Band Subset Selection for Hyperspectral Image Classification
Abstract
:1. Introduction
2. LCMV Criterion for BSS
3. Band Subset Selection
4. LCMV-BSS Algorithms
4.1. SQ LCMV-BSS
| Algorithm 1 SQ LCMV-BSS-1 |
| Step 1: Initial conditions |
|
| Step 2: Outer loop For do |
| Step 3: Inner loop Compute For do Find an index j* by with which specifies the band to be replaced by the lth band Bl. Such a band is now denoted by . A new set of bands is then produced by letting and for |
| Algorithm 2 SQ LCMV-BSS-2 |
Step 1: Initial conditions
|
| Step 2: Outer loop For do |
| Step 3: Inner loop For do Find an index j* by with which specifies the band to be replaced by the lth band Bl. Such a band is now denoted by . A new set of bands is then produced by letting and for |
4.2. SC LCMV-BSS
| Algorithm 3 SC LCMV-BSS |
Step 1: Initial conditions
|
| Step 2: Outer loop For do |
| Step 3: Inner loop For do Find where , . |
| Step 4: Output the final band subset, . |
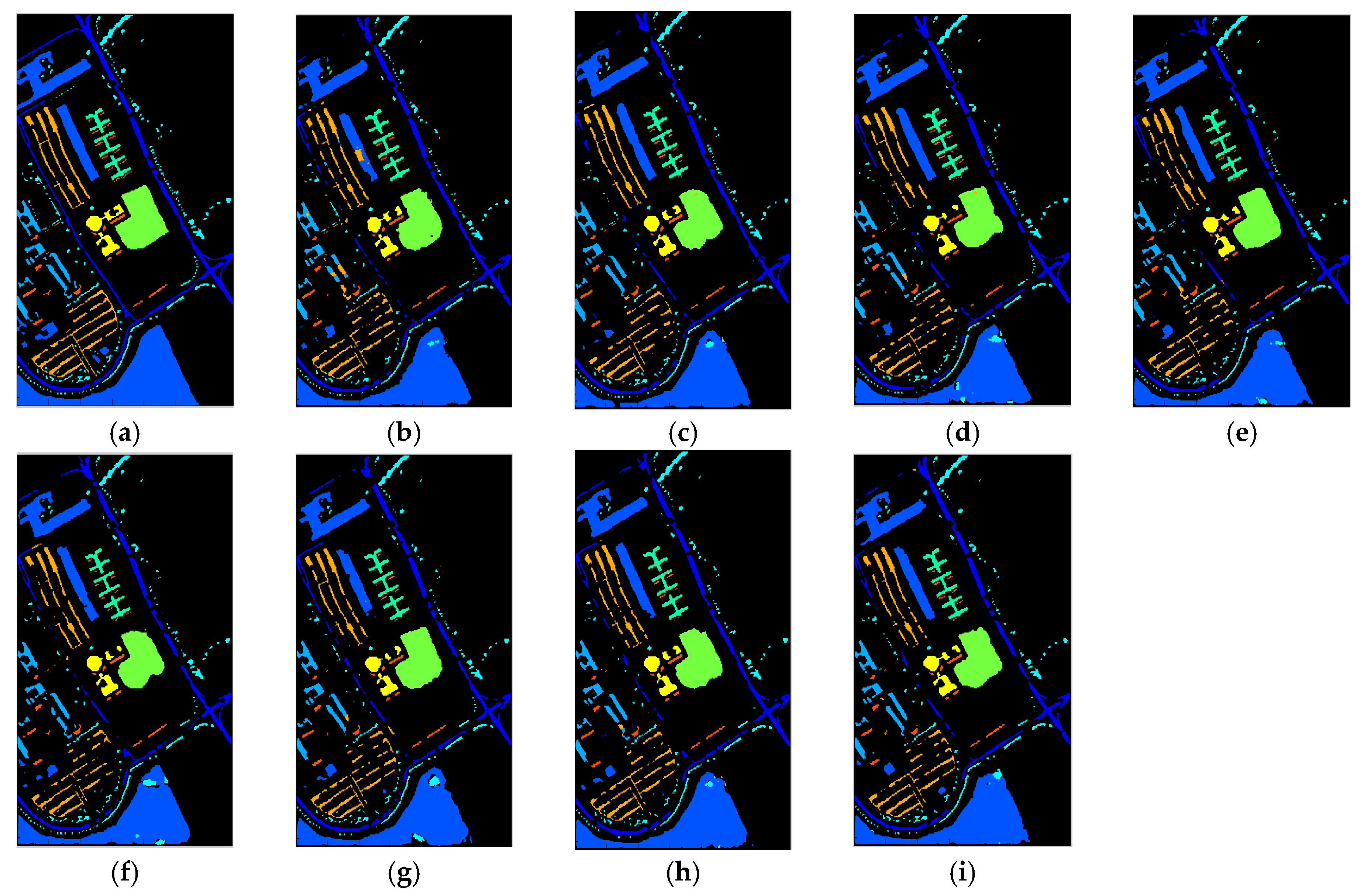
5. Real Image Experiments
5.1. Purdue Indiana Indian Pines Scene
5.2. Salinas
5.3. ROSIS Data
- Uniform band selection (UBS): According to our extensive experiments, UBS is a reasonably good BS method which is also reported in the literature. It does not require any prior knowledge or BS criterion. It is the simplest BS method.
- MEAC: This uses the minimum covariance derived from the estimated abundance matrix, which is similar to the minimum variance in (5). In addition, it can also represent the category of SQMBS methods.
- MDPP and DSEBS: Both represent the category of SMMBS methods. They make use of graph representations to specify band groups. Most importantly, these two methods were compared with CEM/LCMV-based methods in [26] and both are also based on the LCMV formulation specified by (2).
- LCMV-BSS developed in this paper: This represents the category of BSS methods using the LCMV formulation in (2).
- Unlike most supervised classifiers used for HSIC which require training samples, ILCMV only needs the knowledge of the class signatures D, which can be obtained by either prior knowledge or class sample means. Specifically, the class signatures in D are not necessarily real data samples.
- Also, unlike most supervised classifiers used for HSIC which require test and training data samples from the same class, the test samples for ILCMV can be selected from any arbitrary class including the BKG class, and are not necessarily limited to the same class trained by the training samples. This is a crucial difference between ILCMV and existing hyperspectral image classification algorithms reported in the literature. For more details, we refer to [23,76].
- It is very obvious to note that BSS did improve ILCMV classification results. Such an improvement cannot be found in the four EPF-based methods, where the classification results of the four EPF-based methods using band subsets could only get worse compared with the results using full bands. This may be due to the fact that the four EPF-based methods used principal component analysis (PCA) to compress the original data in preprocessing which retains some crucial information provided by full bands.
- According to Table 7, Table 8 and Table 9, ILCMV performed slightly better than the four EPF-based methods in POA but significantly better in PR for Purdue’s data and Salinas. The scene of the University of Pavia is interesting, as shown in Table 13, Table 14 and Table 15. The four EPF-based methods performed very well in POA but did very poorly in PR with about only 20%. Furthermore, POA produced by ILCMV may not be as good as those produced by the four EPF-based methods (about 10% less) but the PR produced by ILCMV were around 96% which is nearly 4.8 times better than the 20% produced by the four EPF-based methods. These experiments demonstrated that the BKG issue is critical in data analysis of the University of Pavia and cannot be ignored or discarded in data processing. Unfortunately, this BKG issue has never been investigated in the past.
- Unlike the four EPF-based methods, which performed well in POA but very poorly in PR, ILCMV consistently performs well in both POA and PR, and even better when it is implemented in conjunction with BSS—a case that the EPF-based methods actually failed, as shown in Table 7, Table 8, Table 9, Table 10, Table 11, Table 12, Table 13, Table 14 and Table 15.
- Last but not least, BS is heavily determined by three factors: the data to be processed, the BS method selected, and the classifier used. Unfortunately, most works on BS for hyperspectral image classification have been focused on the design and development of BS methods but very little has been reported on performance evaluation of different classifiers which use the same set of bands selected by a BS method. For example, as shown in Table 7, Table 8, Table 9, Table 10, Table 11, Table 12, Table 13, Table 14 and Table 15, if the four EPF methods were implemented by BS, their classification results could not be improved, but those of ILCMV could.
- It should be noted that PD results are not included in Table 7, Table 8, Table 9, Table 10, Table 11, Table 12, Table 13, Table 14 and Table 15 due to two reasons. One is that the results of PD using full bands are already available in [23,76]. The other is that EPF-based methods using partial bands did not perform better than their counterparts using full bands. So, it does not make sense to include their results in tables. Besides this, due to limited space, there is no need to include their results.
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Benediktsson, J.A.; Palmason, J.A.; Sveinsson, J.R. Classification of hyperspectral data from urban areas based on extended morphological profiles. IEEE Trans. Geosci. Remote Sens. 2005, 43, 480–491. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Bruzzone, L. Kernel-based methods for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2005, 43, 1351–1362. [Google Scholar] [CrossRef]
- Tarabalka, Y.; Benediktsson, J.A.; Chanussot, J. Spectral-spatial classification of hyperspectral imagery based on partitional clustering techniques. IEEE Trans. Geosci. Remote Sens. 2009, 47, 2973–2987. [Google Scholar] [CrossRef]
- Tarabalka, Y.; Fauvel, M.; Chanussot, J.; Benediktsson, J.A. SVM- and MRF-based method for accurate classification of hyperspectral images. IEEE Geosci. Remote Sens. Lett. 2010, 9, 736–740. [Google Scholar] [CrossRef]
- Li, J.; Bioucas-Dias, J.M.; Plaza, A. Semisupervised hyperspectral image segmentation using multinominal logistic regression model with active learning. IEEE Trans. Geosci. Remote Sens. 2010, 48, 4085–4098. [Google Scholar]
- Zhang, B.; Li, S.; Jia, X.; Gao, L.; Peng, M. Adpative hyperspectral Markov random field approach for classification of hyperspectral imagery. IEEE Geosci. Remote Sens. Lett. 2011, 8, 4085–4098. [Google Scholar] [CrossRef]
- Li, J.; Bioucas-Dias, J.M.; Plaza, A. Hyperspectral image segmentation using a new Bayesion approach with active learning. IEEE Trans. Geosci. Remote Sens. 2011, 49, 3947–3960. [Google Scholar] [CrossRef]
- Chen, Y.; Nasabardi, N.; Tran, T.D. Hyperspectral image segmentation using dictionary-based sparse representation. IEEE Trans. Geosci. Remote Sens. 2011, 49, 3973–3985. [Google Scholar] [CrossRef]
- Tarabalka, Y.; Fauvel, M.; Chanussot, J.; Benediktsson, J.A. Segmentation and classification of hyperspectral images using minimum spanning forest grown from automatically selected makers. IEEE Trans. Syst. Man Cybern. Part B Cybern. 2011, 40, 1267–1279. [Google Scholar] [CrossRef] [PubMed]
- Fauvel, M.; Tarabalka, Y.; Benediktsson, J.A.; Chanussot, J.; Tilton, J. Advances in spectral-spatial classification of hyperspectral images. Proc. IEEE 2013, 101, 652–675. [Google Scholar] [CrossRef]
- Kang, K.; Li, S.; Benediktsson, J.A. Spectral-spatial hyperspectral image classification with edge-preserving filtering. IEEE Trans. Geosci. Remote Sens. 2014, 52, 2666–2677. [Google Scholar] [CrossRef]
- Fu, W.; Li, S.; Fang, L.; Kang, X.; Benediktsson, J.A. Hyperspectral image classification via shape-adaptive joint sparse representation. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 556–567. [Google Scholar] [CrossRef]
- Kang, K.; Li, S.; Fang, L.; Li, M.; Benediktsson, J.A. Extended random walker-based classification of hyperspectral images. IEEE Trans. Geosci. Remote Sens. 2015, 53, 144–153. [Google Scholar] [CrossRef]
- Lu, T.; Li, S.; Fang, L.; Bruzzone, L.; Benediktsson, J.A. Set-to-set distance-based spectral spatial classification of hyperspectral images. IEEE Trans. Geosci. Remote Sens. 2016, 54, 7122–7134. [Google Scholar] [CrossRef]
- Guo, X.; Huang, X.; Zhang, L.; Plaza, A.; Benedikisson, J.A. Support tensor machines for classification of hyperspectral remote sesning imagery. IEEE Trans. Geosci. Remote Sens. 2016, 54, 3248–3264. [Google Scholar] [CrossRef]
- Ma, X.; Wang, H.; Wang, J. Semisupervised classification of hyperspectral image based on multi- decision labeling and deep feature learning. ISPRS J. Photogramm. Remote Sens. 2016, 120, 99–107. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Gomez-Chova, L.; Munoz-Mari, J.; Vila-Frances, J.; Calpe-Maravilla, J. CKs for hyperspectral image classification. IEEE Geosci. Remote Sens. Lett. 2006, 3, 93–97. [Google Scholar] [CrossRef]
- Gurram, P.; Kwon, H. Contextual SVM using Hilbert space embedding for hyperspectral classification. IEEE Geosci. Remote Sens. Lett. 2013, 10, 1031–1035. [Google Scholar] [CrossRef]
- Peng, J.; Zhou, Y.; Chen, C. Region-kernel-based support vector machines for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 4810–4824. [Google Scholar] [CrossRef]
- Sun, S.; Zhong, P.; Xiao, H.; Wang, R. Active learning with Gaussian process classifier for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 1746–1760. [Google Scholar] [CrossRef]
- Pullanagari, R.; Kereszturi, G.; Yule, I.J.; Ghamisi, P. Assessing the performance of multiple spectral-spatial features of a hyperspectral image for classification of urban land cover classes using support vector machines and artificial neural network. J. Appl. Remote Sens. 2017, 11, 026009-1–026009-21. [Google Scholar] [CrossRef]
- Xue, B.; Yu, C.; Wang, Y.; Song, M.; Li, S.; Wang, L.; Chen, H.M.; Chang, C.-I. A subpixel target detection approach to hyperpsectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 5093–5114. [Google Scholar] [CrossRef]
- Chang, C.-I. Hyperspectral Data Processing: Signal Processing Algorithm Design and Analysis; Wiley: Hoboken, NJ, USA, 2013. [Google Scholar]
- Chang, C.-I.; Du, Q.; Sun, T.S.; Althouse, M.L.G. A joint band prioritization and band decorrelation approach to band selection for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 1999, 37, 2631–2641. [Google Scholar] [CrossRef]
- Chang, C.-I.; Wang, S. Constrained band selection for hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2006, 44, 1575–1585. [Google Scholar] [CrossRef]
- Huang, R.; He, M. Band selection based on feature weighting for classification of hyperspectral data. IEEE Trans. Geosci. Remote Sens. Lett. 2005, 2, 156–159. [Google Scholar] [CrossRef]
- Keshava, N. Distance metrics and band selection in hyperspectral processing with applications to material identification. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1552–1565. [Google Scholar] [CrossRef]
- Mausel, P.W.; Kramber, W.J.; Lee, J.K. Optimum band selection for supervised classification of multispectral data. Photogramm. Eng. Remote Sens. 1990, 56, 55–60. [Google Scholar]
- Jia, S.; Ji, Z.; Qian, Y.-Y.; Shen, L.-L. Unsupervised band selection for hyperspectral imagery classification without manual band removal. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 531–543. [Google Scholar] [CrossRef]
- Stearns, S.D.; Wilson, B.E.; Peterson, J.R. Dimensionality reduction by optimal band selection for pixel classification of hyperspectral imagery. Proc. SPIE 1993, 2028, 118–127. [Google Scholar]
- Backer, S.; Kempeneers, P.; Debruyn, W.; Scheunders, P. Band selection for hyperspectral remote sensing. Pattern Recognit. 2005, 2, 319–323. [Google Scholar]
- Du, Q.; Yang, H. Similarity-based unsupervised band selection for hyperspectral image analysis. IEEE Geosci. Remote Sens. Lett. 2008, 5, 564–568. [Google Scholar] [CrossRef]
- Yang, H.; Du, Q. An efficient method for supervised hyperspectral band selection. IEEE Geosci. Remote Sens. Lett. 2011, 8, 138–142. [Google Scholar] [CrossRef]
- Su, H.; Du, Q.; Chen, G.; Du, P. Optimized hyperspectral band selection using particle swarm optimization. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2659–2670. [Google Scholar] [CrossRef]
- Su, H.; Yang, B.; Du, Q. Hyperspectral band selection using improved firefly algorithm. IEEE Geosci. Remote Sens. Lett. 2016, 13, 68–72. [Google Scholar] [CrossRef]
- Wei, W.; Du, Q.; Younan, N.H. Fast supervised hyperspectral band selection using graphics processing unit. J. Appl. Remote Sens. 2012, 6, 061504-1–061504-12. [Google Scholar]
- Yuan, Y.; Zhu, G.; Wang, Q. Hyperspectral band selection by multitask sparisty pursuit. IEEE Trans. Geosci. Remote Sens. 2015, 53, 631–644. [Google Scholar] [CrossRef]
- Lu, X.; Li, X.; Mou, L. Semi-supervised multitask learning for scene recognition. IEEE Trans. Cybern. 2015, 45, 1967–1976. [Google Scholar] [PubMed]
- Zhu, G.; Huang, H.; Lei, J.; Bi, Z.; Xu, F. Unsupervised hyperspectral band selection by dominant set extraction. IEEE Trans. Geosci. Remote Sens. 2016, 54, 227–239. [Google Scholar] [CrossRef]
- Yuan, Y.; Lin, J.; Wang, Q. Dual-clustering-based hyperspectral band selection by contextual analysis. IEEE Trans. Geosci. Remote Sens. 2016, 54, 1431–1445. [Google Scholar] [CrossRef]
- Wang, Q.; Lin, J.; Yuan, Y. Salient band selection for hyperspectral image classification via manifold ranking. IEEE Trans. Neural Netw. Learn. Syst. 2016, 27, 1279–1289. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zheng, X.; Lu, X. Discovering diverse subset for unsupervised hyperspectral band selection. IEEE Trans. Image Process. 2017, 26, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Yuan, Y.; Zheng, X. Joint dictionary learning for multispectral change detection. IEEE Trans. Cybern. 2017, 47, 884–897. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zheng, X.; Yuan, Y. Remote sensing scene classification by unsupervised representation learning. IEEE Trans. Geosci. Remote Sens. 2017, 55, 5148–5157. [Google Scholar] [CrossRef]
- Feng, J.; Jiao, L.C.; Zhang, X.; Sun, T. Hyperspectral band selection based on trivariate mutual information and clonal selection. IEEE Trans. Geosci. Remote Sens. 2014, 52, 4092–4105. [Google Scholar] [CrossRef]
- Feng, J.; Jiao, L.C.; Liu, F.; Sun, T.; Zhang, X. Mutual-information0-based semi-supervised hyperspectral band selection with high discrimination, high information and low redundancy. IEEE Trans. Geosci. Remote Sens. 2015, 53, 2956–2969. [Google Scholar] [CrossRef]
- Wang, C.; Gong, M.; Zhang, M.; Chan, Y. Unsupervised hyperspectral image band selection via column subset selection. IEEE Geosci. Remote Sens. Lett. 2015, 12, 1411–1415. [Google Scholar] [CrossRef]
- Geng, X.; Sun, K.; Ji, L. Band selection for target detection in hyperspectral imagery using sparse CEM. Remote Sens. Lett. 2014, 5, 1022–1031. [Google Scholar] [CrossRef]
- Sun, K.; Geng, X.; Ji, L. A new sparsity-based band selection method for target detection of hyperspectral image. IEEE Geosci. Remote Sens. Lett. 2015, 12, 329–333. [Google Scholar]
- Zare, A.; Gader, P. Hyperspectral band selection and endmember detection using sparisty promoting priors. IEEE Geosci. Remote Sens. Lett. 2008, 5, 256–260. [Google Scholar] [CrossRef]
- Ball, J.E.; Bruce, L.E.; Younan, N.H. Hyperspectral pixel unmixing via spectral band selection and DC-insensitive singular value decomposition. IEEE Geosci. Remote Sens. Lett. 2007, 4, 382–386. [Google Scholar] [CrossRef]
- Sun, K.; Geng, X.; Ji, L.; Lu, Y. A new band selection method for hyperspectral image based on data quality. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2697–2703. [Google Scholar]
- Sun, K.; Geng, X.; Ji, L. Exemplar component analysis: A fast band selection method for hyperspectral imagery. IEEE Geosci. Remote Sens. Lett. 2015, 12, 998–1002. [Google Scholar]
- Koonsanit, K.; Jaruskulchai, C.; Eiumnoh, A. Band selection for dimension reduction in hyper spectral image using integrated information gain and principal components analysis technique. Int. J. Mach. Learn. Comput. 2012, 2, 248–251. [Google Scholar] [CrossRef]
- Xia, W.; Wang, B.; Zhang, L. Band selection for hyperspectral imagery: A new approach based on complex networks. IEEE Geosci. Remote Sens. Lett. 2013, 10, 1229–1233. [Google Scholar] [CrossRef]
- Chang, C.-I.; Liu, K.-H. Progressive band selection for hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2014, 52, 2002–2017. [Google Scholar] [CrossRef]
- Martínez-Usó, A.; Pla, F.; Sotoca, J.M.; García-Sevilla, P. Clustering-based hyperspectral band selection using information measures. IEEE Trans. Geosci. Remote Sens. 2007, 45, 4158–4171. [Google Scholar] [CrossRef]
- Yang, H.; Du, Q.; Sheng, Y. Semisupervised band clustering for dimensionality reduction of hyperspectral imagery. IEEE Geosci. Remote Sens. Lett. 2011, 8, 1135–1139. [Google Scholar]
- Su, H.; Du, Q. Hyperspectral band clustering and band selection for urban land cover classification. Geocarto Int. 2012, 27, 395–411. [Google Scholar] [CrossRef]
- Winter, M.E. N-finder: An algorithm for fast autonomous spectral endmember determination in hyperspectral data. Proc. SPIE 1999, 3753, 266–277. [Google Scholar]
- Wu, C.-C.; Chu, S.; Chang, C.-I. Sequential N-FINDR algorithm. In Proceedings of the SPIE Conference on Imaging Spectrometry XIII, San Diego, CA, USA, 10–14 August 2008. [Google Scholar]
- Xiong, X.; Wu, C.-C.; Chang, C.-I.; Kapalkis, K.; Chen, H.M. Fast algorithms to implement N-FINDR for hyperspectral endmember extraction. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2011, 4, 545–564. [Google Scholar] [CrossRef]
- Chang, C.-I. Maximum Simplex Volume-Based Endmember Extraction Algorithms. U.S. Patent 8,417,748 B2, 9 April 2013. [Google Scholar]
- Chang, C.-I. Real Time Progressive Hyperspectral Image Processing: Endmember Finding and Anomaly Detection; Springer: New York, NY, USA, 2016. [Google Scholar]
- Wang, L.; Chang, C.-I.; Wang, Y.; Xue, B.; Song, M.; Yu, C.; Li, S. Band subset selection for anomaly detection in hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2017, 55, 4887–4898. [Google Scholar] [CrossRef]
- Chang, C.-I.; Lee, L.C.; Xue, B.; Song, M.; Chen, J. Channel capacity approach to band subset selection for hyperspectral imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 4630–4644. [Google Scholar] [CrossRef]
- Wang, L.; Li, H.C.; Xue, B.; Chang, C.-I. Constrained band subset selection for hyperspectral imagery. IEEE Geosci. Remote Sens. Lett. 2017, 14, 2032–2036. [Google Scholar] [CrossRef]
- Chang, C.-I. Hyperspectral Imaging: Techniques for Spectral Detection and Classification; Kluwer Academic/Plenum Publishers: New York, NY, USA, 2003. [Google Scholar]
- Frost, O.L., III. An algorithm for linearly constrained adaptive array processing. Proc. IEEE 1972, 60, 926–935. [Google Scholar] [CrossRef]
- Chang, C.-I. Target signature-constrained mixed pixel classification for hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2002, 40, 1065–1081. [Google Scholar] [CrossRef]
- Harsanyi, J.C. Detection and Classification of Subpixel Spectral Signatures in Hyperspectral Image Sequences; Department of Electrical Engineering, University of Maryland: Baltimore County, MD, USA, 1993. [Google Scholar]
- Pudil, P.; Novovicova, J.; Kittler, J. Floating search methods in feature selection. Pattern Recognit. Lett. 1994, 15, 1119–1125. [Google Scholar] [CrossRef]
- Chang, C.-I.; Du, Q. Estimation of number of spectrally distinct signal sources in hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2004, 42, 608–619. [Google Scholar] [CrossRef]
- Harsanyi, J.C.; Farrand, W.; Chang, C.-I. Detection of subpixel spectral signatures in hyperspectral image sequences. In Proceedings of the American Congress on Surveying & Mapping (ACSM)/American Society of Photogrammetry & Remote Sensing (ASPRS) Annual Converntion and Exposition, Baltimore, MD, USA, 25–28 April 1994; Volume 1, pp. 236–247. [Google Scholar]
- Yu, C.; Xue, B.; Wang, Y.; Song, M.; Wang, L.; Li, S.; Chang, C.-I. Multi-class constrained background suppression approach to hyperspectral image classification. In Proceedings of the 2017 IEEE/GRSS International Geoscience and Remote Sensing Symposium (IGARSS), Fort Worth, TX, USA, 23–28 July 2017. [Google Scholar]





| PF = 10−1 | PF = 10−2 | PF = 10−3 | PF = 10−4 | PF = 10−5 | |
|---|---|---|---|---|---|
| Purdue | 73/21 | 49/19 | 35/18 | 27/18 | 25/17 |
| Salinas | 32/33 | 28/24 | 25/21 | 21/21 | 20/20 |
| Univ. of Pavia | 25/34 | 21/27 | 16/17 | 14/14 | 13/12 |
| Data | Methods | Selected Bands |
|---|---|---|
| Purdue Indian Pines (18 bands) | UBS | 1, 14, 27, 40, 53, 66, 79, 92, 105, 118, 131, 144, 157, 170, 183, 196, 209, 220 |
| MEAC | 159, 3, 92, 96, 82, 36, 39, 55, 41, 1, 2, 33, 206, 38, 163, 17, 204, 9 | |
| MDPP | 10, 39, 59, 75, 79, 85, 92, 130, 140, 146, 147, 149, 150, 152, 160, 164, 175, 193 | |
| DSEBS | 42, 129, 97, 131, 174, 16, 176, 177, 172, 43, 192, 193, 98, 171, 99, 132, 40, 33 | |
| SQ LCMV-BSS-1 | 39, 164, 29, 155, 108, 66, 79, 8, 105, 42, 44, 17, 156, 150, 3, 43, 213, 41 | |
| SQ LCMV-BSS-2 | 38, 109, 29, 52, 163, 66, 158, 8, 164, 219, 43, 78, 157, 220, 3, 49, 218, 2 | |
| SC LCMV-BSS | 54, 156, 42, 159, 53, 41, 79, 91, 105, 57, 51, 43, 157, 48, 107, 160, 115, 163 | |
| Salinas (21 bands) | UBS | 1, 12, 23, 34, 45, 56, 67, 78, 89, 100, 111, 122, 133, 144, 155, 166, 177, 188, 199, 210, 224 |
| MEAC | 107, 148, 203, 149, 5, 8, 105, 3, 28, 12, 18, 10, 44, 36, 25, 17, 51, 32, 110, 68, 58 | |
| MDPP | 1, 8, 11, 22, 27, 28, 50, 57, 58, 65, 90, 99, 105, 119, 123, 134, 142, 157, 175, 191, 204 | |
| DSEBS | 99, 101, 16, 119, 177, 112, 44, 46, 120, 47, 131, 175, 196, 121, 17, 102, 174, 180, 187, 135, 42 | |
| SQ LCMV-BSS-1 | 7, 50, 23, 48, 45, 73, 65, 15, 40, 19, 80, 122, 38, 41, 42, 46, 78, 47, 200, 37, 2 | |
| SQ LCMV-BSS-2 | 7, 42, 56, 28, 45, 58, 67, 15, 41, 19, 50, 122, 38, 34, 36, 47, 224, 46, 183, 37, 172 | |
| SC LCMV-BSS | 18, 39, 41, 31, 45, 44, 67, 78, 90, 101, 40, 91, 42, 141, 46, 48, 102, 185, 47, 86, 50 | |
| Univ. of Pavia (14 bands) | UBS | 1, 9, 17, 25, 33, 41, 49, 57, 65, 73, 81, 89, 97, 103 |
| MEAC | 1, 23, 24, 40, 42, 58, 56, 59, 48, 31, 47, 83, 25, 54 | |
| MDPP | 2, 23, 44, 46, 50, 62, 66, 73, 89, 91, 92, 93, 96, 102 | |
| DSEBS | 86, 102, 64, 20, 21, 63, 65, 6, 19, 22, 7, 66, 95, 67 | |
| SQ LCMV-BSS-1 | 1, 4, 55, 16, 95, 83, 84, 93, 39, 77, 91, 102, 92, 103 | |
| SQ LCMV-BSS-2 | 1, 4, 38, 76, 85, 55, 84, 102, 16, 83, 93, 89, 92, 103 | |
| SC LCMV-BSS | 1, 4, 84, 16, 38, 102, 85, 92, 83, 72, 95, 91, 96, 103 |
| Class | Full Bands | UBS | MEAC | MDPP | DSEBS | SQ LCMV-BSS-1 | SQ LCMV-BSS-2 | SC LCMV-BSS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PD | PR | PD | PR | PD | PR | PD | PR | PD | PR | PD | PR | PD | PR | PD | PR | |
| 1 | 95.65 | 100 | 95.65 | 100 | 93.48 | 100 | 95.65 | 100 | 95.65 | 100 | 95.65 | 100 | 97.83 | 100 | 100 | 100 |
| 2 | 96.01 | 100 | 97.13 | 99.57 | 93.07 | 99.63 | 96.08 | 100 | 96.99 | 100 | 95.59 | 99.71 | 93.78 | 99.85 | 94.89 | 99.85 |
| 3 | 96.99 | 99.88 | 96.51 | 100 | 96.27 | 100 | 97.35 | 100 | 97.23 | 99.88 | 96.39 | 100 | 95.67 | 100 | 94.10 | 100 |
| 4 | 98.73 | 100 | 98.73 | 100 | 98.31 | 100 | 99.58 | 100 | 98.31 | 100 | 97.89 | 100 | 98.31 | 100 | 98.31 | 100 |
| 5 | 89.44 | 100 | 90.68 | 100 | 91.51 | 100 | 92.34 | 100 | 93.58 | 100 | 91.93 | 100 | 92.34 | 100 | 92.96 | 100 |
| 6 | 97.12 | 100 | 97.67 | 100 | 97.40 | 99.58 | 96.71 | 100 | 97.12 | 100 | 96.44 | 100 | 97.95 | 100 | 95.75 | 100 |
| 7 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| 8 | 98.78 | 100 | 98.54 | 100 | 99.16 | 100 | 97.49 | 100 | 97.91 | 100 | 97.91 | 100 | 99.16 | 100 | 98.95 | 100 |
| 9 | 100 | 100 | 100 | 100 | 90.00 | 100 | 100 | 100 | 100 | 100 | 100 | 90.91 | 100 | 95.24 | 100 | 100 |
| 10 | 93.93 | 99.78 | 91.98 | 100 | 93.31 | 100 | 94.65 | 99.78 | 93.00 | 100 | 94.24 | 100 | 91.98 | 99.58 | 91.98 | 100 |
| 11 | 94.70 | 99.87 | 96.13 | 99.96 | 94.55 | 98.22 | 95.48 | 99.87 | 95.85 | 100 | 95.48 | 99.96 | 96.17 | 100 | 95.93 | 99.49 |
| 12 | 95.45 | 100 | 94.94 | 100 | 96.29 | 100 | 96.80 | 100 | 97.30 | 100 | 95.95 | 100 | 95.11 | 100 | 96.63 | 100 |
| 13 | 98.54 | 100 | 98.54 | 100 | 99.02 | 100 | 97.56 | 100 | 96.59 | 100 | 97.56 | 100 | 98.54 | 100 | 98.54 | 100 |
| 14 | 93.52 | 100 | 94.15 | 100 | 94.78 | 100 | 94.70 | 100 | 94.55 | 100 | 95.89 | 100 | 96.05 | 100 | 96.13 | 100 |
| 15 | 90.67 | 100 | 95.60 | 100 | 92.49 | 100 | 96.89 | 100 | 93.52 | 100 | 94.82 | 100 | 94.56 | 100 | 96.11 | 100 |
| 16 | 98.92 | 98.92 | 98.92 | 98.92 | 98.92 | 100 | 98.92 | 98.92 | 98.92 | 100 | 98.92 | 100 | 97.85 | 100 | 95.70 | 97.80 |
| POA | 95.09 | 95.69 | 94.91 | 95.89 | 95.88 | 95.67 | 95.48 | 95.46 | ||||||||
| PR | 97.61 | 97.90 | 97.52 | 98.00 | 97.99 | 97.89 | 97.80 | 97.79 | ||||||||
| Class | Full Bands | UBS | MEAC | MDPP | DSEBS | SQ LCMV-BSS-1 | SQ LCMV-BSS-2 | SC LCMV-BSS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PD | PR | PD | PR | PD | PR | PD | PR | PD | PR | PD | PR | PD | PR | PD | PR | |
| 1 | 95.52 | 100 | 97.16 | 100 | 97.71 | 100 | 97.76 | 100 | 97.16 | 100 | 96.37 | 100 | 97.01 | 100 | 96.91 | 100 |
| 2 | 98.42 | 100 | 98.85 | 100 | 98.44 | 100 | 97.99 | 100 | 99.17 | 100 | 98.79 | 100 | 98.36 | 100 | 98.71 | 100 |
| 3 | 93.78 | 99.70 | 95.50 | 100 | 94.03 | 100 | 93.98 | 100 | 95.65 | 100 | 90.44 | 100 | 95.14 | 100 | 95.95 | 100 |
| 4 | 95.62 | 100 | 94.69 | 98.80 | 94.33 | 97.84 | 97.49 | 98.76 | 94.74 | 99.62 | 96.56 | 98.39 | 95.91 | 99.11 | 92.04 | 94.83 |
| 5 | 96.90 | 100 | 96.45 | 100 | 95.19 | 99.88 | 95.22 | 100 | 96.90 | 99.85 | 95.87 | 100 | 95.94 | 100 | 90.78 | 99.79 |
| 6 | 98.79 | 100 | 98.59 | 100 | 98.56 | 100 | 98.79 | 100 | 98.56 | 100 | 97.95 | 100 | 98.91 | 100 | 97.75 | 100 |
| 7 | 98.63 | 100 | 98.21 | 100 | 98.18 | 100 | 97.99 | 100 | 97.65 | 100 | 98.35 | 100 | 98.44 | 100 | 98.32 | 100 |
| 8 | 96.69 | 98.26 | 95.81 | 99.39 | 97.40 | 99.84 | 95.23 | 99.74 | 96.11 | 99.38 | 95.84 | 99.06 | 97.47 | 100 | 96.61 | 99.42 |
| 9 | 95.87 | 100 | 95.60 | 100 | 94.74 | 100 | 95.29 | 100 | 95.73 | 100 | 94.79 | 100 | 94.89 | 100 | 95.44 | 100 |
| 10 | 96.67 | 100 | 96.37 | 100 | 96.34 | 100 | 96.46 | 100 | 97.25 | 100 | 95.73 | 100 | 96.58 | 100 | 96.77 | 100 |
| 11 | 97.75 | 100 | 97.85 | 100 | 91.10 | 100 | 97.75 | 100 | 98.31 | 100 | 95.79 | 100 | 97.38 | 100 | 97.66 | 100 |
| 12 | 97.15 | 100 | 96.16 | 100 | 95.54 | 100 | 97.46 | 100 | 97.66 | 100 | 96.32 | 100 | 95.39 | 100 | 95.43 | 100 |
| 13 | 96.51 | 100 | 96.94 | 99.44 | 93.35 | 99.88 | 96.40 | 100 | 95.63 | 100 | 87.77 | 100 | 97.38 | 99.78 | 94.00 | 98.97 |
| 14 | 95.89 | 100 | 98.14 | 100 | 97.66 | 99.90 | 97.01 | 100 | 98.04 | 100 | 97.76 | 99.05 | 97.20 | 100 | 96.93 | 99.81 |
| 15 | 94.00 | 98.66 | 95.27 | 98.09 | 96.52 | 100 | 95.42 | 96.70 | 95.25 | 98.84 | 95.42 | 97.73 | 96.27 | 99.86 | 95.84 | 98.60 |
| 16 | 93.30 | 100 | 96.07 | 100 | 93.86 | 100 | 95.07 | 100 | 95.02 | 100 | 95.68 | 100 | 95.13 | 100 | 95.41 | 100 |
| POA | 96.37 | 96.49 | 96.45 | 96.25 | 96.63 | 95.93 | 96.81 | 96.21 | ||||||||
| PR | 98.23 | 98.29 | 98.27 | 98.17 | 98.36 | 98.02 | 98.45 | 98.15 | ||||||||
| Class | Full Bands | UBS | MEAC | MDPP | DSEBS | SQ LCMV-BSS-1 | SQ LCMV-BSS-2 | SC LCMV-BSS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PD | PR | PD | PR | PD | PR | PD | PR | PD | PR | PD | PR | PD | PR | PD | PR | |
| 1 | 86.42 | 99.90 | 87.67 | 99.45 | 86.44 | 99.76 | 87.97 | 99.68 | 87.71 | 99.74 | 84.44 | 99.63 | 88.05 | 99.44 | 88.67 | 99.77 |
| 2 | 73.34 | 99.99 | 84.38 | 99.95 | 83.33 | 99.89 | 82.14 | 99.96 | 84.63 | 99.92 | 84.14 | 99.89 | 85.21 | 99.98 | 86.76 | 99.95 |
| 3 | 79.85 | 96.30 | 78.90 | 100 | 76.66 | 100 | 76.17 | 99.02 | 79.22 | 100 | 76.49 | 100 | 74.71 | 100 | 78.56 | 99.95 |
| 4 | 98.81 | 96.65 | 97.84 | 95.16 | 98.88 | 87.91 | 96.95 | 91.85 | 97.99 | 88.71 | 98.14 | 88.96 | 97.77 | 95.30 | 97.70 | 93.11 |
| 5 | 91.49 | 100 | 89.93 | 100 | 91.33 | 100 | 93.32 | 100 | 87.11 | 100 | 93.50 | 100 | 90.57 | 100 | 90.77 | 100 |
| 6 | 89.10 | 99.98 | 91.35 | 100 | 82.78 | 100 | 87.53 | 100 | 87.44 | 100 | 86.19 | 100 | 90.00 | 100 | 91.13 | 100 |
| 7 | 81.10 | 100 | 83.32 | 100 | 76.26 | 100 | 75.64 | 100 | 76.34 | 100 | 82.84 | 100 | 82.92 | 100 | 82.46 | 100 |
| 8 | 78.46 | 85.20 | 79.09 | 97.37 | 79.51 | 97.20 | 79.83 | 95.71 | 79.30 | 97.44 | 77.09 | 97.16 | 77.09 | 98.45 | 79.30 | 98.96 |
| 9 | 77.24 | 99.87 | 75.86 | 99.47 | 76.32 | 100 | 74.01 | 98.44 | 76.17 | 100 | 80.21 | 99.46 | 78.22 | 99.86 | 77.08 | 99.87 |
| POA | 84.32 | 85.19 | 83.85 | 84.33 | 84.25 | 84.45 | 85.41 | 85.92 | ||||||||
| PR | 96.76 | 96.93 | 96.64 | 96.76 | 96.75 | 96.78 | 96.96 | 96.95 | ||||||||
| MEAC | MDPP | DSEBS | SQ LCMV-BSS-1 | SQ LCMV-BSS-2 | SC LCMV-BSS | |
|---|---|---|---|---|---|---|
| Purdue | 13.70 | 41.14 | 0.58 | 7.00 | 7.10 | 6.93 |
| Salinas | 83.64 | 44.66 | 5.27 | 43.43 | 46.63 | 43.55 |
| University of Pavia | 44.53 | 29.22 | 4.62 | 16.67 | 17.52 | 16.84 |
| Class | EPF-B-g with Full Bands | EPF-B-c with Full Bands | EPF-G-g with Full Bands | EPF-G-c with Full Bands | ILCMV with Full Bands | EPF-B-g-BS | EPF-B-c-BS | EPF-G-g-BS | EPF-G-c-BS | ILCMV-BS |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 100 | 100 | 97.83 | 100 | 95.65 | 100 | 100 | 100 | 100 | 95.65 |
| 2 | 76.47 | 87.82 | 80.60 | 82.98 | 96.01 | 74.79 | 90.76 | 71.99 | 79.90 | 95.59 |
| 3 | 93.49 | 83.98 | 79.64 | 65.42 | 96.99 | 63.37 | 75.66 | 84.10 | 72.77 | 96.39 |
| 4 | 99.16 | 100 | 100 | 96.20 | 98.73 | 100 | 100 | 100 | 97.89 | 97.89 |
| 5 | 93.79 | 94.00 | 97.10 | 94.82 | 89.44 | 89.86 | 97.52 | 96.07 | 94.41 | 91.93 |
| 6 | 100 | 99.59 | 99.59 | 99.45 | 97.12 | 98.22 | 99.86 | 99.73 | 99.32 | 96.44 |
| 7 | 92.86 | 92.86 | 96.43 | 96.43 | 100 | 92.86 | 64.29 | 92.86 | 89.29 | 100 |
| 8 | 100 | 100 | 100 | 100 | 98.78 | 100 | 100 | 100 | 100 | 97.91 |
| 9 | 80.00 | 65.00 | 100 | 100 | 100 | 65.00 | 95.00 | 65.00 | 10.00 | 100 |
| 10 | 90.53 | 91.46 | 87.14 | 93.00 | 93.93 | 75.31 | 73.97 | 84.88 | 76.54 | 94.24 |
| 11 | 90.67 | 92.67 | 86.27 | 88.88 | 94.70 | 78.98 | 70.79 | 64.73 | 80.94 | 95.48 |
| 12 | 98.31 | 96.46 | 93.93 | 92.07 | 95.45 | 47.55 | 42.50 | 79.76 | 57.67 | 95.95 |
| 13 | 99.02 | 99.51 | 99.51 | 99.51 | 98.54 | 100 | 100 | 100 | 100 | 97.56 |
| 14 | 97.71 | 97.00 | 97.87 | 98.26 | 93.52 | 94.86 | 94.86 | 87.67 | 92.57 | 95.89 |
| 15 | 100 | 100 | 99.74 | 82.90 | 90.67 | 95.85 | 96.37 | 95.85 | 97.41 | 94.82 |
| 16 | 97.85 | 100 | 100 | 100 | 98.92 | 98.92 | 100 | 98.92 | 97.85 | 98.92 |
| POA | 92.27 | 93.45 | 90.32 | 89.79 | 95.09 | 81.62 | 82.94 | 81.17 | 84.15 | 95.67 |
| PR | 44.98 | 44.56 | 44.03 | 43.77 | 97.61 | 39.79 | 40.43 | 39.86 | 41.02 | 97.89 |
| Time(s) | 196.58 | 200.84 | 194.09 | 200.87 | 25.37 | 31.27 | 36.77 | 31.14 | 36.16 | 37.25 |
| Class | EPF-B-g with Full Bands | EPF-B-c with Full Bands | EPF-G-g with Full Bands | EPF-G-c with Full Bands | ILCMV with Full Bands | EPF-B-g-BS | EPF-B-c-BS | EPF-G-g-BS | EPF-G-c-BS | ILCMV-BS |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 100 | 100 | 100 | 100 | 95.65 | 100 | 100 | 100 | 100 | 97.83 |
| 2 | 86.83 | 82.49 | 86.62 | 84.52 | 96.01 | 85.78 | 81.86 | 64.22 | 66.60 | 93.78 |
| 3 | 90.84 | 85.90 | 82.53 | 80.72 | 96.99 | 81.93 | 74.58 | 63.98 | 64.82 | 95.67 |
| 4 | 99.16 | 99.16 | 100 | 99.58 | 98.73 | 99.16 | 100 | 99.58 | 100 | 98.31 |
| 5 | 95.03 | 92.96 | 97.72 | 92.34 | 89.44 | 93.17 | 95.03 | 91.72 | 92.13 | 92.34 |
| 6 | 100 | 99.86 | 99.59 | 99.73 | 97.12 | 99.73 | 97.81 | 100 | 99.59 | 97.95 |
| 7 | 89.29 | 89.29 | 96.43 | 96.43 | 100 | 92.86 | 78.57 | 96.43 | 92.86 | 100 |
| 8 | 100 | 100 | 100 | 100 | 98.78 | 100 | 100 | 100 | 100 | 99.16 |
| 9 | 60.00 | 75.00 | 70.00 | 50.00 | 100 | 100 | 95.00 | 65.00 | 35.00 | 100 |
| 10 | 92.59 | 90.33 | 90.74 | 92.49 | 93.93 | 70.27 | 66.26 | 64.71 | 54.42 | 91.98 |
| 11 | 89.12 | 89.33 | 88.51 | 86.44 | 94.70 | 64.89 | 70.26 | 82.12 | 82.24 | 96.17 |
| 12 | 96.46 | 98.31 | 98.15 | 98.99 | 95.45 | 67.45 | 52.78 | 51.10 | 34.74 | 95.11 |
| 13 | 99.02 | 99.02 | 99.51 | 99.51 | 98.54 | 99.51 | 99.51 | 99.51 | 99.51 | 98.54 |
| 14 | 98.81 | 98.18 | 96.36 | 95.02 | 93.52 | 90.59 | 97.79 | 92.81 | 89.41 | 96.05 |
| 15 | 99.74 | 100 | 94.04 | 95.34 | 90.67 | 90.41 | 90.41 | 93.26 | 83.42 | 94.56 |
| 16 | 100 | 100 | 100 | 100 | 98.92 | 95.70 | 98.92 | 100 | 100 | 97.85 |
| POA | 93.37 | 92.17 | 92.10 | 90.96 | 95.09 | 81.49 | 81.28 | 80.01 | 77.66 | 95.48 |
| PR | 45.52 | 44.93 | 44.89 | 44.34 | 97.61 | 39.72 | 39.61 | 39.00 | 37.85 | 97.80 |
| Time(s) | 194.14 | 199.37 | 194.13 | 200.36 | 25.37 | 31.16 | 37.93 | 32.56 | 36.51 | 41.58 |
| Class | EPF-B-g with Full Bands | EPF-B-c with Full Bands | EPF-G-g with Full Bands | EPF-G-c with Full Bands | ILCMV with Full Bands | EPF-B-g-BS | EPF-B-c-BS | EPF-G-g-BS | EPF-G-c-BS | ILCMV-BS |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 100 | 100 | 100 | 97.83 | 95.65 | 100 | 97.83 | 97.83 | 100 | 100 |
| 2 | 83.26 | 83.26 | 83.05 | 78.92 | 96.01 | 48.25 | 42.09 | 48.74 | 39.92 | 94.89 |
| 3 | 79.16 | 69.64 | 80.48 | 66.87 | 96.99 | 62.05 | 55.18 | 60.36 | 44.10 | 94.10 |
| 4 | 100 | 98.31 | 100 | 99.58 | 98.73 | 100 | 100 | 100 | 99.58 | 98.31 |
| 5 | 93.79 | 93.17 | 95.86 | 93.79 | 89.44 | 91.93 | 94.00 | 93.58 | 93.79 | 92.96 |
| 6 | 99.73 | 99.73 | 98.08 | 99.32 | 97.12 | 99.32 | 99.45 | 94.93 | 99.45 | 95.75 |
| 7 | 96.43 | 92.86 | 96.43 | 89.29 | 100 | 85.71 | 71.43 | 92.86 | 64.29 | 100 |
| 8 | 100 | 99.79 | 100 | 100 | 98.78 | 100 | 100 | 99.79 | 100 | 98.95 |
| 9 | 20.00 | 75.00 | 75.00 | 65.00 | 100 | 20.00 | 45.00 | 35.00 | 0 | 100 |
| 10 | 75.31 | 84.16 | 84.47 | 89.61 | 93.93 | 59.05 | 48.97 | 47.22 | 49.90 | 91.98 |
| 11 | 91.65 | 91.00 | 87.98 | 91.73 | 94.70 | 76.25 | 78.09 | 70.22 | 72.87 | 95.93 |
| 12 | 96.29 | 92.07 | 96.12 | 97.47 | 95.45 | 70.83 | 60.20 | 44.86 | 58.85 | 96.63 |
| 13 | 99.51 | 99.51 | 99.51 | 99.51 | 98.54 | 100 | 100 | 100 | 100 | 98.54 |
| 14 | 92.89 | 98.34 | 96.36 | 96.68 | 93.52 | 98.34 | 93.04 | 97.23 | 97.39 | 96.13 |
| 15 | 100 | 100 | 98.19 | 99.48 | 90.67 | 93.26 | 89.12 | 79.27 | 67.36 | 96.11 |
| 16 | 100 | 100 | 100 | 100 | 98.92 | 100 | 100 | 93.55 | 95.70 | 95.70 |
| POA | 90.06 | 90.42 | 90.56 | 90.37 | 95.09 | 77.37 | 74.12 | 72.31 | 71.25 | 95.46 |
| PR | 43.90 | 44.08 | 44.15 | 44.05 | 97.61 | 37.72 | 36.13 | 35.25 | 34.73 | 97.79 |
| Time(s) | 187.76 | 203.60 | 195.20 | 201.21 | 25.37 | 32.01 | 38.78 | 31.92 | 38.07 | 42.73 |
| Class | EPF-B-g with Full Bands. | EPF-B-c with Full Bands | EPF-G-g with Full Bands | EPF-G-c with Full Bands | ILCMV with Full Bands | EPF-B-g-BS | EPF-B-c-BS | EPF-G-g-BS | EPF-G-c-BS | ILCMV-BS |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 100 | 100 | 100 | 100 | 95.52 | 100 | 99.75 | 100 | 100 | 96.37 |
| 2 | 100 | 100 | 100 | 100 | 98.42 | 99.97 | 100 | 99.97 | 100 | 98.79 |
| 3 | 100 | 100 | 100 | 100 | 93.78 | 100 | 100 | 100 | 100 | 90.44 |
| 4 | 100 | 100 | 100 | 99.93 | 95.62 | 99.93 | 99.86 | 99.93 | 100 | 96.56 |
| 5 | 99.37 | 99.25 | 98.69 | 99.25 | 96.90 | 98.92 | 98.95 | 98.92 | 98.95 | 95.87 |
| 6 | 100 | 100 | 100 | 100 | 98.79 | 99.97 | 99.97 | 99.97 | 99.97 | 97.95 |
| 7 | 100 | 99.92 | 100 | 100 | 98.63 | 99.66 | 99.72 | 99.66 | 99.83 | 98.35 |
| 8 | 90.92 | 90.40 | 89.26 | 90.61 | 96.69 | 87.11 | 91.94 | 87.11 | 91.70 | 95.84 |
| 9 | 99.98 | 100 | 99.97 | 99.97 | 95.87 | 99.58 | 100 | 99.58 | 99.97 | 94.79 |
| 10 | 96.95 | 98.08 | 98.60 | 98.51 | 96.67 | 96.19 | 98.57 | 96.19 | 98.60 | 95.73 |
| 11 | 99.91 | 99.91 | 99.91 | 100 | 97.75 | 99.91 | 99.91 | 99.91 | 99.81 | 95.79 |
| 12 | 100 | 100 | 100 | 100 | 97.15 | 100 | 100 | 100 | 100 | 96.32 |
| 13 | 99.89 | 99.56 | 99.02 | 99.89 | 96.51 | 98.47 | 99.13 | 98.47 | 99.56 | 87.77 |
| 14 | 99.91 | 99.25 | 99.63 | 100 | 95.89 | 98.97 | 99.35 | 98.97 | 98.04 | 97.76 |
| 15 | 89.01 | 85.65 | 85.65 | 87.08 | 94.00 | 90.66 | 82.21 | 90.66 | 85.99 | 95.42 |
| 16 | 99.83 | 100 | 99.45 | 99.83 | 93.30 | 99.67 | 100 | 99.67 | 100 | 95.68 |
| POA | 96.40 | 95.89 | 95.64 | 96.17 | 96.37 | 95.64 | 95.73 | 95.64 | 96.19 | 95.93 |
| PR | 46.97 | 46.26 | 46.95 | 46.85 | 98.23 | 46.60 | 46.64 | 46.60 | 46.86 | 98.02 |
| Time(s) | 1060.77 | 741.84 | 1082.51 | 1134.06 | 167.80 | 75.17 | 78.73 | 75.17 | 104.31 | 134.96 |
| Class | EPF-B-g with Full Bands | EPF-B-c with Full Bands | EPF-G-g with Full Bands | EPF-G-c with Full Bands | ILCMV with Full Bands | EPF-B-g-BS | EPF-B-c-BS | EPF-G-g-BS | EPF-G-c-BS | ILCMV-BS |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 100 | 100 | 100 | 100 | 95.52 | 100 | 100 | 100 | 100 | 97.01 |
| 2 | 100 | 100 | 100 | 100 | 98.42 | 100 | 100 | 100 | 99.97 | 98.36 |
| 3 | 100 | 100 | 100 | 100 | 93.78 | 100 | 100 | 99.95 | 100 | 95.14 |
| 4 | 100 | 100 | 100 | 99.78 | 95.62 | 100 | 99.93 | 100 | 100 | 95.91 |
| 5 | 99.48 | 98.95 | 98.58 | 99.14 | 96.90 | 98.66 | 98.73 | 99.14 | 98.84 | 95.94 |
| 6 | 100 | 100 | 100 | 100 | 98.79 | 99.95 | 100 | 100 | 100 | 98.91 |
| 7 | 100 | 99.89 | 100 | 99.89 | 98.63 | 99.92 | 99.97 | 99.89 | 99.80 | 98.44 |
| 8 | 88.63 | 91.98 | 87.90 | 90.86 | 96.69 | 87.84 | 89.50 | 90.61 | 91.86 | 97.47 |
| 9 | 99.90 | 99.98 | 99.95 | 100 | 95.87 | 99.97 | 99.94 | 99.60 | 100 | 94.89 |
| 10 | 97.56 | 98.90 | 97.04 | 97.50 | 96.67 | 99.33 | 99.51 | 99.21 | 98.29 | 96.58 |
| 11 | 100 | 99.91 | 99.91 | 99.72 | 97.75 | 100 | 99.91 | 99.72 | 100 | 97.38 |
| 12 | 100 | 100 | 100 | 100 | 97.15 | 100 | 100 | 100 | 100 | 95.39 |
| 13 | 99.24 | 99.89 | 98.80 | 99.13 | 96.51 | 99.02 | 100 | 98.36 | 99.13 | 97.38 |
| 14 | 99.91 | 99.91 | 99.91 | 99.25 | 95.89 | 100 | 100 | 99.91 | 99.35 | 97.20 |
| 15 | 88.66 | 85.43 | 94.25 | 91.43 | 94.00 | 79.35 | 93.04 | 83.94 | 86.13 | 96.27 |
| 16 | 99.94 | 100 | 99.61 | 99.50 | 93.30 | 100 | 100 | 100 | 100 | 95.13 |
| POA | 95.91 | 96.24 | 96.42 | 96.69 | 96.37 | 94.56 | 96.77 | 95.71 | 96.24 | 96.81 |
| PR | 46.73 | 46.89 | 46.97 | 47.11 | 98.23 | 46.07 | 47.15 | 46.63 | 46.89 | 98.45 |
| Time(s) | 1128.55 | 755.52 | 1050.34 | 722.76 | 167.80 | 73.49 | 101.80 | 71.57 | 98.16 | 159.26 |
| Class | EPF-B-g with Full Bands | EPF-B-c with Full Bands | EPF-G-g with Full Bands | EPF-G-c with Full Bands | ILCMV with Full Bands | EPF-B-g-BS | EPF-B-c-BS | EPF-G-g-BS | EPF-G-c-BS | ILCMV-BS |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 100 | 100 | 100 | 100 | 95.52 | 100 | 100 | 100 | 100 | 96.91 |
| 2 | 100 | 100 | 100 | 100 | 98.42 | 99.95 | 99.84 | 99.92 | 99.97 | 98.71 |
| 3 | 100 | 100 | 100 | 100 | 93.78 | 99.90 | 100 | 100 | 99.90 | 95.95 |
| 4 | 100 | 100 | 100 | 100 | 95.62 | 100 | 99.57 | 100 | 99.86 | 92.04 |
| 5 | 99.07 | 99.22 | 99.10 | 98.92 | 96.90 | 98.88 | 99.55 | 99.33 | 98.99 | 90.78 |
| 6 | 100 | 100 | 100 | 100 | 98.79 | 100 | 100 | 100 | 99.90 | 97.75 |
| 7 | 100 | 99.97 | 100 | 99.97 | 98.63 | 99.61 | 99.89 | 100 | 99.94 | 98.32 |
| 8 | 90.18 | 89.51 | 89.40 | 91.38 | 96.69 | 90.88 | 89.02 | 90.68 | 90.30 | 96.61 |
| 9 | 100 | 99.98 | 99.81 | 99.94 | 95.87 | 99.95 | 99.95 | 99.97 | 100 | 95.44 |
| 10 | 97.47 | 97.71 | 97.41 | 99.48 | 96.67 | 98.90 | 99.24 | 98.11 | 98.29 | 96.77 |
| 11 | 100 | 100 | 100 | 100 | 97.75 | 100 | 99.72 | 99.34 | 99.72 | 97.66 |
| 12 | 100 | 100 | 100 | 100 | 97.15 | 100 | 100 | 100 | 100 | 95.43 |
| 13 | 100 | 99.89 | 98.91 | 99.89 | 96.51 | 99.34 | 99.89 | 99.24 | 99.13 | 94.00 |
| 14 | 100 | 100 | 100 | 99.81 | 95.89 | 99.35 | 99.63 | 98.79 | 98.32 | 96.93 |
| 15 | 88.99 | 84.26 | 88.08 | 86.90 | 94.00 | 89.74 | 93.35 | 78.92 | 88.92 | 95.84 |
| 16 | 99.28 | 99.89 | 100 | 99.94 | 93.30 | 100 | 100 | 100 | 99.94 | 95.41 |
| POA | 96.25 | 95.52 | 95.95 | 96.34 | 96.37 | 96.50 | 96.70 | 95.02 | 96.26 | 96.21 |
| PR | 46.89 | 46.53 | 46.75 | 46.94 | 98.23 | 47.02 | 47.11 | 46.29 | 46.90 | 98.15 |
| Time(s) | 1139.99 | 1106.95 | 1154.78 | 1089.61 | 167.80 | 73.06 | 94.17 | 69.92 | 91.50 | 147.30 |
| Class | EPF-B-g with Full Bands | EPF-B-c with Full Bands | EPF-G-g with Full Bands | EPF-G-c with Full Bands | ILCMV with Full Bands | EPF-B-g-BS | EPF-B-c-BS | EPF-G-g-BS | EPF-G-c-BS | ILCMV-BS |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 98.04 | 98.04 | 98.08 | 97.81 | 77.24 | 96.15 | 92.47 | 96.34 | 97.29 | 80.21 |
| 2 | 98.66 | 99.39 | 97.79 | 98.28 | 86.42 | 94.31 | 94.84 | 97.39 | 98.06 | 84.44 |
| 3 | 91.09 | 93.52 | 95.00 | 94.33 | 73.34 | 92.62 | 94.81 | 95.57 | 95.33 | 84.14 |
| 4 | 93.47 | 95.27 | 92.92 | 98.01 | 79.85 | 97.52 | 97.45 | 97.00 | 96.87 | 76.49 |
| 5 | 100 | 100 | 100 | 99.85 | 98.81 | 100 | 100 | 100 | 100 | 98.14 |
| 6 | 99.98 | 100 | 100 | 100 | 91.49 | 99.64 | 98.15 | 99.64 | 100 | 93.50 |
| 7 | 100 | 99.32 | 99.92 | 99.77 | 89.10 | 99.92 | 99.32 | 99.40 | 100 | 86.19 |
| 8 | 99.02 | 99.00 | 97.80 | 99.78 | 81.10 | 95.60 | 96.22 | 96.41 | 96.28 | 82.84 |
| 9 | 100 | 100 | 100 | 100 | 78.46 | 100 | 99.89 | 100 | 100 | 77.09 |
| POA | 98.12 | 98.67 | 97.80 | 98.46 | 84.32 | 95.96 | 97.49 | 97.49 | 98.01 | 84.45 |
| PR | 20.24 | 20.35 | 20.17 | 20.31 | 96.76 | 19.79 | 19.71 | 20.11 | 20.21 | 96.82 |
| Time(s) | 225.93 | 265.79 | 232.05 | 252.50 | 401.08 | 51.16 | 83.46 | 52.90 | 78.32 | 1387.01 |
| Class | EPF-B-g with Full Bands | EPF-B-c with Full Bands | EPF-G-g with Full Bands | EPF-G-c with Full Bands | ILCMV with Full Bands | EPF-B-g-BS | EPF-B-c-BS | EPF-G-g-BS | EPF-G-c-BS | ILCMV-BS |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 94.89 | 97.80 | 97.84 | 98.21 | 77.24 | 95.43 | 95.96 | 88.76 | 92.01 | 78.22 |
| 2 | 98.58 | 97.63 | 99.25 | 98.01 | 86.42 | 96.43 | 94.88 | 92.59 | 93.13 | 88.05 |
| 3 | 94.90 | 93.57 | 92.57 | 95.19 | 73.34 | 92.19 | 92.47 | 95.52 | 93.62 | 85.21 |
| 4 | 95.63 | 94.97 | 93.02 | 98.83 | 79.85 | 98.69 | 96.02 | 98.37 | 98.47 | 74.71 |
| 5 | 100 | 100 | 100 | 99.85 | 98.81 | 100 | 100 | 100 | 100 | 97.77 |
| 6 | 100 | 100 | 100 | 100 | 91.49 | 100 | 99.52 | 99.72 | 99.94 | 90.57 |
| 7 | 100 | 100 | 99.40 | 99.77 | 89.10 | 100 | 99.92 | 100 | 100 | 90.00 |
| 8 | 96.93 | 98.89 | 98.94 | 98.48 | 81.10 | 93.97 | 94.70 | 95.22 | 93.56 | 82.92 |
| 9 | 100 | 100 | 100 | 100 | 78.46 | 100 | 100 | 100 | 100 | 77.09 |
| POA | 97.76 | 97.85 | 98.36 | 98.39 | 84.32 | 96.74 | 95.97 | 94.25 | 94.78 | 85.41 |
| PR | 20.16 | 20.18 | 20.29 | 20.29 | 96.76 | 19.95 | 19.79 | 19.44 | 19.55 | 96.94 |
| Time(s) | 219.10 | 249.76 | 226.85 | 238.81 | 401.08 | 48.52 | 82.95 | 53.53 | 79.05 | 971.56 |
| Class | EPF-B-g with Full Bands | EPF-B-c with Full Bands | EPF-G-g with Full Bands | EPF-G-c with Full Bands | ILCMV with Full Bands | EPF-B-g-BS | EPF-B-c-BS | EPF-G-g-BS | EPF-G-c-BS | ILCMV-BS |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 99.43 | 97.16 | 97.95 | 97.98 | 77.24 | 95.93 | 92.73 | 98.27 | 97.99 | 77.08 |
| 2 | 98.19 | 99.15 | 98.80 | 98.79 | 86.42 | 95.44 | 94.31 | 97.00 | 96.23 | 88.67 |
| 3 | 99.24 | 95.09 | 94.85 | 93.57 | 73.34 | 93.52 | 91.04 | 92.38 | 94.14 | 86.76 |
| 4 | 93.93 | 94.65 | 94.61 | 98.27 | 79.85 | 97.52 | 96.74 | 98.27 | 96.96 | 78.56 |
| 5 | 100 | 99.85 | 100 | 99.93 | 98.81 | 100 | 100 | 100 | 100 | 97.70 |
| 6 | 99.96 | 99.34 | 99.68 | 100 | 91.49 | 99.70 | 98.11 | 99.05 | 100 | 90.77 |
| 7 | 100 | 100 | 99.70 | 99.77 | 89.10 | 99.62 | 99.47 | 99.92 | 99.55 | 91.13 |
| 8 | 96.85 | 98.13 | 97.77 | 99.51 | 81.10 | 92.88 | 92.61 | 92.99 | 92.97 | 82.46 |
| 9 | 100 | 100 | 100 | 100 | 78.46 | 100 | 100 | 100 | 100 | 79.30 |
| POA | 98.37 | 98.32 | 98.28 | 98.67 | 84.32 | 96.22 | 94.85 | 97.21 | 96.92 | 85.92 |
| PR | 20.29 | 20.28 | 20.27 | 20.35 | 96.76 | 19.85 | 19.56 | 20.05 | 19.99 | 97.09 |
| Time(s) | 238.01 | 270.24 | 234.01 | 259.33 | 401.08 | 51.13 | 83.41 | 49.37 | 76.48 | 998.17 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, C.; Song, M.; Chang, C.-I. Band Subset Selection for Hyperspectral Image Classification. Remote Sens. 2018, 10, 113. https://doi.org/10.3390/rs10010113
Yu C, Song M, Chang C-I. Band Subset Selection for Hyperspectral Image Classification. Remote Sensing. 2018; 10(1):113. https://doi.org/10.3390/rs10010113
Chicago/Turabian StyleYu, Chunyan, Meiping Song, and Chein-I Chang. 2018. "Band Subset Selection for Hyperspectral Image Classification" Remote Sensing 10, no. 1: 113. https://doi.org/10.3390/rs10010113
APA StyleYu, C., Song, M., & Chang, C.-I. (2018). Band Subset Selection for Hyperspectral Image Classification. Remote Sensing, 10(1), 113. https://doi.org/10.3390/rs10010113





