Novel Mutation in the Acetohydroxyacid Synthase (AHAS), Gene Confers Imidazolinone Resistance in Chickpea Cicer arietinum L. Plants
Abstract
:1. Introduction
2. Results
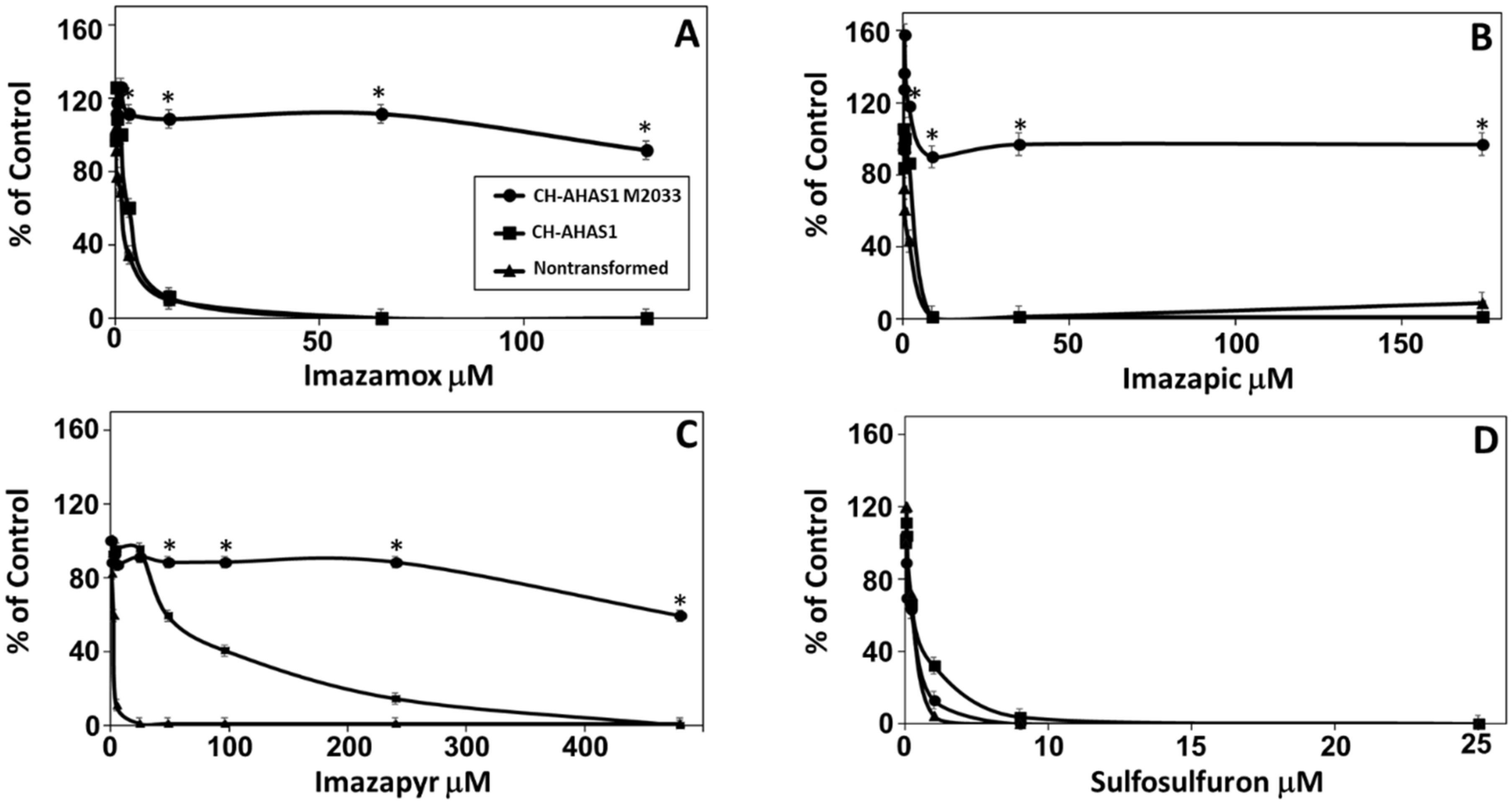
2.1. Cross-Resistance of Line M2033 to AHAS Inhibitors
2.2. Line M2033 Contains a Novel Mutation in the AHAS1 Gene
2.3. The Novel Mutation Confers AHAS Resistance in Transgenic Tobacco Plants
2.4. Mutation Inheritance
2.5. Mutation Changes AHAS1 Structure
3. Discussion
4. Materials and Methods
4.1. Mutagenesis and Characterization of M2033 Mutant
4.1.1. Mutagenesis
4.1.2. Screening for Imazamox Resistance
4.1.3. DNA Extraction and PCR Amplification
4.1.4. Genotype Determination
4.1.5. Determination of Cross-Resistance to AHAS Inhibitors
4.1.6. Determination of Mutation Inheritance
4.2. Confirmation of the Resistance Mechanism on Transgenic Plants
4.2.1. Construction of Chickpea AHAS Genes and Agrobacterium Transformation
4.2.2. Plant Transformation
4.2.3. Examination of IMI Resistance in Transgenic Plants
4.3. Mutation Changes AHAS1 Structure
4.3.1. Ligand–Protein Contact Analysis
4.3.2. Hydrophobic Cluster Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boydston, R.A.; Nelson, H.; Chaves-Cordoba, B. Tolerance of chickpeas to postemergence broadleaf herbicides. Weed Technol. 2018, 32, 190–194. [Google Scholar] [CrossRef]
- Kilinç, O. Aclonifen: The identikit of a widely used herbicide. Afr. J. Agric. Res. 2011, 6, 2411–2419. [Google Scholar]
- Regan, K.L.; Siddique, K.H.; Martin, L.D. Response of kabuli chickpea (Cicer artietinum) to sowing rate in Mediterranean–type environments of southwestern Australia. J. Exp. Agric. 2003, 43, 87–97. [Google Scholar] [CrossRef]
- Rizwan, M.; Akhtar, A.; Aslam, M.; Asghar, M.J. Development of herbicide crops through induced mutations. Adv. Life Sci. 2015, 3, 1–8. [Google Scholar]
- Duggleby, R.G.; Pang, S.S. Acetohydroxyacid synthase. J. Biochem. Mol. Biol. 2000, 33, 1–36. [Google Scholar]
- Iwakami, S.; Uchito, A.; Watanabe, H.; Yamasue, Y.; Inamura, T. Isolation and expression of genes for acetolactate synthase and acetyl-CoA carboxylase in Echinochloa phyllopogon, a polyploid weed species. Pest Manag. Sci. 2012, 68, 1098–1106. [Google Scholar] [CrossRef] [PubMed]
- Owen, M.J.; Goddin, D.E.; Powles, S.B. Identification of resistance to either paraquat or ALS-inhibiting herbicides in two Western Australian Hordeum leporinum biotypes. Pest Manag. Sci. 2012, 68, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Schloss, J.V. Recent advances in understanding the mechanism and inhibition of acetolactate synthase. In Herbicides Inhibiting Branch Chain Amino Acid Biosynthesis; Setter, J., Ed.; Springer: New York, NY, USA, 1995; pp. 4–11. [Google Scholar]
- Barak, Z.; Chipman, D.M. Allosteric regulation in acetohydroxyacid synthases (AHASs)–different structures and kinetic behavior in isozymes in the same organisms. Arch. Biochem. Biophys. 2012, 519, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Duggleby, R.G.; McCourt, J.A.; Guddat, L.W. Structure and mechanism of inhibition of plant acetohydroxyacid synthase. Plant Physiol. Biochem. 2008, 46, 309–324. [Google Scholar] [CrossRef]
- Senseman, S.A. Herbicide Handbook, 9th ed.; Weed Science Society of America: Lawrence, KS, USA, 2007; pp. 47–125. [Google Scholar]
- Han, X.J.; Dong, Y.; Sung, X.; Li, F.; Zheng, M. Molecular basis of resistance to tribenuron-methyl in Descurainia sophia (L.) populations from China. Pesticide Biochem. Physiol. 2012, 104, 77–81. [Google Scholar] [CrossRef]
- McCourt, J.A.; Pang, S.S.; King-Scott, J.; Guddat, L.W.; Duggleby, R.G. Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase. Proc. Natl. Acad. Sci. USA 2006, 103, 569–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, J.M. Review of glyphosate and ALS-inhibiting herbicide crop resistance and resistant weed management. Weed Technol. 2007, 21, 547–558. [Google Scholar] [CrossRef]
- Tan, S.; Evans, R.R.; Dahmer, M.L.; Singh, B.K.; Shaner, D.L. Imidazolinone-tolerant crops: History, current status, and future. Pest Manage. Sci. 2005, 61, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Westwood, J.H.; Whaley, C.M.; Wilson, H.M. A new mutation in plant ALS confers resistance to five classes of ALS-inhibiting herbicides. Weed Sci. 2007, 55, 83–90. [Google Scholar]
- Hershenhorn, J.; Goldwasser, Y.; Plakhine, D.; Ali, R.; Blumenfeld, T.; Bucsbaum, H.; Herzlinger, G.; Golan, S.; Chilf, T.; Eizenberg, H.; et al. Orobanche aegyptiaca control in tomato fields with sulfonylurea herbicides. Weed Res. 1998, 38, 343–349. [Google Scholar] [CrossRef]
- Hershenhorn, J.; Goldwasser, Y.; Plakhine, D.; Lavan, Y.; Herzlinger, G.; Golan, S.; Chilf, T.; Kleifeld, Y. Effect of sulfonylurea herbicides on Egyptian broomrape (Orobanche aegyptiaca) in tomato (Lycopersicon esculentum) under greenhouse conditions. Weed Technol. 1998, 12, 115–120. [Google Scholar] [CrossRef]
- Hershenhorn, J.; Plakhine, D.; Goldwasser, Y.; Westwood, J.H.; Foy, C.L.; Kleifeld, Y. Effect of sulfonylurea herbicides on early development of Egyptian broomrape (Orobanche aegyptiaca) in tomato (Lycopersicon esculentum). Weed Technol. 1998, 12, 108–114. [Google Scholar] [CrossRef]
- Eizenberg, H.; Hershenhorn, J.; Graph, S.; Manor, H. Orobanche aegyptiaca control in tomato with sulfonylurea herbicides. Acta Hort. 2003, 613, 205–208. [Google Scholar] [CrossRef]
- Eizenberg, H.; Lande, T.; Achdari, G.; Roichman, A.; Hershenhorn, J. Effect of Egyptian broomrape (Orobanche aegyptiaca) burial depth on parasitism dynamics and chemical control in tomato. Weed Sci. 2007, 51, 152–156. [Google Scholar] [CrossRef]
- Eizenberg, H.; Hershenhorn, J.; Ephrath, J.H.; Kanampiu, F. Chemical control. In Parasitic Orobanchaceae: Parasitic Mechanisms and Control Strategies; Joel, D.M., Gresse, J., Musselman, L.J., Eds.; Springer: New York, NY, USA; Berlin/Heidelberg, Germany, 2013; pp. 415–432. [Google Scholar]
- Rubiales, D.; Fernandez-Aparico, M. Innovations in parasitic weeds management in legume crops. A review. Agron. Sustain. Dev. 2012, 32, 433–449. [Google Scholar] [CrossRef]
- Lee, Y.-T.; Singh, K.B. Winter chickpea: Problems and potential in the Mediterranean region. Options Mediterr. Ser. A Semin. Mediterr. 1990, 9, 25–34. [Google Scholar]
- Rubiales, D.; Alcantara, C.; Perez-de-Luque, A.; Gil, J.; Sillero, J.C. Infection by broomrape (Orobanche crenata) in chickpea (Cicer arietinum) as influenced by sowing date and weather conditions. Agronomie 2003, 23, 359–362. [Google Scholar] [CrossRef]
- Galili, S.; Hovav, R.; Dor, E.; Hershenhornb, J.; Harel, A.; Amir-Segev, O.; Bellaloua, A.; Badani, H.; Smirnov, E.; Achdari, G. The history of chickpea cultivation and breeding in Israel. Isr. J. Plant Sci. 2018, 65, 186–193. [Google Scholar] [CrossRef]
- Kantar, F.; Elkoca, E.; Zengin, H. Chemical and agronomical weed control in chickpea (Cicer arietinum L. cv. Aziziye-94). Turk. J. Agric. For. 1999, 23, 631–635. [Google Scholar]
- Goud, V.V.; Murade, N.B.; Khakre, M.S.; Patil, A.N. Efficacy of imazethapyr and quizalofop-ethyl herbicides on growth and yield of chickpea. Bioscan 2013, 8, 1015–1018. [Google Scholar]
- Vasilakoglou, I.; Vlachostergios, D.; Dhima, K.; Lithourgidis, A. Response of vetch, lentil, chickpea and red pea to pre- or post-emergence applied herbicides. Span. J. Agric. Res. 2013, 11, 1101–1111. [Google Scholar] [CrossRef] [Green Version]
- Tranel, P.J.; Wright, T.R.; Heap, I.M. Mutations in Herbicide-Resistant Weeds to Inhibition of Acetolactate Synthase. Available online: http://weedscience.org/mutations/mutationdisplayall.aspx (accessed on 12 June 2021).
- Haughn, G.W.; Somerville, C.R. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 1986, 204, 430–434. [Google Scholar] [CrossRef]
- Wright, T.; Penner, D. Cell selection and inheritance of imidazolinone resistance in sugar beet (Beta vulgaris). Theor. Appl. Genet. 1998, 96, 612–620. [Google Scholar] [CrossRef]
- Hart, S.E.; Saunders, J.W.; Penner, D. Chlorsulfuron resistant sugar beet: Cross-resistance and physiological basis of resistance. Weed Sci. 1992, 40, 378–383. [Google Scholar] [CrossRef]
- Swanson, E.B.; Harrgesel, M.J.; Arnoldo, M.J.; Sippell, M.; Wong, R.S.V. Microspore mutagenesis and selection: Canola plants with field resistance to imidazolinone. Theor. Appl. Genet. 1989, 78, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, S.A.; Fader, G.M.; Ulrich, J.F.; Forney, D.R.; Chaleff, R.S. Semidominant soybean mutation for resistance to sulfonylurea herbicide. Crop Sci. 1989, 29, 1403–1408. [Google Scholar] [CrossRef]
- Chaleff, R.S.; Ray, T.B. Herbicide resistant mutants from tobacco culture. Science 1984, 223, 1148–1151. [Google Scholar] [CrossRef] [PubMed]
- Rajasekaran, K.; Grula, J.W.; Anderson, D.M. Selection and characterization of mutant cotton (Gossypium hirsutum L.) cell lines resistant to sulfonylurea and imidazolinone herbicides. Plant Sci. 1996, 199, 115–124. [Google Scholar] [CrossRef]
- Croughan, T.P. Inventor. Herbicide Resistant Rice. Board of Supervisors of Louisiana State University. US Patent 5,773,704, 30 June 1998. [Google Scholar]
- Pozniak, C.J.; Hucl, P.J. Genetic analysis of imidazolinone resistance in mutation-derived lines of common wheat. Crop Sci. 2004, 44, 23–30. [Google Scholar]
- Newhouse, K.; Smith, W.A.; Starrett, M.A.; Schaefer, T.J.; Singh, B.K. Tolerance to imidazolinone herbicides in wheat. Plant Physiol. 1992, 100, 882–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dor, E.; Smirnov, E.; Galili, S.; Achdari, G.; Hershenhorn, J. Characterization of the novel tomato mutant HRT1, resistant to acetolactate synthase-inhibiting herbicides. Weed Sci. 2016, 64, 348–360. [Google Scholar] [CrossRef]
- Tranel, P.J.; Wright, T.R. Resistance to ALS-inhibiting herbicides: What have we learned? Weed Sci. 2002, 50, 700–712. [Google Scholar] [CrossRef]
- Walsh, D.T.; Babiker, E.M.; Burke, I.C.; Hulbert, S.H. Camelina mutants resistant to acetolactate synthase inhibitor herbicides. Mol. Breed. 2012, 30, 1053–1063. [Google Scholar] [CrossRef]
- Sobolev, V.; Sorokine, A.; Prilusky, J.; Abola, E.E.; Edelman, M. Automated analysis of interatomic contacts in proteins. Bioinformatics 1999, 15, 327–332. [Google Scholar] [CrossRef]
- Thompson, C.; Tar’an, B. Genetic characterization of the acetohydroxyacid synthase (AHAS) gene responsible for resistance to imidazolinone in chickpea (Cicer arietinum L.). Theor. Appl. Genet. 2014, 127, 1583–1591. [Google Scholar] [CrossRef]
- Jain, P.; Tar’an, B. Analysis of acetohydroxyacid synthase1 gene in chickpea conferring resistance to imazamox herbicide. Genome 2014, 57, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Pang, S.S.; Guddat, L.W.; Duggleby, R.G. Molecular basis of sulfonylurea herbicide inhibition of acetohydroxyacid synthase. J. Biol. Chem. 2003, 278, 7639–7644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolkman, J.M.; Slabaugh, M.B.; Bruniard, J.M.; Berry, S.; Bushman, B.S.; Olungu, C.; Maes, N.; Abratti, G.; Zambelli, A.; Miller, J.F.; et al. Acetohydroxyacid synthase mutations conferring resistance to imidazolinone or sulfonylurea herbicides in sunflower. Theor. Appl. Genet. 2004, 109, 1147–1159. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.-L.; Pu, H.-M.; Gao, J.-Q.; Long, W.H.; Chen, F.; Zhou, X.-Y.; Zhang, W.; Peng, Q.; Chen, S.; Zhang, J.-F. Inheritance and molecular characterization of resistance to AHAS-inhibiting herbicides in rapeseed. J. Integr. Agric. 2017, 16, 2421–2433. [Google Scholar] [CrossRef] [Green Version]
- Shoba, D.; Raveendran, M.; Manonmani, S.; Utharasu, S.; Dhivyapriya, D.; Subhasini, G.; Ramchandar, S.; Valarmathi, R.; Nitasha, G.; Krishnan, S.G.; et al. Development and genetic characterization of a novel herbicide (Imazethapyr) tolerant mutant in rice (Oryza sativa L.). Rice 2017, 10, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fulton, T.M.; Chunwongse, J.; Tanksley, S.D. Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol. Biol. Rep. 1995, 13, 207–209. [Google Scholar] [CrossRef]
- Schreiber, G.; Reuveni, M.; Evenor, D.; Oren-Shamir, M.; Ovadia, R.; Sapir-Mir, M.; Bootbool-Man, A.; Nahon, S.; Shlomo, H.; Chen, L. ANTHOCYANIN1 from Solanum chilense is more efficient in accumulating anthocyanin metabolites than its Solanum lycopersicum counterpart in association with the anthocyanin fruit phenotype of tomato. Theor. Appl. Genet. 2012, 124, 295–307. [Google Scholar] [CrossRef] [PubMed]
- You, F.M.; Huo, N.; Gu, Y.Q.; Luo, M.C.; Ma, Y.; Hane, D.; Lazo, G.R.; Dvorak, J.; Anderson, O.D. BatchPrimer3: A high throughput web application for PCR and sequencing primer design. BMC Bioinform. 2008, 9, 253. [Google Scholar] [CrossRef] [Green Version]
- Dahiya, B.S. Crossing without emasculation in chickpea. Indian J. Gen. Plant Breed. 1974, 34, 206–207. [Google Scholar]
- Callebaut, I.; Labesse, G.; Durand, P.; Poupon, A.; Canard, L.; Chomilier, J.; Henrissat, B.; Mornon, J.P. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): Current status and perspectives. Cell Mol. Life Sci. 1997, 53, 621–645. [Google Scholar] [CrossRef]







| Herbicide | Chickpea Line | R2 | p | b | x0 | y0 |
|---|---|---|---|---|---|---|
| A. Imazamox | WT | 0.99 | <0.0001 | 1.02 | 21.04 | 3.99 |
| M2033 | 0.98 | <0.0001 | 1.30 | 53.60 | 3.54 | |
| B. Imazapic | WT | 0.99 | <0.0001 | 0.70 | 18.50 | 0.1 |
| M2033 | 0.97 | <0.0001 | 0.95 | 121.24 | 1.54 | |
| C. Imazapyr | WT | 0.99 | <0.0001 | 1.31 | 29.11 | 3.1 |
| M2033 | 0.95 | <0.0001 | 1.57 | 109.62 | 1.47 | |
| D. Trifloxysulfuron | WT | 0.99 | <0.0001 | 0.69 | 0.27 | 0.00 |
| M2033 | 0.99 | <0.0001 | 0.65 | 0.20 | 0.00 | |
| E. Chlorsulfuron | WT | 0.96 | <0.0001 | 0.77 | 0.05 | 0.00 |
| M2033 | 0.98 | <0.0001 | 0.75 | 0.05 | 0.00 | |
| F. Sulfosulfuron | WT | 0.99 | <0.0001 | 0.58 | 1.77 | 3.3 |
| M2033 | 0.99 | <0.0001 | 0.48 | 1.40 | 3.6 | |
| G. Foramsulfuron | WT | 0.98 | <0.0001 | 1.03 | 40.23 | 2.33 |
| M2033 | 0.98 | <0.0001 | 0.98 | 36.33 | 4.16 | |
| H. Propoxycar-bazone sodium | WT | 0.99 | <0.0001 | 1.11 | 57.25 | 3.42 |
| M2033 | 0.99 | <0.0001 | 1.16 | 52.46 | 3.88 | |
| I. Pyrithiobac sodium | WT | 0.99 | <0.0001 | 1.15 | 11.42 | 1.03 |
| M2033 | 0.99 | <0.0001 | 1.06 | 8.42 | 2.57 |
| Primer Set | Gene | Forward Primer (5′→3′) | Reverse Primer (5′→3′) | Product Size (bp) | Amplified Region |
|---|---|---|---|---|---|
| 1 | 1 | TAAACTCGAACTCCATCATTCA | GAATATACCGCCTTGTTCGTGA | 443 | 0–414 |
| 2 | 1 | CCACCGCACCATCCTCCATAAC | AACGCAAACTTCATGTCAGCAC | 582 | 173–755 |
| 3 | 1 | TCCTAGGGTTGTTAGAGAGGCTTT | AACGCAAACTTCATGTCAGCAC | 564 | 659–1223 |
| 4 | 1 | TGATTCGGCTGAAATTGGGA | TCAACAACAACAGCATCAGGG | 413 | 1155–1568 |
| 5 | 1 | GACAATGGTTAACTTCGGGTGGAC | CTGCCCAATCAATCAGTAACTCC | 521 | 1471–1992 |
| 6 | 2 | AAACAATAGAGATTTTAAAGGCC | TTCGAGGTTCGTCAAGGGCA | 250 | 0–250 |
| 7 | 2 | ACTCCCCTCCCCTCAACCGAACAA | TTCTAAATGTGACTCAGAAGGTGA | 636 | 192–828 |
| 8 | 2 | GGCTAGGTTACCAAAGTCACCTTC | TCCTATTAATCCCCCTCAAAGCC | 443 | 788–1231 |
| 9 | 2 | GTGTCAGTTTGTGGGGATTTA | CTCTGTTGTGGTTGACATCGA | 383 | 1182–1565 |
| 10 | 2 | ATGTGGTCTGCTCAATTTTATAGT | ACATAATCGGCATCAAGATAAACC | 582 | 1425–2007 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galili, S.; Hershenhorn, J.; Edelman, M.; Sobolev, V.; Smirnov, E.; Amir-Segev, O.; Bellalou, A.; Dor, E. Novel Mutation in the Acetohydroxyacid Synthase (AHAS), Gene Confers Imidazolinone Resistance in Chickpea Cicer arietinum L. Plants. Plants 2021, 10, 2791. https://doi.org/10.3390/plants10122791
Galili S, Hershenhorn J, Edelman M, Sobolev V, Smirnov E, Amir-Segev O, Bellalou A, Dor E. Novel Mutation in the Acetohydroxyacid Synthase (AHAS), Gene Confers Imidazolinone Resistance in Chickpea Cicer arietinum L. Plants. Plants. 2021; 10(12):2791. https://doi.org/10.3390/plants10122791
Chicago/Turabian StyleGalili, Shmuel, Joseph Hershenhorn, Marvin Edelman, Vladimir Sobolev, Evgeny Smirnov, Orit Amir-Segev, Aharon Bellalou, and Evgenia Dor. 2021. "Novel Mutation in the Acetohydroxyacid Synthase (AHAS), Gene Confers Imidazolinone Resistance in Chickpea Cicer arietinum L. Plants" Plants 10, no. 12: 2791. https://doi.org/10.3390/plants10122791
APA StyleGalili, S., Hershenhorn, J., Edelman, M., Sobolev, V., Smirnov, E., Amir-Segev, O., Bellalou, A., & Dor, E. (2021). Novel Mutation in the Acetohydroxyacid Synthase (AHAS), Gene Confers Imidazolinone Resistance in Chickpea Cicer arietinum L. Plants. Plants, 10(12), 2791. https://doi.org/10.3390/plants10122791





