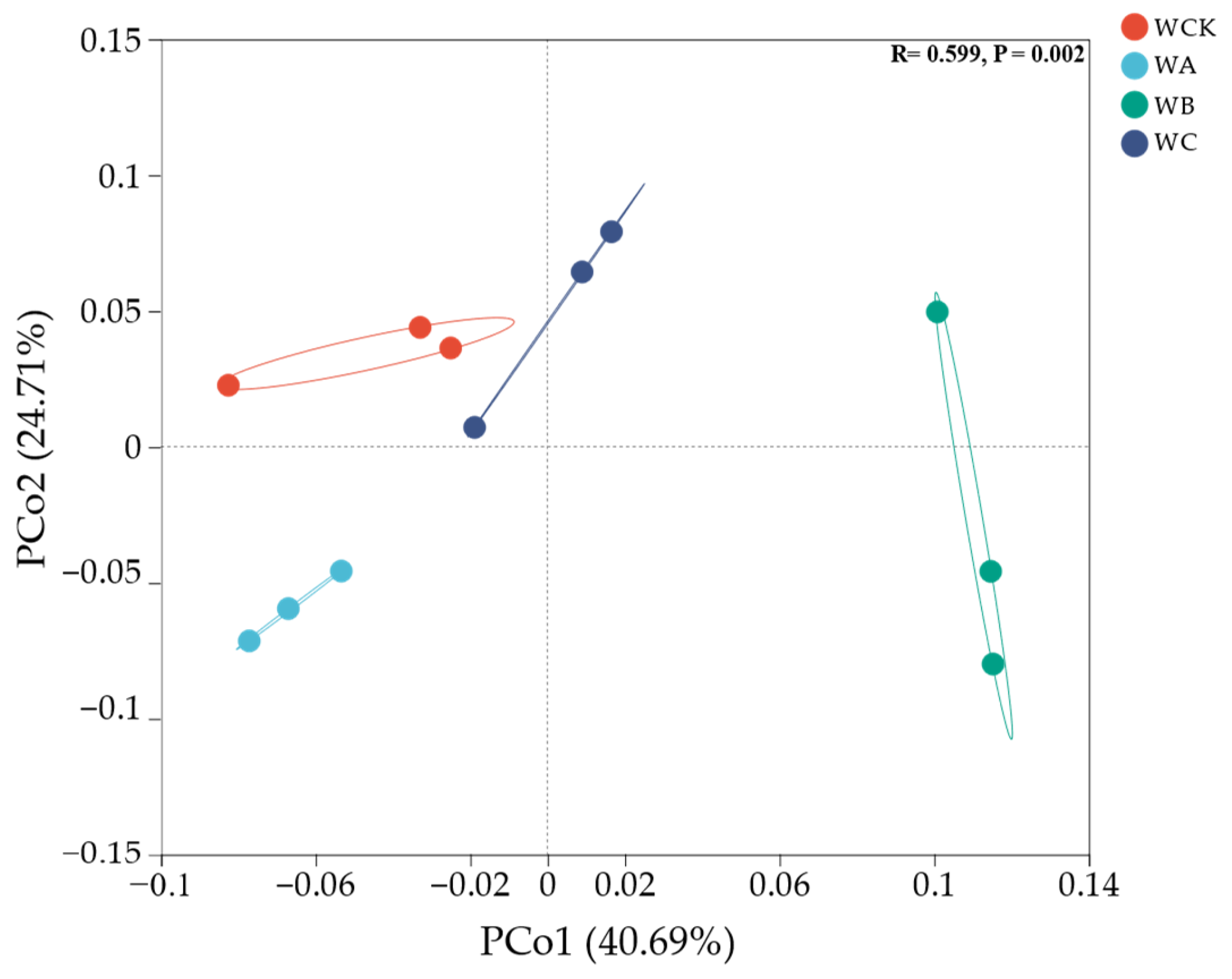
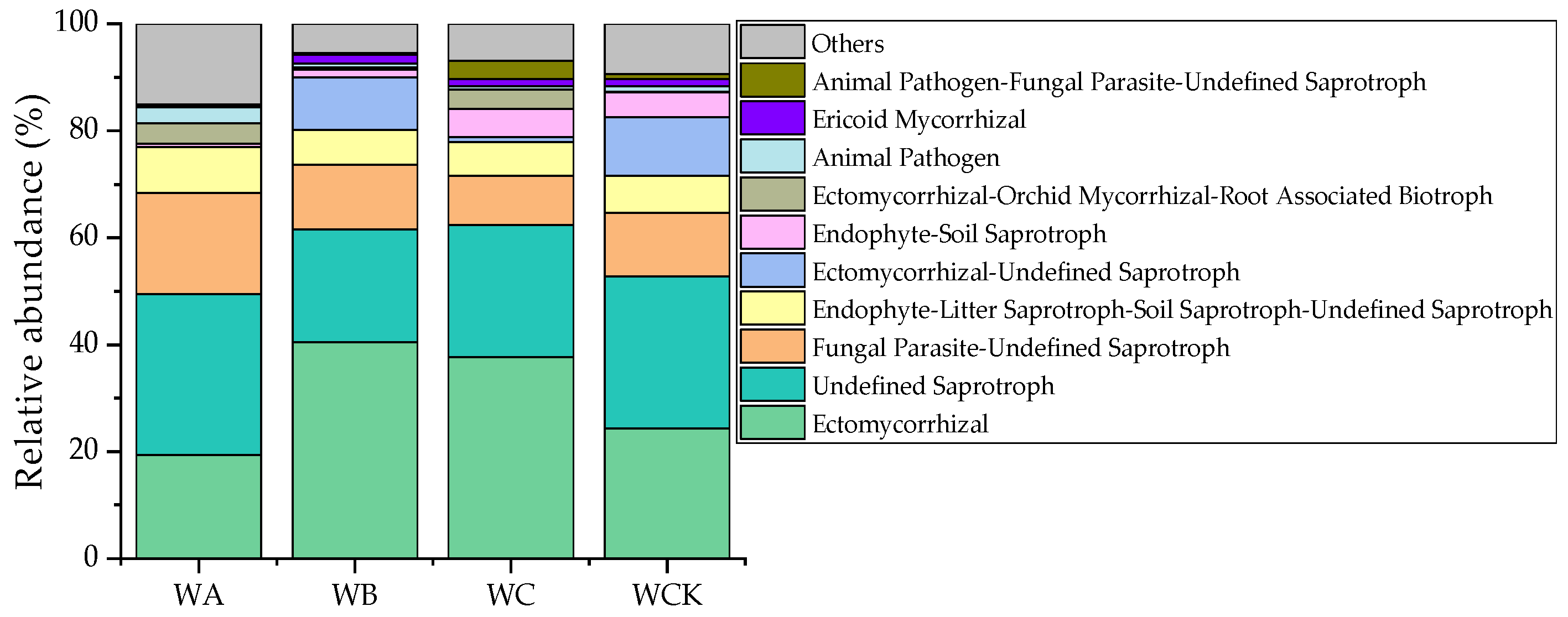
3.2. Soil Fungal-Community Composition, Diversity, and Functional Groups
3.3. Network of Soil Fungal Community
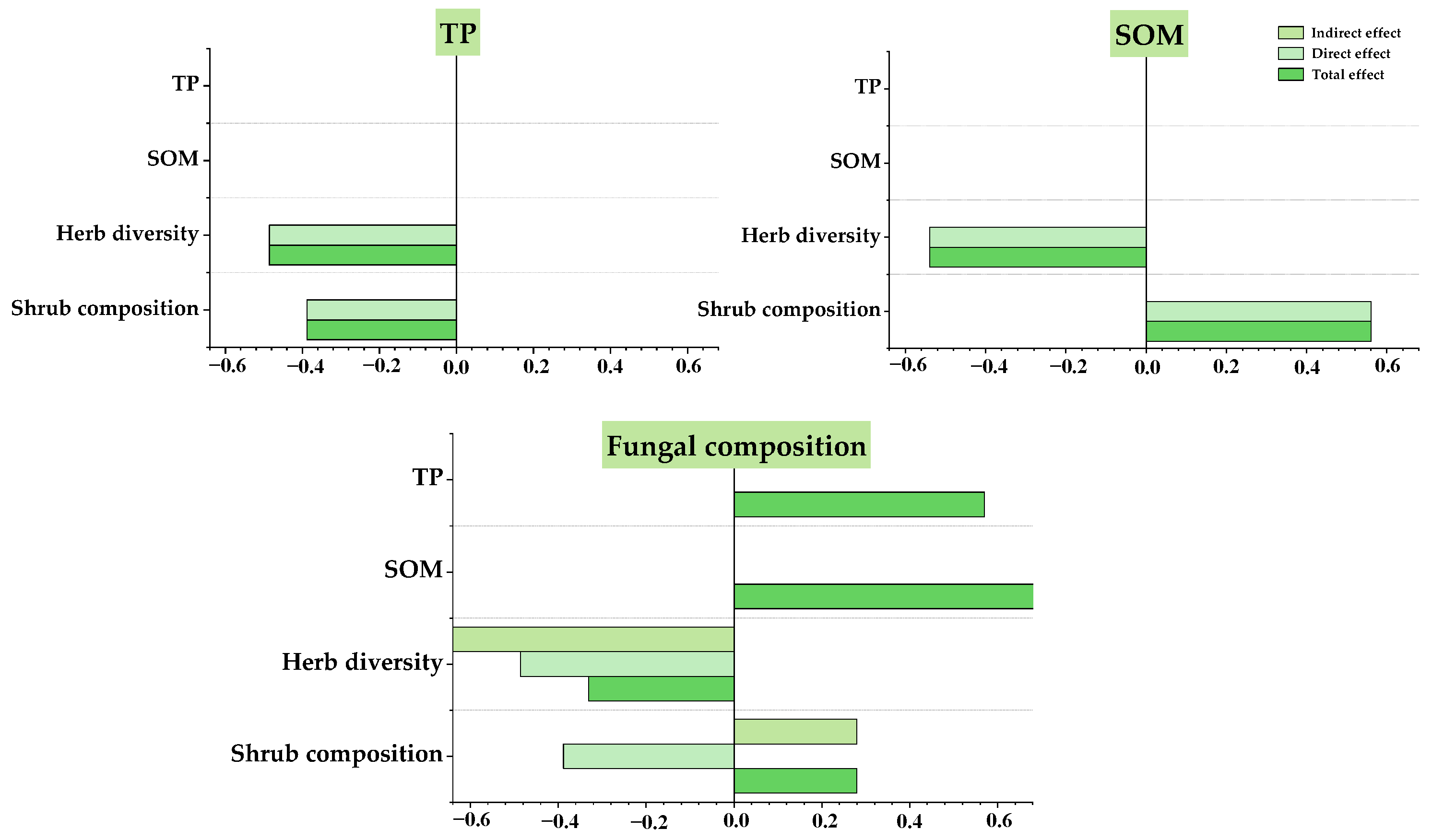
3.4. Relationship between Understory Vegetation, Soil Physicochemical Properties, and Fungal Communities
4. Discussion
4.1. Responses of Understory Plant Diversity to Crop-Tree Thinning
4.2. Responses of Soil Fungal-Community Structure to Crop-Tree Thinning
4.3. Relationships between Plants, Soil Factors, and Fungal Community
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ding, X.; Liu, G.; Fu, S.; Chen, H.Y.H. Tree Species Composition and Nutrient Availability Affect Soil Microbial Diversity and Composition across Forest Types in Subtropical China. Catena 2021, 201, 105224. [Google Scholar] [CrossRef]
- Zhou, L.; Cai, L.; He, Z.; Wang, R.; Wu, P.; Ma, X. Thinning Increases Understory Diversity and Biomass, and Improves Soil Properties without Decreasing Growth of Chinese Fir in Southern China. Environ. Sci. Pollut. Res. 2016, 23, 24135–24150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hao, M.; Yu, Q.; Dun, X.; Xu, J.; Gao, P. The Effect of Thinning Intensity on the Soil Carbon Pool Mediated by Soil Microbial Communities and Necromass Carbon in Coastal Zone Protected Forests. Sci. Total Environ. 2023, 881, 163492. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dong, Z.; Chen, D.; Zhao, S.; Zhou, F.; Cao, X.; Fang, K. Growth Decline of Pinus Massoniana in Response to Warming Induced Drought and Increasing Intrinsic Water Use Efficiency in Humid Subtropical China. Dendrochronologia 2019, 57, 125609. [Google Scholar] [CrossRef]
- Ward, J.S. Twenty-Five Year Response of Non-Crop Trees to Partial Release during Precommercial Crop Tree Management. For. Ecol. Manag. 2017, 387, 12–18. [Google Scholar] [CrossRef]
- Idol, T.W.; Morales, R.M.; Friday, J.B.; Scowcroft, P.G. Precommercial Release Thinning of Potential Acacia Koa Crop Trees Increases Stem and Crown Growth in Dense, 8-Year-Old Stands in Hawaii. For. Ecol. Manag. 2017, 392, 105–114. [Google Scholar] [CrossRef]
- Lyu, Q.; Shen, Y.; Li, X.; Chen, G.; Li, D.; Fan, C. Early Effects of Crop Tree Management on Undergrowth Plant Diversity and Soil Physicochemical Properties in a Pinus Massoniana Plantation. PeerJ 2021, 9, e11852. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial Diversity Drives Multifunctionality in Terrestrial Ecosystems. Nat. Commun. 2016, 7, 10541. [Google Scholar] [CrossRef] [PubMed]
- Hooper, D.U.; Bignell, D.E.; Brown, V.K.; Brussaard, L.; Dangerfield, J.M.; Wall, D.H.; Wardle, D.A.; Coleman, D.C.; Giller, K.E.; Lavelle, P.; et al. Interactions between Aboveground and Belowground Biodiversity in Terrestrial Ecosystems: Patterns, Mechanisms, and Feedbacks: We Assess the Evidence for Correlation between Aboveground and Belowground Diversity and Conclude That a Variety of Mechanisms Co. Bioscience 2000, 50, 1049–1061. [Google Scholar] [CrossRef]
- Zheng, J.; Mao, X.; Jan van Groenigen, K.; Zhang, S.; Wang, M.; Guo, X.; Yu, W.; Luo, L.; Chang, J.; Shi, Z.; et al. Decoupling of Soil Carbon Mineralization and Microbial Community Composition across a Climate Gradient on the Tibetan Plateau. Geoderma 2024, 441, 116736. [Google Scholar] [CrossRef]
- Mukhtar, S.; Mirza, B.S.; Mehnaz, S.; Mirza, M.S.; Mclean, J.; Malik, K.A. Impact of Soil Salinity on the Microbial Structure of Halophyte Rhizosphere Microbiome. World J. Microbiol. Biotechnol. 2018, 34, 136. [Google Scholar] [CrossRef] [PubMed]
- Gil-Martínez, M.; López-García, Á.; Domínguez, M.T.; Kjøller, R.; Navarro-Fernández, C.M.; Rosendahl, S.; Marañón, T. Soil Fungal Diversity and Functionality Are Driven by Plant Species Used in Phytoremediation. Soil Biol. Biochem. 2021, 153, 108102. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, J.; Hui, D.; Wang, Y.P.; Li, J.; Chen, J.; Chen, G.; Zhu, Y.; Zhang, L.; Zhang, D.; et al. Mycorrhizal Fungi Alleviate Acidification-Induced Phosphorus Limitation: Evidence from a Decade-Long Field Experiment of Simulated Acid Deposition in a Tropical Forest in South China. Glob. Chang. Biol. 2022, 28, 3605–3619. [Google Scholar] [CrossRef]
- Toju, H.; Peay, K.G.; Yamamichi, M.; Narisawa, K.; Hiruma, K.; Naito, K.; Fukuda, S.; Ushio, M.; Nakaoka, S.; Onoda, Y.; et al. Core Microbiomes for Sustainable Agroecosystems. Nat. Plants 2018, 4, 247–257. [Google Scholar] [CrossRef]
- Shi, Y.; Delgado-Baquerizo, M.; Li, Y.; Yang, Y.; Zhu, Y.G.; Peñuelas, J.; Chu, H. Abundance of Kinless Hubs within Soil Microbial Networks Are Associated with High Functional Potential in Agricultural Ecosystems. Environ. Int. 2020, 142, 105869. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, J.A. Microbial Community Structure and Its Functional Implications. Nature 2009, 459, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Shen, F.; Liu, Y.; Yang, Y.; Wang, J.; Purahong, W.; Yang, L. Contrasting Altitudinal Patterns and Co-Occurrence Networks of Soil Bacterial and Fungal Communities along Soil Depths in the Cold-Temperate Montane Forests of China. Catena 2022, 209, 105844. [Google Scholar] [CrossRef]
- De Vries, F.T.; Griffiths, R.I.; Bailey, M.; Craig, H.; Girlanda, M.; Gweon, H.S.; Hallin, S.; Kaisermann, A.; Keith, A.M.; Kretzschmar, M.; et al. Soil Bacterial Networks Are Less Stable under Drought than Fungal Networks. Nat. Commun. 2018, 9, 3033. [Google Scholar] [CrossRef] [PubMed]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The Ecology of the Microbiome: Networks, Competition, and Stability. Science (80-) 2015, 350, 663–666. [Google Scholar] [CrossRef]
- Neutel, A.M.; Heesterbeek, J.A.P.; De Ruiter, P.C. Stability in Real Food Webs: Weak Links in Long Loops. Science (80-) 2002, 296, 1120–1123. [Google Scholar] [CrossRef]
- Xiao, W.; Chen, C.; Chen, X.; Huang, Z.; Chen, H.Y.H. Functional and Phylogenetic Diversity Promote Litter Decomposition across Terrestrial Ecosystems. Glob. Ecol. Biogeogr. 2020, 29, 2261–2272. [Google Scholar] [CrossRef]
- Wan, X.; Huang, Z.; He, Z.; Yu, Z.; Wang, M.; Davis, M.R.; Yang, Y. Soil C:N Ratio Is the Major Determinant of Soil Microbial Community Structure in Subtropical Coniferous and Broadleaf Forest Plantations. Plant Soil 2015, 387, 103–116. [Google Scholar] [CrossRef]
- Qiang, W.; Gunina, A.; Kuzyakov, Y.; Luo, R.; Zhang, Y.; Liu, B.; Pang, X. Shifts of Understory Vegetation Induced by Thinning Drive the Expansion of Soil Rare Fungi. J. Environ. Manag. 2023, 342, 118119. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, Q.; Xie, L.; Yin, C. Interspecific Plant-Plant Interactions Increase the Soil Microbial Network Stability, Shift Keystone Microbial Taxa, and Enhance Their Functions in Mixed Stands. For. Ecol. Manag. 2023, 533, 120851. [Google Scholar] [CrossRef]
- Huang, L.; Pan, X.; Ma, J.; Jian, R.; Zhang, H.; Bai, K.; Mo, Y.; Zhang, Q.; Yang, Z. Assembly of Understory Woody Communities during the Close-to-Nature Restoration of a Pinus Massoniana Lamb. Plantation in the Southern Subtropical Region of China: From Environmental Filtering to Competitive Exclusion. For. Ecol. Manage. 2023, 541, 121060. [Google Scholar] [CrossRef]
- Yin, H.; Su, Y.; Liu, S.; Li, X.; Li, X.; Fan, C.; Guan, P.; Xie, Z.; Wang, S.; Scheu, S.; et al. Consistent Response of Nematode Communities to Management of Coniferous Plantations. For. Ecosyst. 2022, 9, 100045. [Google Scholar] [CrossRef]
- Ren, C.; Chen, J.; Deng, J.; Zhao, F.; Han, X.; Yang, G.; Tong, X.; Feng, Y.; Shelton, S.; Ren, G. Response of Microbial Diversity to C:N:P Stoichiometry in Fine Root and Microbial Biomass Following Afforestation. Biol. Fertil. Soils 2017, 53, 457–468. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, W.; Zhang, X.; Liu, Y.; Wang, S.; Liu, Y. Effects of Reforestation on Plant Species Diversity on the Loess Plateau of China: A Case Study in Danangou Catchment. Sci. Total Environ. 2019, 651, 979–989. [Google Scholar] [CrossRef] [PubMed]
- Pruden, G.; Kalembasa, S.J.; Jenkinson, D.S. Reduction of Nitrate Prior to Kjeldahl Digestion. J. Sci. Food Agric. 1985, 36, 71–73. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An Open Annotation Tool for Parsing Fungal Community Datasets by Ecological Guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Fernández-Guisuraga, J.M.; Ansola, G.; Pinto, R.; Marcos, E.; Calvo, L.; Sáenz de Miera, L.E. Resistance of Soil Bacterial Communities from Montane Heathland Ecosystems in the Cantabrian Mountains (NW Spain) to a Gradient of Experimental Nitrogen Deposition. Sci. Total Environ. 2024, 920, 171079. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Gui, H.; Breed, M.; Li, Y.; Xu, Q.; Yang, J.; Wanasinghe, D.N.; Li, Y.; Xu, J.; Mortimer, P. Continental-Scale Insights into the Soil Microbial Co-Occurrence Networks of Australia and Their Environmental Drivers. Soil Biol. Biochem. 2023, 186, 109177. [Google Scholar] [CrossRef]
- Jin, X.; Liu, Y.; Hu, W.; Wang, G.; Kong, Z.; Wu, L.; Ge, G. Soil Bacterial and Fungal Communities and the Associated Nutrient Cycling Responses to Forest Conversion after Selective Logging in a Subtropical Forest of China. For. Ecol. Manag. 2019, 444, 308–317. [Google Scholar] [CrossRef]
- Landuyt, D.; De Lombaerde, E.; Perring, M.P.; Hertzog, L.R.; Ampoorter, E.; Maes, S.L.; De Frenne, P.; Ma, S.; Proesmans, W.; Blondeel, H.; et al. The Functional Role of Temperate Forest Understorey Vegetation in a Changing World. Glob. Chang. Biol. 2019, 25, 3625–3641. [Google Scholar] [CrossRef] [PubMed]
- Marialigeti, S.; Tinya, F.; Bidlo, A.; Odor, P. Environmental Drivers of the Composition and Diversity of the Herb Layer in Mixed Temperate Forests in Hungary. Plant Ecol. 2016, 217, 549–563. [Google Scholar] [CrossRef]
- Chen, Y.L.; Xu, T.L.; Veresoglou, S.D.; Hu, H.W.; Hao, Z.P.; Hu, Y.J.; Liu, L.; Deng, Y.; Rillig, M.C.; Chen, B.D. Plant Diversity Represents the Prevalent Determinant of Soil Fungal Community Structure across Temperate Grasslands in Northern China. Soil Biol. Biochem. 2017, 110, 12–21. [Google Scholar] [CrossRef]
- Cho, S.; Myeong, H.-H.; Choung, Y. Promotion of Plant Species Diversity of Artificial Plantations in Korean National Parks through Thinning. J. Asia Pac. Biodivers. 2020, 13, 631–636. [Google Scholar] [CrossRef]
- Von Oheimb, G.; Härdtle, W. Selection Harvest in Temperate Deciduous Forests: Impact on Herb Layer Richness and Composition. Biodivers. Conserv. 2009, 18, 271–287. [Google Scholar] [CrossRef]
- Gallet, S.; Roze, F. Resistance of Atlantic Heathlands to Trampling in Brittany (France): Influence of Vegetation Type, Season and Weather Conditions. Biol. Conserv. 2001, 97, 189–198. [Google Scholar] [CrossRef]
- Li, X.; Lu, Y.; Huang, A.; Yuan, R.; Li, J.; Hu, D.; Zhong, Q.; Cheng, D. Light Response Model Fitting and Photosynthetic Characteristics of Ten Different Fern Species in Subtropics. ACTA Ecol. Sin. 2022, 42, 3333–3344. [Google Scholar] [CrossRef]
- Qu, Z.L.; Santalahti, M.; Köster, K.; Berninger, F.; Pumpanen, J.; Heinonsalo, J.; Sun, H. Soil Fungal Community Structure in Boreal Pine Forests: From Southern to Subarctic Areas of Finland. Front. Microbiol. 2021, 12, 653896. [Google Scholar] [CrossRef] [PubMed]
- Caihong, Z.; Nier, S.; Hao, W.; Honglin, X.; Hailong, S.; Ling, Y. Effects of Thinning on Soil Nutrient Availability and Fungal Community Composition in a Plantation Medium-Aged Pure Forest of Picea Koraiensis. Sci. Rep. 2023, 13, 2492. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Zeng, X.; Li, M.; Weng, X.; Frey, B.; Yang, L.; Li, M. Influence of Different Vegetation Types on Soil Physicochemical Parameters and Fungal Communities. Microorganisms 2022, 10, 829. [Google Scholar] [CrossRef] [PubMed]
- Arunrat, N.; Sansupa, C.; Sereenonchai, S.; Hatano, R. Short-Term Response of Soil Bacterial and Fungal Communities to Fire in Rotational Shifting Cultivation, Northern Thailand. Appl. Soil Ecol. 2024, 196, 105303. [Google Scholar] [CrossRef]
- Yao, F.; Yang, S.; Wang, Z.; Wang, X.; Ye, J.; Wang, X.; DeBruyn, J.M.; Feng, X.; Jiang, Y.; Li, H. Microbial Taxa Distribution Is Associated with Ecological Trophic Cascades along an Elevation Gradient. Front. Microbiol. 2017, 8, 2071. [Google Scholar] [CrossRef] [PubMed]
- Högberg, M.N.; Bååth, E.; Nordgren, A.; Arnebrant, K.; Högberg, P. Contrasting Effects of Nitrogen Availability on Plant Carbon Supply to Mycorrhizal Fungi and Saprotrophs—A Hypothesis Based on Field Observations in Boreal Forest. New Phytol. 2003, 160, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Prashar, P.; Kapoor, N.; Sachdeva, S. Rhizosphere: Its Structure, Bacterial Diversity and Significance. Rev. Environ. Sci. Biotechnol. 2014, 13, 63–77. [Google Scholar] [CrossRef]
- Orumaa, A.; Agan, A.; Anslan, S.; Drenkhan, T.; Drenkhan, R.; Kauer, K.; Köster, K.; Tedersoo, L.; Metslaid, M. Long-Term Effects of Forest Fires on Fungal Community and Soil Properties along a Hemiboreal Scots Pine Forest Fire Chronosequence. Sci. Total Environ. 2022, 851, 158173. [Google Scholar] [CrossRef] [PubMed]
- Mushinski, R.M.; Gentry, T.J.; Boutton, T.W. Organic Matter Removal Associated with Forest Harvest Leads to Decade Scale Alterations in Soil Fungal Communities and Functional Guilds. Soil Biol. Biochem. 2018, 127, 127–136. [Google Scholar] [CrossRef]
- Shigyo, N.; Hirao, T. Saprotrophic and Ectomycorrhizal Fungi Exhibit Contrasting Richness Patterns along Elevational Gradients in Cool-Temperate Montane Forests. Fungal Ecol. 2021, 50, 101036. [Google Scholar] [CrossRef]
- Khalid, M.; ur Rahman, S.; Liu, X.; Rehman, A.; Jumpponen, A.; Johan Kotze, D.; Setälä, H.; Hui, N. Exploring the Impact of Urbanization and Vegetation Type on Fungal Communities: Insights into Divergent, Mycorrhizal, and Saprophytic Associations Driven by Climate Patterns. Catena 2024, 238, 107860. [Google Scholar] [CrossRef]
- Li, D.H.; Han, Z.L.; Wu, Q.G.; Luo, G.R.; Liang, M.X.; Chen, J. Review of Functional Characteristics of Saprophytic Fungi Affecting the Decomposition of Fine Roots in Forest Ecosystem. World For. Res. 2021, 34, 26–31. [Google Scholar]
- Banerjee, S.; Walder, F.; Büchi, L.; Meyer, M.; Held, A.Y.; Gattinger, A.; Keller, T.; Charles, R.; van der Heijden, M.G.A. Agricultural Intensification Reduces Microbial Network Complexity and the Abundance of Keystone Taxa in Roots. ISME J. 2019, 13, 1722–1736. [Google Scholar] [CrossRef]
- Ma, B.; Wang, H.; Dsouza, M.; Lou, J.; He, Y.; Dai, Z.; Brookes, P.C.; Xu, J.; Gilbert, J.A. Geographic Patterns of Co-Occurrence Network Topological Features for Soil Microbiota at Continental Scale in Eastern China. ISME J. 2016, 10, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Faust, K.; Raes, J. Microbial Interactions: From Networks to Models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef]
- Banerjee, S.; Kirkby, C.A.; Schmutter, D.; Bissett, A.; Kirkegaard, J.A.; Richardson, A.E. Network Analysis Reveals Functional Redundancy and Keystone Taxa amongst Bacterial and Fungal Communities during Organic Matter Decomposition in an Arable Soil. Soil Biol. Biochem. 2016, 97, 188–198. [Google Scholar] [CrossRef]
- Peter, D.H.; Harrington, T.B. Effects of Forest Harvesting, Logging Debris, and Herbicides on the Composition, Diversity and Assembly of a Western Washington, USA Plant Community. For. Ecol. Manag. 2018, 417, 18–30. [Google Scholar] [CrossRef]
- Likulunga, L.E.; Rivera Pérez, C.A.; Schneider, D.; Daniel, R.; Polle, A. Tree Species Composition and Soil Properties in Pure and Mixed Beech-Conifer Stands Drive Soil Fungal Communities. For. Ecol. Manag. 2021, 502, 119709. [Google Scholar] [CrossRef]
- Nakashizuka, T. Species Coexistence in Temperate, Mixed Deciduous Forests. Trends Ecol. Evol. 2001, 16, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, H.; Dong, B.; Zhou, M.; Ma, L.; Jia, Z.; Duan, J. Regeneration Response to Canopy Gap Size in a Chinese Pine Plantation: Species Diversity Patterns, Size Structures and Spatial Distributions. For. Ecol. Manag. 2017, 397, 97–107. [Google Scholar] [CrossRef]
- Awad, A.; Majcherczyk, A.; Schall, P.; Schröter, K.; Schöning, I.; Schrumpf, M.; Ehbrecht, M.; Boch, S.; Kahl, T.; Bauhus, J.; et al. Ectomycorrhizal and Saprotrophic Soil Fungal Biomass Are Driven by Different Factors and Vary among Broadleaf and Coniferous Temperate Forests. Soil Biol. Biochem. 2019, 131, 9–18. [Google Scholar] [CrossRef]
- Tan, C.; Ferguson, D.K.; Tang, Z.; Yang, Y. Distribution and Conservation of the Lauraceae in China. Glob. Ecol. Conserv. 2023, 46, e02566. [Google Scholar] [CrossRef]
- Liu, L.; Zhu, K.; Krause, S.M.B.; Li, S.; Wang, X.; Zhang, Z.; Shen, M.; Yang, Q.; Lian, J.; Wang, X.; et al. Changes in Assembly Processes of Soil Microbial Communities during Secondary Succession in Two Subtropical Forests. Soil Biol. Biochem. 2021, 154, 108144. [Google Scholar] [CrossRef]
- Ceci, A.; Pinzari, F.; Russo, F.; Maggi, O.; Persiani, A.M. Saprotrophic Soil Fungi to Improve Phosphorus Solubilisation and Release: In Vitro Abilities of Several Species. Ambio 2018, 47, 30–40. [Google Scholar] [CrossRef]
- Niu, K.; He, J.S.; Zhang, S.; Lechowicz, M.J. Grazing Increases Functional Richness but Not Functional Divergence in Tibetan Alpine Meadow Plant Communities. Biodivers. Conserv. 2016, 25, 2441–2452. [Google Scholar] [CrossRef]
- Liu, Z.; Li, J.; Bayaerta; Niu, K. Biodiversity in Mosaic Communities: Soil Microbial Diversity Associates with Plant Functional Groups Relating to Soil Available Phosphorus in Tibetan Alpine Meadow. Eur. J. Soil Biol. 2023, 116, 103479. [Google Scholar] [CrossRef]
- Bridge, P.; Spooner, B. Soil Fungi: Diversity and Detection. Plant Soil 2001, 232, 147–154. [Google Scholar] [CrossRef]
- Hagenbo, A.; Kyaschenko, J.; Clemmensen, K.E.; Lindahl, B.D.; Fransson, P. Fungal Community Shifts Underpin Declining Mycelial Production and Turnover across a Pinus Sylvestris Chronosequence. J. Ecol. 2018, 106, 490–501. [Google Scholar] [CrossRef]
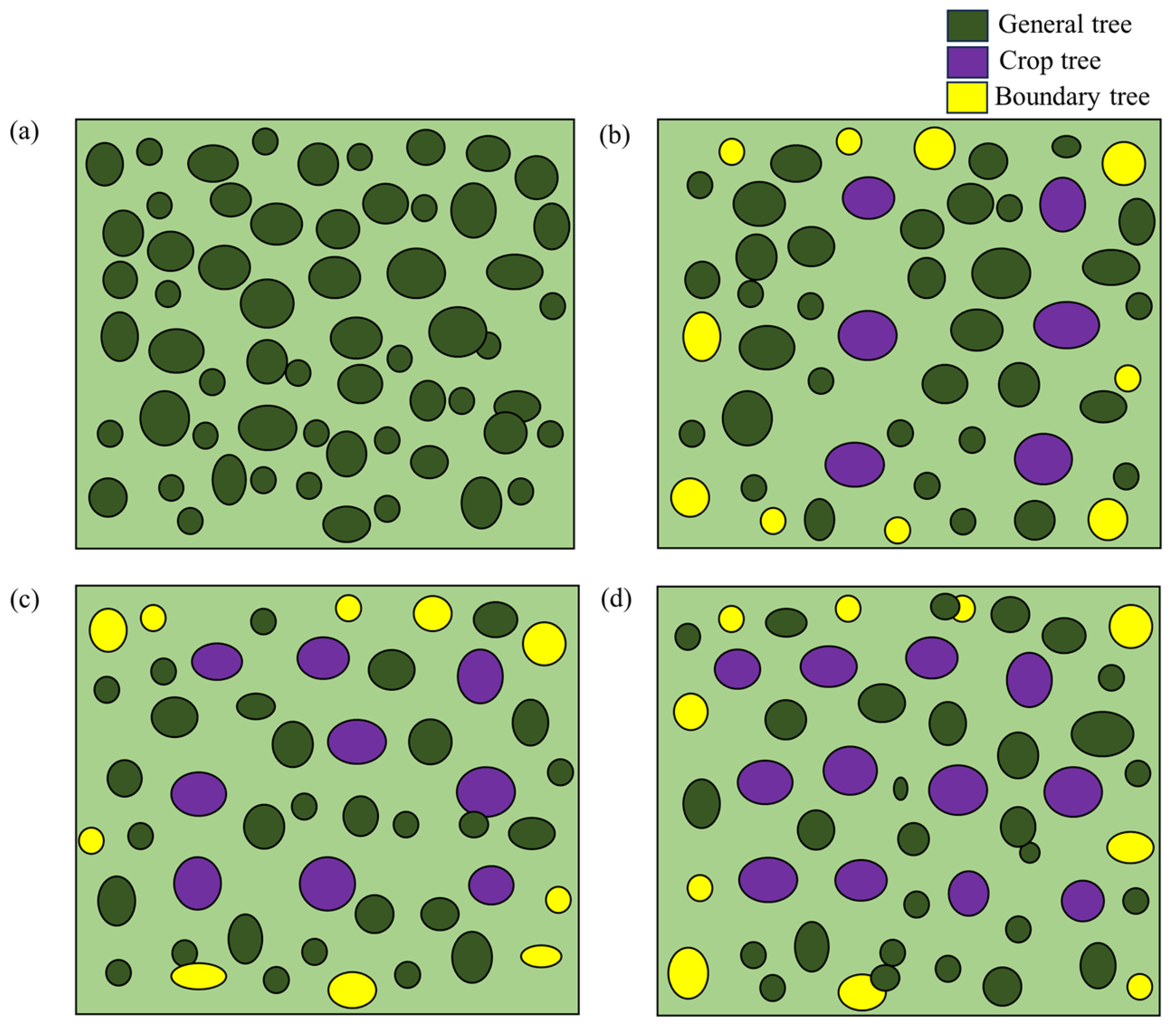


| Plot | Number of Crop Trees (N/667 m2) | Number of Crop Trees (N/ha) | Stand Density (trees/ha) | DBH (cm) | Average Height (m) | Altitude (m) | Slope (°) | Slope Aspect |
|---|---|---|---|---|---|---|---|---|
| WA | 6 | 100 | 1000 | 32.7 ± 6.5 | 14.5 ± 1.8 | 507 | 29 | South by west |
| WB | 9 | 150 | 1014 | 33.4 ± 4.5 | 12.9 ± 1.1 | 529 | 32 | North by west |
| WC | 12 | 200 | 1005 | 27.6 ± 5.2 | 13.0 ± 1.3 | 508 | 34 | North by west |
| WCK | 0 | 0 | 1252 | 26.1 ± 1.4 | 12.3 ± 0.2 | 479 | 27 | South by west |
| Plot | R1 | D1 | H1 | J1 | R2 | D2 | H2 | J2 |
|---|---|---|---|---|---|---|---|---|
| WA | 22.67 ± 0.58 c | 0.91 ± 0.002 b | 1.17 ± 0.01 c | 0.86 ± 0.01 b | 14.00 ± 1.00 b | 0.83 ± 0.02 c | 0.91 ± 0.05 c | 0.80 ± 0.02 b |
| WB | 30.00 ± 1.73 b | 0.95 ± 0.001 a | 1.34 ± 0.03 b | 0.90 ± 0.04 ab | 16.33 ± 0.58 a | 0.92 ± 0.002 a | 1.14 ± 0.04 a | 0.94 ± 0.02 a |
| WC | 32.67 ± 1.53 a | 0.96 ± 0.003 a | 1.42 ± 0.03 a | 0.94 ± 0.01 a | 17.33 ± 0.58 a | 0.91 ± 0.01 a | 1.14 ± 0.02 a | 0.92 ± 0.01 a |
| WCK | 18.67 ± 1.53 d | 0.91 ± 0.01 b | 1.12 ± 0.03 c | 0.89 ± 0.03 b | 12.33 ± 0.58 c | 0.89 ± 0.01 b | 1.01 ± 0.03 b | 0.93 ± 0.01 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyu, Q.; Yang, H.; Yin, B.; Xiang, Y.; Zhao, K.; Hou, G.; Chen, G.; Fan, C.; Li, X. Response of Soil Fungal-Community Structure to Crop-Tree Thinning in Pinus massoniana Plantation. Forests 2024, 15, 743. https://doi.org/10.3390/f15050743
Lyu Q, Yang H, Yin B, Xiang Y, Zhao K, Hou G, Chen G, Fan C, Li X. Response of Soil Fungal-Community Structure to Crop-Tree Thinning in Pinus massoniana Plantation. Forests. 2024; 15(5):743. https://doi.org/10.3390/f15050743
Chicago/Turabian StyleLyu, Qian, Huiqin Yang, Biran Yin, Yongqi Xiang, Kuangji Zhao, Guirong Hou, Gang Chen, Chuan Fan, and Xianwei Li. 2024. "Response of Soil Fungal-Community Structure to Crop-Tree Thinning in Pinus massoniana Plantation" Forests 15, no. 5: 743. https://doi.org/10.3390/f15050743





