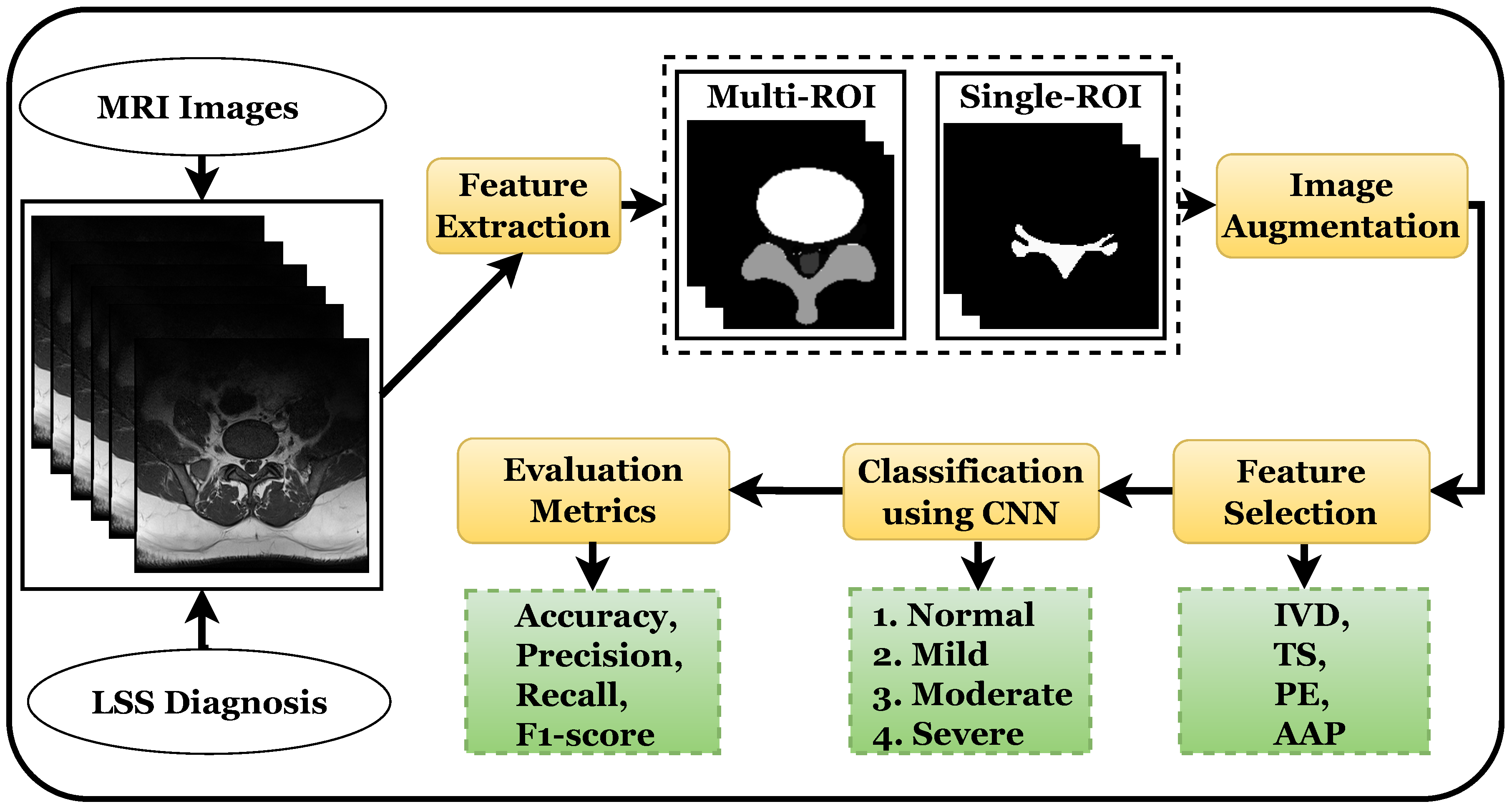
This section provides details of the experimental setup, results of the proposed approach, and discussions of the results.
3.2. Data Preparation
The initial step in using deep learning models is to prepare the training data for the classifiers. The deep learning approach is tremendously data-hungry because it also incorporates representation learning [
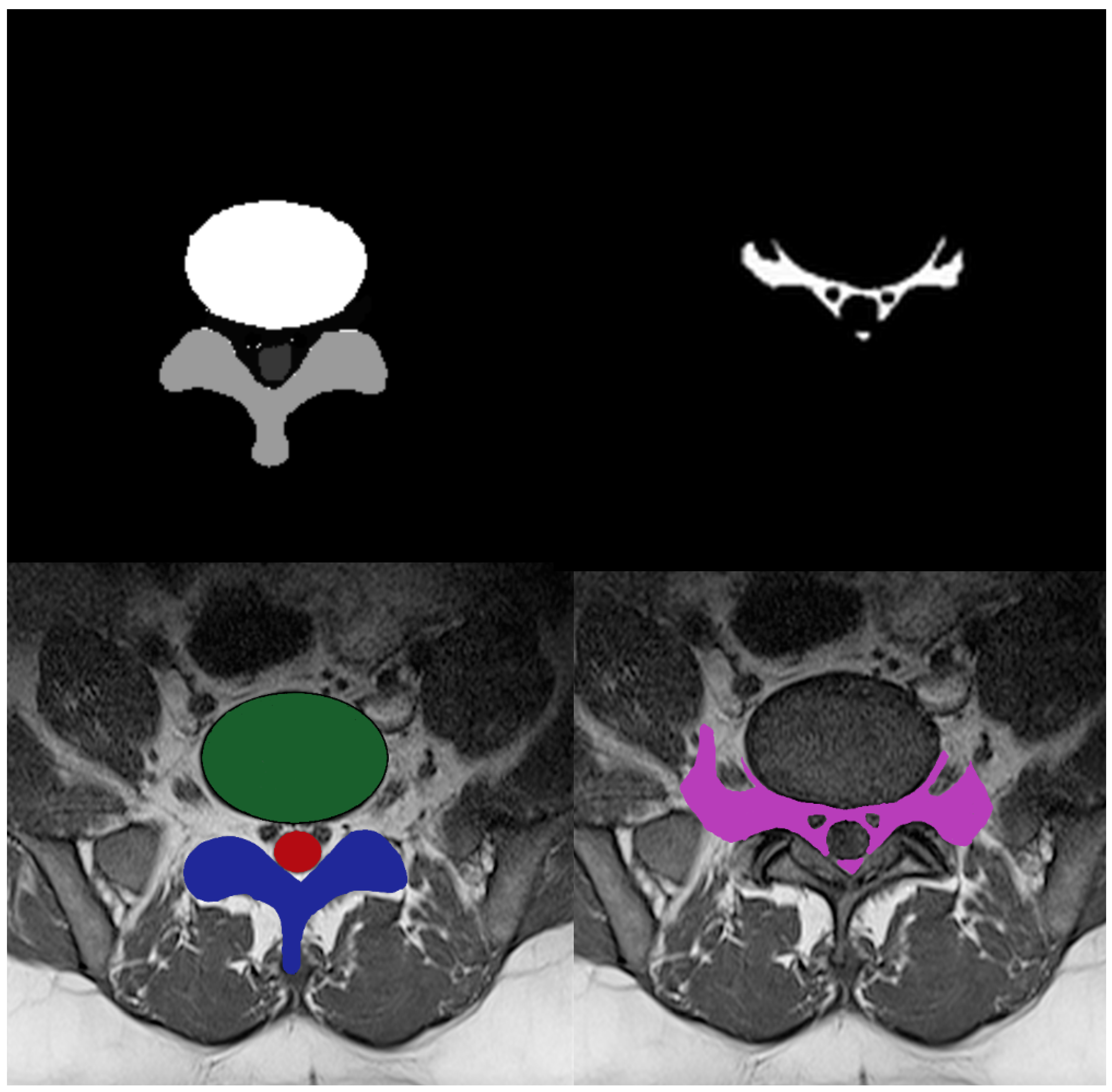
33]. Multi-ROI and single-ROI training datasets were employed in our experiment. The first mask shown in
Figure 3, contained IVD, PE, and TS, but the second mask only included the AAP region, so stenosis diagnosis could be performed after measuring this region.
To improve model performance, training was performed using various augmented sets of data including 5 k, 10 k, and 12.5 k, and testing was performed on 330 labeled images. The training was carried out using an 80:20 ratio; the classifier was trained on 80% and tested on 20% of the data. The training and testing ratio was chosen randomly, which was found to be most effective in prior research [
10].
The model takes modest steps to reduce the negative gradient of the loss function, which is specified as the categorical cross-entropy probability distribution of each class. The learning rate parameter, which is 0.001, alters the step size, and the Adam optimizer is utilized. In the 12.5 k dataset, the batch size utilized for training was 256 per image, for a total of 34 epochs. A batch size of 128 with 34 epochs was applied in the 10 k dataset, with a batch size of 64 with 200 epochs in the case of the 5 k dataset.
The proposed model outperformed the compared models because its pruning neural networks reduced computational complexity and training inputs to some extent. The model was trained using 34 epochs instead of 100 epochs with a GPU, as the model took less than 12 h to complete the training process and produce the results, whereas prior models [
34] took two or more days to complete the training process. After these epochs, the model showed no improvement in performance.
3.4. Results Using 10 K Dataset
The CNN accuracy plot in
Figure 5 shows the training process of the model for 34 epochs for both the multi-ROI and single-ROI datasets. The model learns gradually as training progresses, although its accuracy graphs in both datasets exhibit variance towards the conclusion. Multi-ROI data training and validation accuracy fluctuate, whereas the accuracy of single-ROI training and validation increases steadily as the number of epochs increases. The model traverses the same epochs to compute model loss from the plot. The validation loss of the model was initially substantial, but as the number of epochs increased, the quantity of loss dropped. The results of the proposed model are varied in comparison to previously reported results.
Table 3 displays multi-ROI and single-ROI classification reports of classes using the CNN, which shows a precision of 0.92, recall of 0.96, and F1 score of 0.94, for the moderate class using multi-ROI data. The precision for the severe class is 0.94, while the F1 score for the mild and moderate classes are also 0.94. The precision, recall, and F1 score for the normal class are 0.98, 1.00 and 0.99, respectively. The average accuracy for the multi-ROI data using 10 k data is 0.96, which is slightly lower than that achieved using 12.5 k data.
The weighted average for precision, recall, and F1 scores is 0.98 each for single-ROI data using 10 k data. The precision for the normal class is 0.97, while the recall and F1 scores for the severe class and F1 score for normal class is 0.98. For the moderate class, precision, recall, and F1 scores are 0.96 each. The CNN model achieves a 0.98 accuracy score for a single-ROI dataset with 10 k data.
3.5. Results Using 5 K Dataset
Figure 6 shows the training and validation accuracy and loss for the models using 200 epochs for both multi-ROI and single-ROI datasets. In this scenario, the model yields the lowest results compared to the other datasets. The model learns slowly as training proceeds, with the number of epochs increasing. With the increase in epochs, the training accuracy outperforms validation accuracy. Both datasets have a validation accuracy of 86.30% for multi-ROI and 93.15% for single-ROI. To depict the model loss, it went through the same number of epochs. The validation loss of the model was initially significant, but as the number of epochs increased, the amount of loss decreased slightly.
Table 4 illustrates the multi-ROI and single-ROI classification report produced by the CNN model, yielding a precision of 0.74 and an F1 score of 0.79 for the moderate class. The precision, recall, and F1 scores for the mild class are all 0.81, whereas those for normal and moderate classes are 0.84 and 0.85, respectively. The precision and recall for severe and normal classes are 0.86, 0.89, and 0.96 and 0.84, respectively.
Results for the single-ROI dataset indicate that for the normal and moderate classes, the recall and precision score is 0.86. The precision for the severe class is 0.89, while the recall and F1 scores are 0.99 and 0.94, respectively. In terms of accuracy, the CNN model achieves 0.86 and 0.92 accuracy scores for all multi-ROI and single-ROI classes, respectively. This performance is substantially lower compared to results achieved using a 12.5 k dataset.
3.9. Discussion
Over the last decade, several CAD approaches [
41,
42,
43,
44,
45] have been investigated for their potential to address the challenges of spinal MRI interpretation and full automation the LSS diagnostic procedure, which could help to improve detection accuracy. In this regard, the CNN model is often employed with medical imaging modalities such as MRI and computed tomography (CT), with a high success rate. Several previously proposed approaches for neural foraminal stenosis disease detection using binary and multigraded (normal, mild, moderate, and severe) classification are discussed herein.
Among the most current diagnostic frameworks for LSS is that proposed by Natalia et al. [
20], who used the SegNet model to automatically assess the area between the anterior and posterior (AAP) diameter and foraminal widths in MRI-, T1-, and T2-weighted composite images. Six ROIs were extracted after semantic segmentation, including intervertebral disc (IVD), posterior element (PE), and thecal sac (TS), as well as auxiliary ROIs, such as AAP and others. The contour evaluation technique was used to increase the accuracy of the segmentation result in specified ROIs. The results demonstrate a 96.7% diameter agreement with the expert. Similarly, Sartoretti’s classification [
21] is based on a six-point grading system for detecting lumbar foraminal stenosis (FS) on MRI images of high resolution. Grade A has no FS. The superior, posterior, inferior, and anterior boundaries of the lumbar foramen are graded B, C, D, and E, respectively, indicating nerve root contact with surrounding anatomical structures. The existence of FS in the nerve root with morphological changes was graded F in this research, in which we employed sagittal high-resolution T1-weighted and T2-weighted MRI data from 101 subjects.
A study in regard to grading of CAD systems by Salehi et al. [
22] showed that a CNN can be utilized to diagnose disc herniation using MRI images. A performance evaluation was carried out for normal, bulge, protrusion, and extrusion images. The experiment was performed on 2329 axial-view lumbar MRI datasets collected from a local medical center. Experimental results reported an 87.75% accuracy with data augmentation. Lu et al. [
38] used the U-Net architecture of the CNN model to grade central and FS as normal, mild, moderate, or severe based on both sagittal and axial MRI images. A large-scale dataset of 22,796 was used, which included data from 4075 patients. An accuracy of 94% was reported for this study.
A different technique proposed by Han et al. [
40] localizes six vertebrae and disc T12 to S1 using a deep multiscale multitask learning network (DMML-Net) that integrated into a full convolution network that grades the lumbar neural FS into normal and abnormal cases. The experimental setup included a dataset comprising 200 T1- and T2-weighted MRI images from 200 patients, achieving an accuracy of 84.5% using the proposed approach. An approach recently proposed by Hallinan et al. [
35] is to classify neural foraminal stenosis into normal, mild, moderate, or severe classes using a deep learning CNN model that achieved 84.5% accuracy using a dataset of T2-weighted axial MRI images and T1-weighted sagittal MRI images from 446 patients.
Using a deep learning ResNet-50 model, multitask classification was performed in [
36], which demonstrated the automated grading of lumbar disc herniation (LDH), lumbar central canal stenosis (LCCS), and lumbar nerve roots compression (LNRC) in lumbar axial MRIs. An internal test dataset and an external test dataset were used for classification systems with four graded levels (grade 0, grade 1, grade 2, and grade 3). A total of 1115 patients (1015 patients from the internal dataset and 100 patients from the external test dataset) were evaluated, and the best MRI slices were obtained. The efficiency of the model on the given datasets was evaluated using precision, accuracy, sensitivity, specificity, F1 scores, confusion matrices, receiver operating characteristics, and inter-rater agreement (Gwet k). On the internal test dataset, the overall grading accuracy for LDH, LCCS, and LNRC were 84.17%, 86.99%, and 81.21%, respectively. For the external test data, 74.16%, 79.65%, and 81.21% accuracy are reported for LDH, LCCS, and LNRC, respectively.
Bharadwaj et al. [
46] utilized a V-Net model to segment the dural sac and IVD and localize the facet and foramen. Big transfer (BiT) models were trained for classification tasks. Multievaluation metrics including Cohen’s Kappa score were used for the dural sac and IVD. The authors used axial T2-weighted MRI images of the lumbar spine obtained between 2008 and 2019. The area under the receiver operator characteristic curve (AUROC) values used for the binary classification of facet and neural foraminal stenosis were 0.92 and 0.93, respectively. Sinan et al. [
37] proposed an LSS-VGG16 and U-Net model that detects LSS in MR and CT images and achieved 87.70% classification accuracy on VGG16. A total of 1560 MR images were used with U-Net, with a 0.93 DICE score.
The authors of [
47] a 3D LSS segmentation framework that enables the complete determination of the regions of the body that cannot be fully opened during LSS surgeries, particularly in the nerve roots. The spinal disc, canal, thecal sac, posterior element, and other regions and backgrounds in the image that are crucial for LSS were all segmented and divided into a total of six classes in MRI images. The intersection over union (IoU) metric was deployed for each class to assess the success of segmentation, since the canal had an IoU value of 0.61. The study employed T2 sequence lumbar MRI images of 300 LSS patients in the digital imaging and communications in medicine (DICOM) format.
Abhinav et al. [
48] also recently presented a U-Net-dependent CNN model to segment the IVD, PE, TS, and AAP regions of LSS on an axial MRI dataset [
10] and performed binary classification. The performance of the model was evaluated by IoU metrics. Since IVD is the simplest region to label and PE has a particular shape that resembles the letter Y, the values of regions like IVD, PE, and IoU vary between 0.80 and 1.0. And because AAP was the most challenging to identify, its IoU metric value is 0.6568, which is lower than that of the other regions.
Another innovative study [
39] compared conventional and ultrafast methods and analyzed sagittal T1-weighted, T2-weighted, short-TI inversion recovery, and axial T2-weighted MRI images of 58 patients. Cohen’s kappa metrics were used to assess foraminal stenosis in axial images, and the results were provided in a multigraded classification. The accuracy obtained using this method was 95%.
In this study, we investigated LSS detection using a customized CNN model. We evaluated the algorithm’s performance using a variety of metrics. Experiments were conducted using two datasets and with and without augmentation techniques using different data values. Multi-ROI and single-ROI datasets with 5 k achieved the lowest results in terms of accuracy scores: 0.85 and 0.92, respectively. The cure of model accuracy shows that the model could be trained more to prevent underfitting and inflection because the model was not overlearned for the training set. Due to inadequate training, the model loss exhibits a divergence from the training curve, which indicates why the overall loss is large in the results of both datasets.
The two 10 k datasets achieved accuracy scores of 0.96 and 0.98, respectively.
Figure 6 demonstrates that the trained model fit well; however, the validation curve is slightly unsatisfactory, owing to underfitting for the multi-ROI dataset, requiring more training data samples to improve accuracy. The model loss curve shows that training significantly decreased the loss, although it remains high during the initial epochs.
Table 5 illustrates that the accuracy is similar between the two 12.5 k datasets: 0.97 and 0.98, respectively. The model accuracy curve show in
Figure 4 indicates that while model training performs well on a single-ROI dataset, results on a multi-ROI dataset might be further enhanced by adding more training data and by further reducing model loss.
Table 8 shows an analytical summary of the discussed research works. It can be observed that for LSS detection and segmentation, the models suffer from low accuracy. The CNN model and its variants were tested, yet no CNN technique was able to more accurately categorize LSS disease, necessitating the development of automatic methods that better classify the disease.