The Antioxidant Drug Edaravone Binds to the Aryl Hydrocarbon Receptor (AHR) and Promotes the Downstream Signaling Pathway Activation
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Purified OPC Cultures
2.3. Transcriptome Analysis
2.4. RNA Extraction and Quantitative (q)PCR
2.5. Preparation of Proteins for Docking
2.6. Preparation of Ligands for Docking
2.7. Docking Studies
2.8. Molecular Dynamics
2.9. SH-SY5Y Cell Cultures and Treatments
2.10. Protein Extract Preparation and Western Blotting
2.11. Drug Treatments on Fish
2.12. Statistical Analysis
3. Results
3.1. Edaravone Increases the Expression of AHR-Related Target Genes in Primary Mouse OPCs
3.2. Edaravone Is Predicted to Be an AHR Ligand
3.3. Edaravone Induces AHR Nuclear Translocation and AHR Target Gene Expression in the SH-SY5Y Neuroblastoma Cell Line
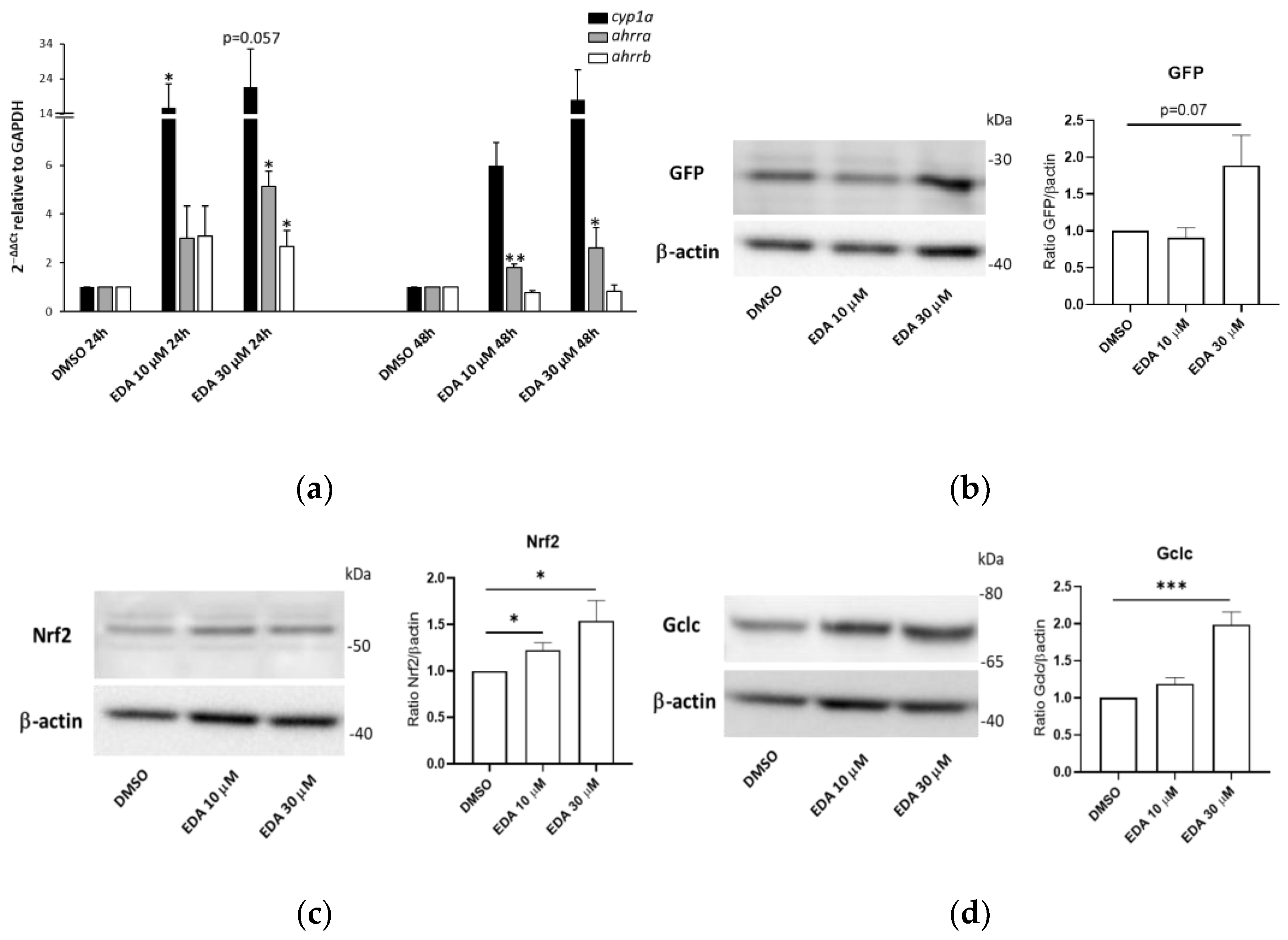
3.4. Edaravone Promotes Activation of the AHR and NRF2 Pathways and Olig2 Transgene Expression in Zebrafish Larvae
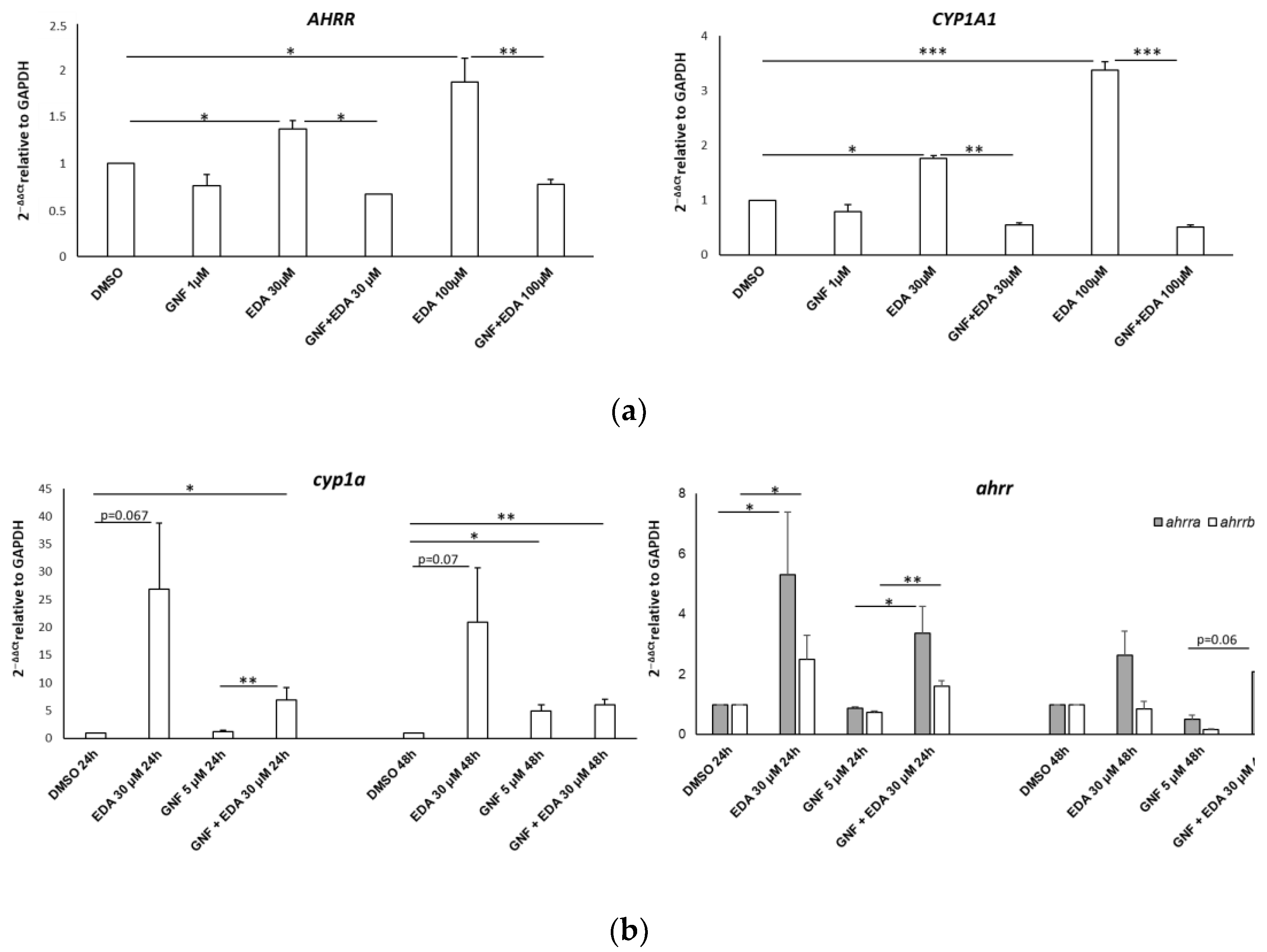
3.5. Edaravone-Mediated Induction of CYP Genes Is Dampened by the AHR Antagonist GNF-351 in SH-SY5Y Cells and Zebrafish
3.6. GNF-351 Competes with Edaravone for the Same AHR Binding Site
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cho, H.; Shukla, S. Role of Edaravone as a Treatment Option for Patients with Amyotrophic Lateral Sclerosis. Pharmaceuticals 2020, 14, 29. [Google Scholar] [CrossRef] [PubMed]
- Michaličková, D.; Kübra Öztürk, H.; Hroudová, J.; L’Upták, M.; Kučera, T.; Hrnčíř, T.; Kutinová Canová, N.; Šíma, M.; Slanař, O. Edaravone Attenuates Disease Severity of Experimental Auto-Immune Encephalomyelitis and Increases Gene Expression of Nrf2 and HO-1. Physiol. Res. 2022, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiari, M.; Ghasemi, N.; Salehi, H.; Amirpour, N.; Kazemi, M.; Mardani, M. Evaluation of Edaravone Effects on the Differentiation of Human Adipose Derived Stem Cells into Oligodendrocyte Cells in Multiple Sclerosis Disease in Rats. Life Sci. 2021, 282, 119812. [Google Scholar] [CrossRef]
- Li, B.; Yu, D.; Xu, Z. Edaravone Prevents Neurotoxicity of Mutant L166P DJ-1 in Parkinson’s Disease. J. Mol. Neurosci. 2013, 51, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shao, R.; Li, M.; Yang, G. Edaravone Protects Neurons in the Rat Substantia Nigra Against 6-Hydroxydopamine-Induced Oxidative Stress Damage. Cell Biochem. Biophys. 2014, 70, 1247–1254. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.-S.; Yao, X.-Q.; Liu, Y.-H.; Wang, Q.-H.; Zeng, F.; Lu, J.-J.; Liu, J.; Zhu, C.; Shen, L.-L.; Liu, C.-H.; et al. Edaravone Alleviates Alzheimer’s Disease-Type Pathologies and Cognitive Deficits. Proc. Natl. Acad. Sci. USA 2015, 112, 5225–5230. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, L.; Chen, X.; Li, R.; Yu, B.; Pan, M.; Fang, L.; Li, J.; Cui, X.; Yao, M.; et al. Neuroprotective Effect and Possible Mechanism of Edaravone in Rat Models of Spinal Cord Injury: A Protocol for a Systematic Review and Meta-Analysis. Syst. Rev. 2023, 12, 177. [Google Scholar] [CrossRef] [PubMed]
- Oosthoek, M.; Lili, A.; Almeida, A.; Van Loosbroek, O.; Van Der Geest, R.; De Greef-van Der Sandt, I.; Van Bokhoven, P.; Sikkes, S.A.M.; Teunissen, C.E.; Vijverberg, E.G.B. ASURE Clinical Trial Protocol: A Randomized, Placebo-Controlled, Proof-of-Concept Study Aiming to Evaluate Safety and Target Engagement Following Administration of TW001 in Early Alzheimer’s Disease Patients. J. Prev. Alzheimer’s Dis. 2023, 10, 669–674. [Google Scholar] [CrossRef]
- Watanabe, K.; Tanaka, M.; Yuki, S.; Hirai, M.; Yamamoto, Y. How Is Edaravone Effective against Acute Ischemic Stroke and Amyotrophic Lateral Sclerosis? J. Clin. Biochem. Nutr. 2018, 62, 20–38. [Google Scholar] [CrossRef] [PubMed]
- Olufunmilayo, E.O.; Gerke-Duncan, M.B.; Holsinger, R.M.D. Oxidative Stress and Antioxidants in Neurodegenerative Disorders. Antioxidants 2023, 12, 517. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, C.; Meng, X.; Li, Z.; Lv, C.; Cao, P. Neuroprotection of Edaravone on the Hippocampus of Kainate-Induced Epilepsy Rats through Nrf2/HO-1 Pathway. Neurochem. Int. 2018, 112, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Shou, L.; Bei, Y.; Song, Y.; Wang, L.; Ai, L.; Yan, Q.; He, W. Nrf2 Mediates the Protective Effect of Edaravone after Chlorpyrifos-induced Nervous System Toxicity. Environ. Toxicol. 2019, 34, 626–633. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Guo, Y.; Wang, H.; Zhao, L.; Ma, Z.; Li, T.; Liu, J.; Sun, M.; Jian, Y.; Yao, L.; et al. Edaravone Reduces Aβ-Induced Oxidative Damage in SH-SY5Y Cells by Activating the Nrf2/ARE Signaling Pathway. Life Sci. 2019, 221, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Jangra, A.; Kwatra, M.; Singh, T.; Pant, R.; Kushwah, P.; Ahmed, S.; Dwivedi, D.; Saroha, B.; Lahkar, M. Edaravone Alleviates Cisplatin-Induced Neurobehavioral Deficits via Modulation of Oxidative Stress and Inflammatory Mediators in the Rat Hippocampus. Eur. J. Pharmacol. 2016, 791, 51–61. [Google Scholar] [CrossRef]
- Okuyama, S.; Morita, M.; Sawamoto, A.; Terugo, T.; Nakajima, M.; Furukawa, Y. Edaravone Enhances Brain-Derived Neurotrophic Factor Production in the Ischemic Mouse Brain. Pharmaceuticals 2015, 8, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Zhu, W.; Kong, W.; Li, T.; Zou, P.; Chen, H. Edaravone Attenuates Neuronal Apoptosis in Hippocampus of Rat Traumatic Brain Injury Model via Activation of BDNF/TrkB Signaling Pathway. Arch. Med. Sci. 2021, 17, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yi, J.; Pan, M.; Hu, B.; Duan, H. Edaravone Alleviated Propofol-Induced Neural Injury in Developing Rats by BDNF/TrkB Pathway. J. Cell Mol. Med. 2021, 25, 4974–4987. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-T.; Yu, Y.; Li, X.-X.; Lang, X.-Y.; Gu, R.-Z.; Fan, S.-R.; Fang, X.; Bai, J.-P.; Lan, R.; Qin, X.-Y. Edaravone Attenuates H2O2 or Glutamate-Induced Toxicity in Hippocampal Neurons and Improves AlCl3/D-Galactose Induced Cognitive Impairment in Mice. NeuroToxicology 2021, 85, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Eleuteri, C.; Olla, S.; Veroni, C.; Umeton, R.; Mechelli, R.; Romano, S.; Buscarinu, M.; Ferrari, F.; Calò, G.; Ristori, G.; et al. A Staged Screening of Registered Drugs Highlights Remyelinating Drug Candidates for Clinical Trials. Sci. Rep. 2017, 7, 45780. [Google Scholar] [CrossRef] [PubMed]
- Takase, H.; Liang, A.C.; Miyamoto, N.; Hamanaka, G.; Ohtomo, R.; Maki, T.; Pham, L.-D.D.; Lok, J.; Lo, E.H.; Arai, K. Protective Effects of a Radical Scavenger Edaravone on Oligodendrocyte Precursor Cells against Oxidative Stress. Neurosci. Lett. 2018, 668, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Xu, H.; Xu, L.; Jiang, W.; Chen, C.; Chang, Y.; Liu, C.; Tian, Z.; Qiu, X.; Xie, C.; et al. Remyelination in Neuromyelitis Optica Spectrum Disorder Is Promoted by Edaravone through MTORC1 Signaling Activation. Glia 2023, 71, 284–304. [Google Scholar] [CrossRef] [PubMed]
- Colombo, E.; Olla, S.; Minnelli, C.; Formato, A.; Veroni, C.; Corbisiero, S.; Pericolo, M.; Siguri, C.; Mobbili, G.; Agresti, C.; et al. Synthesis and Characterization of Edaravone Analogues as Remyelinating Agents and Putative Mechanistic Probes. Molecules 2023, 28, 6928. [Google Scholar] [CrossRef] [PubMed]
- Agresti, C.; D’Urso, D.; Levi, G. Reversible Inhibitory Effects of Interferon-Gamma and Tumour Necrosis Factor-Alpha on Oligodendroglial Lineage Cell Proliferation and Differentiation In Vitro. Eur. J. Neurosci. 1996, 8, 1106–1116. [Google Scholar] [CrossRef] [PubMed]
- Gruszczyk, J.; Grandvuillemin, L.; Lai-Kee-Him, J.; Paloni, M.; Savva, C.G.; Germain, P.; Grimaldi, M.; Boulahtouf, A.; Kwong, H.-S.; Bous, J.; et al. Cryo-EM Structure of the Agonist-Bound Hsp90-XAP2-AHR Cytosolic Complex. Nat. Commun. 2022, 13, 7010. [Google Scholar] [CrossRef]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Banks, J.L.; Beard, H.S.; Cao, Y.; Cho, A.E.; Damm, W.; Farid, R.; Felts, A.K.; Halgren, T.A.; Mainz, D.T.; Maple, J.R.; et al. Integrated Modeling Program, Applied Chemical Theory (IMPACT). J. Comput. Chem. 2005, 26, 1752–1780. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert. Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Abel, R.; Zhu, K.; Cao, Y.; Zhao, S.; Friesner, R.A. The VSGB 2.0 Model: A next Generation Energy Model for High Resolution Protein Structure Modeling. Proteins 2011, 79, 2794–2812. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Gasteiger, J.; Marsili, M. A New Model for Calculating Atomic Charges in Molecules. Tetrahedron Lett. 1978, 19, 3181–3184. [Google Scholar] [CrossRef]
- Sallem, M.A.S.; Sousa, S.A.J.D. AutoGrid: Towards an Autonomic Grid Middleware. In Proceedings of the 16th IEEE International Workshops on Enabling Technologies: Infrastructure for Collaborative Enterprises (WETICE 2007), Paris, France, 18–20 June 2007; IEEE: Evry Cedex, France, 2007; pp. 223–228. [Google Scholar] [CrossRef]
- Moitessier, N.; Englebienne, P.; Lee, D.; Lawandi, J.; Corbeil, C.R. Towards the Development of Universal, Fast and Highly Accurate Docking/Scoring Methods: A Long Way to Go. Br. J. Pharmacol. 2008, 153, S7–S26. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr. Comput.-Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the ACM/IEEE SC 2006 Conference (SC’06), Tampa, FL, USA, 11–17 November 2006; p. 43. [Google Scholar]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical Dynamics: Equilibrium Phase-Space Distributions. Phys. Rev. A 1985, 31, 1695–1697. [Google Scholar] [CrossRef] [PubMed]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant Pressure Molecular Dynamics Algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Frey, B.J.; Dueck, D. Clustering by Passing Messages Between Data Points. Science 2007, 315, 972–976. [Google Scholar] [CrossRef] [PubMed]
- Mallozzi, C.; Pepponi, R.; Visentin, S.; Chiodi, V.; Lombroso, P.J.; Bader, M.; Popoli, P.; Domenici, M.R. The Activity of the Striatal-enriched Protein Tyrosine Phosphatase in Neuronal Cells Is Modulated by Adenosine A2A Receptor. J. Neurochem. 2020, 152, 284–298. [Google Scholar] [CrossRef] [PubMed]
- Morel, Y.; Mermod, N.; Barouki, R. An Autoregulatory Loop Controlling CYP1A1 Gene Expression: Role of H2O2 and NFI. Mol. Cell. Biol. 1999, 19, 6825–6832. [Google Scholar] [CrossRef] [PubMed]
- Nebert, D.W.; Dalton, T.P.; Okey, A.B.; Gonzalez, F.J. Role of Aryl Hydrocarbon Receptor-Mediated Induction of the CYP1 Enzymes in Environmental Toxicity and Cancer. J. Biol. Chem. 2004, 279, 23847–23850. [Google Scholar] [CrossRef] [PubMed]
- Imran, S.; Ferretti, P.; Vrzal, R. Different Regulation of Aryl Hydrocarbon Receptor-Regulated Genes in Response to Dioxin in Undifferentiated and Neuronally Differentiated Human Neuroblastoma SH-SY5Y Cells. Toxicol. Mech. Methods 2015, 25, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Miao, W.; Hu, L.; Scrivens, P.J.; Batist, G. Transcriptional Regulation of NF-E2 P45-Related Factor (NRF2) Expression by the Aryl Hydrocarbon Receptor-Xenobiotic Response Element Signaling Pathway. J. Biol. Chem. 2005, 280, 20340–20348. [Google Scholar] [CrossRef] [PubMed]
- Badenetti, L.; Manzoli, R.; Rubin, M.; Cozza, G.; Moro, E. Monitoring Nrf2/ARE Pathway Activity with a New Zebrafish Reporter System. Int. J. Mol. Sci. 2023, 24, 6804. [Google Scholar] [CrossRef] [PubMed]
- Sekhar, K.R.; Crooks, P.A.; Sonar, V.N.; Friedman, D.B.; Chan, J.Y.; Meredith, M.J.; Starnes, J.H.; Kelton, K.R.; Summar, S.R.; Sasi, S.; et al. NADPH Oxidase Activity Is Essential for Keap1/Nrf2-Mediated Induction of GCLC in Response to 2-Indol-3-Yl-Methylenequinuclidin-3-Ols. Cancer Res. 2003, 63, 5636–5645. [Google Scholar] [PubMed]
- Shackleford, G.; Sampathkumar, N.K.; Hichor, M.; Weill, L.; Meffre, D.; Juricek, L.; Laurendeau, I.; Chevallier, A.; Ortonne, N.; Larousserie, F.; et al. Involvement of Aryl Hydrocarbon Receptor in Myelination and in Human Nerve Sheath Tumorigenesis. Proc. Natl. Acad. Sci. USA 2018, 115, E1319–E1328. [Google Scholar] [CrossRef]
- Nellessen, A.; Nyamoya, S.; Zendedel, A.; Slowik, A.; Wruck, C.; Beyer, C.; Fragoulis, A.; Clarner, T. Nrf2 Deficiency Increases Oligodendrocyte Loss, Demyelination, Neuroinflammation and Axonal Damage in an MS Animal Model. Metab. Brain Dis. 2020, 35, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Park, H.-C.; Topczewska, J.M.; Mawdsley, D.J.; Appel, B. Neural Cell Fate Analysis in Zebrafish Using Olig2 BAC Transgenics. Methods Cell Sci. 2003, 25, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.J.; Murray, I.A.; Tanos, R.; Tellew, J.; Boitano, A.E.; Bisson, W.H.; Kolluri, S.K.; Cooke, M.P.; Perdew, G.H. Identification of a High-Affinity Ligand That Exhibits Complete Aryl Hydrocarbon Receptor Antagonism. J. Pharmacol. Exp. Ther. 2011, 338, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Avilla, M.N.; Malecki, K.M.C.; Hahn, M.E.; Wilson, R.H.; Bradfield, C.A. The Ah Receptor: Adaptive Metabolism, Ligand Diversity, and the Xenokine Model. Chem. Res. Toxicol. 2020, 33, 860–879. [Google Scholar] [CrossRef]
- Rothhammer, V.; Quintana, F.J. The Aryl Hydrocarbon Receptor: An Environmental Sensor Integrating Immune Responses in Health and Disease. Nat. Rev. Immunol. 2019, 19, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Chen, R.; Chen, X.; Huang, B.; Lin, R.; Xie, X.; Chen, J.; Jiang, J.; Deng, Y.; Wen, J. AhR Regulates the Expression of Human Cytochrome P450 1A1 ( CYP1A1 ) by Recruiting Sp1. FEBS J. 2019, 286, 4215–4231. [Google Scholar] [CrossRef] [PubMed]
- Großkopf, H.; Walter, K.; Karkossa, I.; Von Bergen, M.; Schubert, K. Non-Genomic AhR-Signaling Modulates the Immune Response in Endotoxin-Activated Macrophages after Activation by the Environmental Stressor BaP. Front. Immunol. 2021, 12, 620270. [Google Scholar] [CrossRef] [PubMed]
- Stading, R.; Chu, C.; Couroucli, X.; Lingappan, K.; Moorthy, B. Molecular Role of Cytochrome P4501A Enzymes in Oxidative Stress. Curr. Opin. Toxicol. 2020, 20–21, 77–84. [Google Scholar] [CrossRef]
- Wang, G.; Zeng, X.; Gong, S.; Wang, S.; Ge, A.; Liu, W.; Ge, J.; He, Q. Exploring the Mechanism of Edaravone for Oxidative Stress in Rats with Cerebral Infarction Based on Quantitative Proteomics Technology. Evid. Based Complement. Altern. Med. 2022, 2022, 8653697. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.R.; Patel, R.; Kossack, M.E.; Tian, L.; Camarillo, M.A.; Cintrón-Rivera, L.G.; Gawdzik, J.C.; Yue, M.S.; Nwagugo, F.O.; Elemans, L.M.H.; et al. Proper Modulation of AHR Signaling Is Necessary for Establishing Neural Connectivity and Oligodendrocyte Precursor Cell Development in the Embryonic Zebrafish Brain. Front. Mol. Neurosci. 2022, 15, 1032302. [Google Scholar] [CrossRef] [PubMed]
- Juricek, L.; Carcaud, J.; Pelhaitre, A.; Riday, T.T.; Chevallier, A.; Lanzini, J.; Auzeil, N.; Laprévote, O.; Dumont, F.; Jacques, S.; et al. AhR-Deficiency as a Cause of Demyelinating Disease and Inflammation. Sci. Rep. 2017, 7, 9794. [Google Scholar] [CrossRef] [PubMed]
- Majumder, S.; Kono, M.; Lee, Y.T.; Byrnes, C.; Li, C.; Tuymetova, G.; Proia, R.L. A Genome-Wide CRISPR/Cas9 Screen Reveals That the Aryl Hydrocarbon Receptor Stimulates Sphingolipid Levels. J. Biol. Chem. 2020, 295, 4341–4349. [Google Scholar] [CrossRef] [PubMed]
- Grishanova, A.Y.; Perepechaeva, M.L. Aryl Hydrocarbon Receptor in Oxidative Stress as a Double Agent and Its Biological and Therapeutic Significance. Int. J. Mol. Sci. 2022, 23, 6719. [Google Scholar] [CrossRef] [PubMed]
- Tsaktanis, T.; Beyer, T.; Nirschl, L.; Linnerbauer, M.; Grummel, V.; Bussas, M.; Tjon, E.; Mühlau, M.; Korn, T.; Hemmer, B.; et al. Aryl Hydrocarbon Receptor Plasma Agonist Activity Correlates with Disease Activity in Progressive MS. Neurol. Neuroimmunol. Neuroinflamm 2021, 8, e933. [Google Scholar] [CrossRef]
- Rothhammer, V.; Kenison, J.E.; Li, Z.; Tjon, E.; Takenaka, M.C.; Chao, C.-C.; Alves De Lima, K.; Borucki, D.M.; Kaye, J.; Quintana, F.J. Aryl Hydrocarbon Receptor Activation in Astrocytes by Laquinimod Ameliorates Autoimmune Inflammation in the CNS. Neurol. Neuroimmunol. Neuroinflamm 2021, 8, e946. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, E.F.; Saili, K.S.; Koch, D.C.; Kopparapu, P.R.; Farrer, D.; Bisson, W.H.; Mathew, L.K.; Sengupta, S.; Kerkvliet, N.I.; Tanguay, R.L.; et al. The Anti-Inflammatory Drug Leflunomide Is an Agonist of the Aryl Hydrocarbon Receptor. PLoS ONE 2010, 5, e13128. [Google Scholar] [CrossRef] [PubMed]
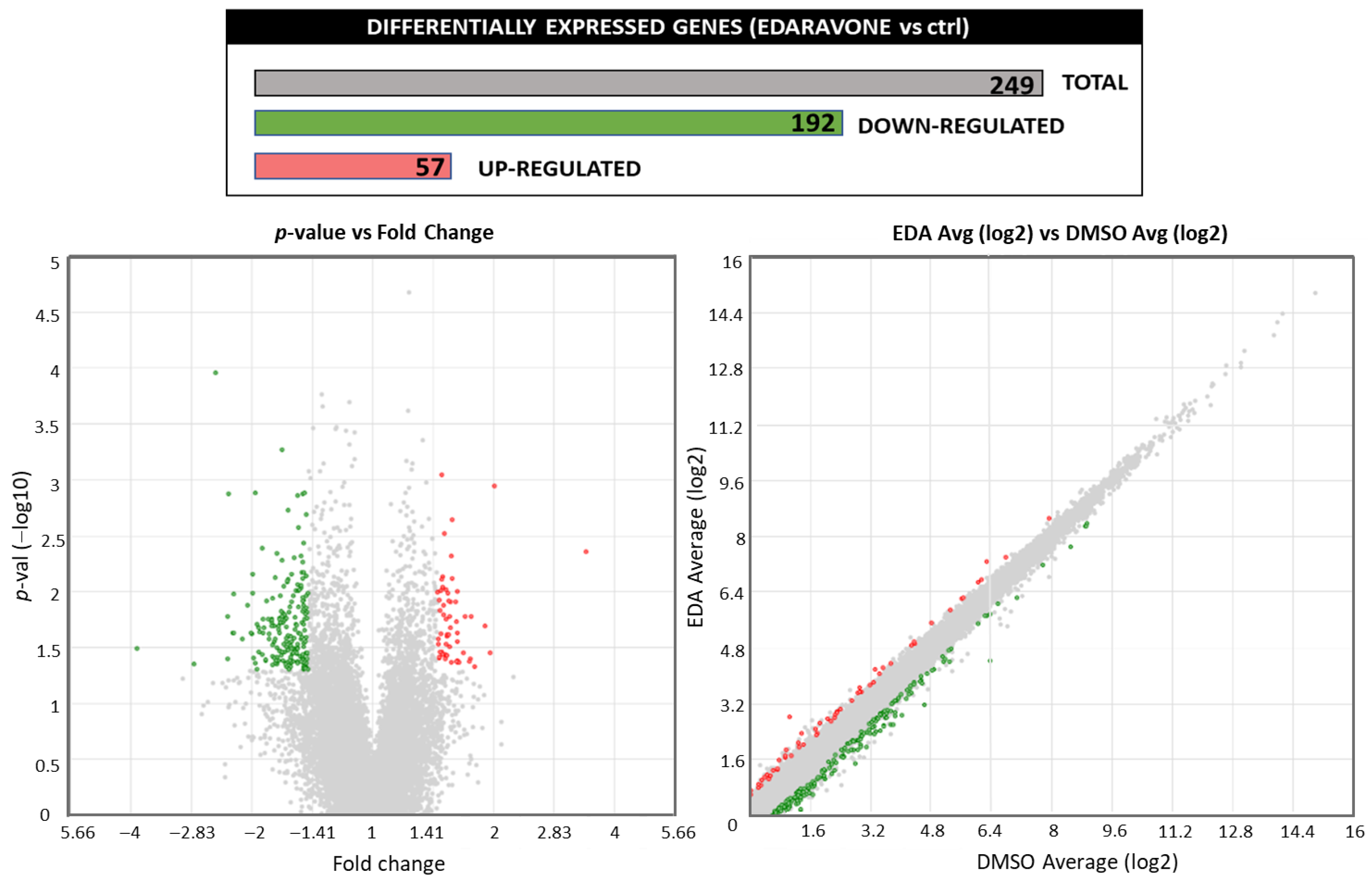





| Category | Term | Count | % | p-Value | |
|---|---|---|---|---|---|
| UP-REGULATED | REACTOME_PATHWAY | Cytochrome P450—arranged by substrate type | 3 | 5.5 | 1.1 × 10−2 |
| REACTOME_PATHWAY | Synthesis of epoxy (EET) and dihydroxyeicosatrienoic acids (DHET) | 2 | 3.6 | 1.5 × 10−2 | |
| REACTOME_PATHWAY | Synthesis of (16-20)-hydroxyeicosatetraenoic acids (HETE) | 2 | 3.6 | 1.8 × 10−2 | |
| REACTOME_PATHWAY | Phase I—Functionalization of compounds | 3 | 5.5 | 2.3 × 10−2 | |
| DOWN-REGULATED | KEGG_PATHWAY | Phagosome | 5 | 2.9 | 4.7 × 10−2 |
| KEGG_PATHWAY | PI3K-Akt signaling pathway | 7 | 4.1 | 5 × 10−2 | |
| REACTOME_PATHWAY | Mitotic prometaphase | 8 | 4.7 | 1.3 × 10−3 | |
| REACTOME_PATHWAY | Metabolism of water-soluble vitamins and cofactors | 5 | 2.9 | 1.4 × 10−2 | |
| REACTOME_PATHWAY | Metabolism of vitamins and cofactors | 6 | 3.5 | 1.5 × 10−2 | |
| REACTOME_PATHWAY | Nucleotide catabolism | 3 | 1.7 | 3.8 × 10−2 | |
| REACTOME_PATHWAY | Organelle biogenesis and maintenance | 6 | 3.5 | 4.5 × 10−2 | |
| REACTOME_PATHWAY | M Phase | 8 | 4.7 | 4.9 × 10−2 | |
| WIKIPATHWAYS | Translation factors | 4 | 2.3 | 6.4 × 10−3 | |
| WIKIPATHWAYS | Focal adhesion: PI3K-Akt-mTOR signaling pathway | 7 | 4.1 | 3.7 × 10−2 |
| Compound | XP-GScore Glide XP kcal/mol | MMGBSA_dGbind Prime kcal/mol | Binding Energy (BE) Autodock kcal/mol |
|---|---|---|---|
| Indirubin | −11.33 | −67.57 | −9.08 |
| Leflunomide | −9.074 | −51.34 | −7.17 |
| Edaravone | −7.55 | −45.03 | −5.97 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veroni, C.; Olla, S.; Brignone, M.S.; Siguri, C.; Formato, A.; Marra, M.; Manzoli, R.; Macario, M.C.; Ambrosini, E.; Moro, E.; et al. The Antioxidant Drug Edaravone Binds to the Aryl Hydrocarbon Receptor (AHR) and Promotes the Downstream Signaling Pathway Activation. Biomolecules 2024, 14, 443. https://doi.org/10.3390/biom14040443
Veroni C, Olla S, Brignone MS, Siguri C, Formato A, Marra M, Manzoli R, Macario MC, Ambrosini E, Moro E, et al. The Antioxidant Drug Edaravone Binds to the Aryl Hydrocarbon Receptor (AHR) and Promotes the Downstream Signaling Pathway Activation. Biomolecules. 2024; 14(4):443. https://doi.org/10.3390/biom14040443
Chicago/Turabian StyleVeroni, Caterina, Stefania Olla, Maria Stefania Brignone, Chiara Siguri, Alessia Formato, Manuela Marra, Rosa Manzoli, Maria Carla Macario, Elena Ambrosini, Enrico Moro, and et al. 2024. "The Antioxidant Drug Edaravone Binds to the Aryl Hydrocarbon Receptor (AHR) and Promotes the Downstream Signaling Pathway Activation" Biomolecules 14, no. 4: 443. https://doi.org/10.3390/biom14040443
APA StyleVeroni, C., Olla, S., Brignone, M. S., Siguri, C., Formato, A., Marra, M., Manzoli, R., Macario, M. C., Ambrosini, E., Moro, E., & Agresti, C. (2024). The Antioxidant Drug Edaravone Binds to the Aryl Hydrocarbon Receptor (AHR) and Promotes the Downstream Signaling Pathway Activation. Biomolecules, 14(4), 443. https://doi.org/10.3390/biom14040443








