Optimization of Callus Induction and Shoot Regeneration from Tomato Cotyledon Explants
Abstract
1. Introduction
2. Results
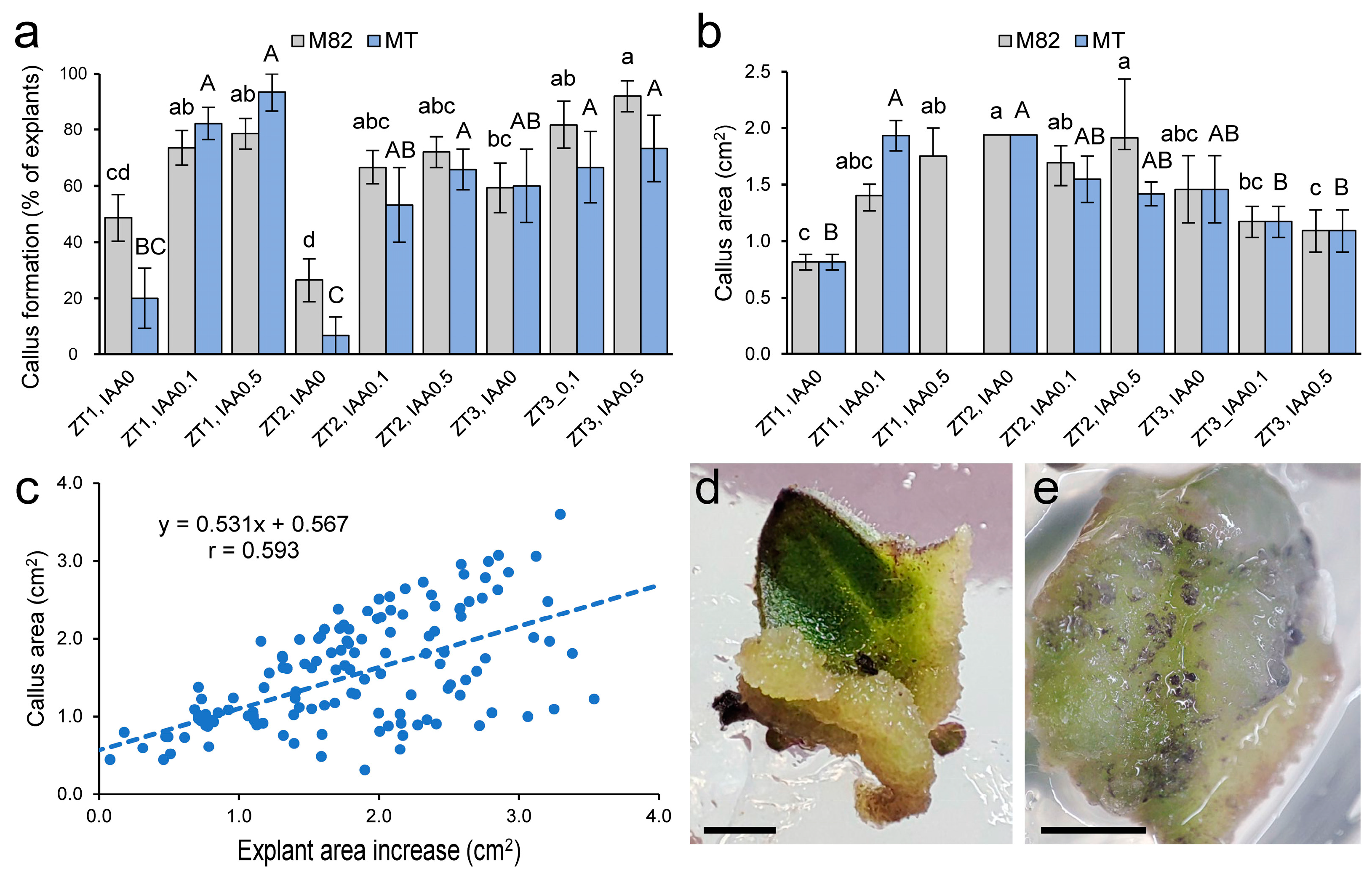
2.1. Optimization of Callus and Shoot Formation in M82 and MT Cotyledon Explants
2.2. Evaluation of the Regenerative Potential of Explants in Different Tomato Genotypes
2.3. Cellular Features of De Novo Organ Formation in Tomato Cotyledon Explants
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Statistical Analyses
4.3. Microscopic Observation and EdU Staining
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Available online: https://www.fao.org/faostat/en (accessed on 1 July 2023).
- Kumar, L.; Chhogyel, N.; Gopalakrishnan, T.; Hasan, M.K.; Jayasinghe, S.L.; Kariyawasam, C.S.; Kogo, B.K.; Ratnayake, S. Climate change and future of agri-food production. In Future Foods: Global Trends, Opportunities, and Sustainability Challenges; Academic Press: Cambridge, MA, USA, 2022; pp. 49–79. [Google Scholar]
- Van Vu, T.; Das, S.; Tran, M.T.; Hong, J.C.; Kim, J.-Y. Precision genome engineering for the breeding of tomatoes: Recent progress and future perspectives. Front. Genome Ed. 2020, 2, 612137. [Google Scholar]
- Long, Y.; Yang, Y.; Pan, G.; Shen, Y. New insights into tissue culture plant-regeneration mechanisms. Front. Plant Sci. 2022, 13, 926752. [Google Scholar] [CrossRef] [PubMed]
- Martí, E.; Gisbert, C.; Bishop, G.J.; Dixon, M.S.; García-Martínez, J.L. Genetic and Physiological characterization of tomato cv. Micro-Tom. J. Exp. Bot. 2006, 57, 2037–2047. [Google Scholar] [CrossRef] [PubMed]
- Shikata, M.; Ezura, H. Micro-Tom tomato as an alternative plant model system: Mutant collection and efficient transformation. Methods Mol. Biol. 2016, 1363, 47–55. [Google Scholar]
- Rothan, C.; Diouf, I.; Causse, M. Trait discovery and editing in tomato. Plant J. 2019, 97, 73–90. [Google Scholar]
- Mata-Nicolás, E.; Montero-Pau, J.; Gimeno-Paez, E.; Garcia-Carpintero, V.; Ziarsolo, P.; Menda, N.; Mueller, L.A.; Blanca, J.; Cañizares, J.; van der Knaap, E.; et al. Exploiting the diversity of tomato: The development of a phenotypically and genetically detailed germplasm collection. Hortic. Res. 2020, 7, 66. [Google Scholar]
- Sánchez-López, J.; Atarés, A.; Jáquez-Gutiérrez, M.; Ortiz-Atienza, A.; Capel, C.; Pineda, B.; García-Sogo, B.; Yuste-Lisbona, F.J.; Lozano, R.; Moreno, V. Approaching the genetic dissection of indirect adventitious organogenesis process in tomato explants. Plant Sci. 2021, 302, 110721. [Google Scholar]
- de Siqueira Pinto, M.; Abeyratne, C.R.; Benedito, V.A.; Peres, L.E.P. Genetic and physiological characterization of three natural allelic variations affecting the organogenic capacity in tomato (Solanum lycopersicum cv. Micro-Tom). Plant Cell Tissue Organ Cult. 2017, 129, 89–103. [Google Scholar] [CrossRef]
- Khuong, T.T.H.; Crété, P.; Robaglia, C.; Caffarri, S. Optimisation of tomato Micro-Tom regeneration and selection on glufosinate/Basta and dependency of gene silencing on transgene copy number. Plant Cell Rep. 2013, 32, 1441–1454. [Google Scholar] [CrossRef]
- Hashmi, M.H.; Saeed, F.; Demirel, U.; Bakhsh, A. Establishment of highly efficient and reproducible Agrobacterium-mediated transformation system for tomato (Solanum lycopersicum L.). Vitr. Cell. Dev. Biol.—Plant 2022, 58, 1066–1076. [Google Scholar] [CrossRef]
- Gupta, S.; Van Eck, J. Modification of plant regeneration medium decreases the time for recovery of Solanum lycopersicum cultivar M82 stable transgenic lines. Plant Cell Tissue Organ Cult. 2016, 127, 417–423. [Google Scholar] [CrossRef]
- Prihatna, C.; Chen, R.; Barbetti, M.J.; Barker, S.J. Optimisation of regeneration parameters improves transformation efficiency of recalcitrant tomato. Plant Cell Tissue Organ Cult. 2019, 137, 473–483. [Google Scholar]
- Vats, S.; Shivaraj, S.M.; Sonah, H.; Patil, G.; Roy, J.; Sharma, T.R.; Deshmukh, R. Efficient regeneration and Agrobacterium-mediated transformation method for cultivated and wild tomato. Plant Mol. Biol. Rep. 2023, 1–12. [Google Scholar] [CrossRef]
- Saeed, W.; Naseem, S.; Gohar, D.; Ali, Z. Efficient and reproducible somatic embryogenesis and micropropagation in tomato via novel structures—Rhizoid tubers. PLoS ONE 2019, 14, e0215929. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Lee, J.; Jie, E.Y.; Choi, S.H.; Jiang, L.; Ahn, W.S.; Kim, C.Y.; Kim, S.W. Temporal and spatial expression analysis of shoot-regeneration regulatory genes during the adventitious shoot formation in hypocotyl and cotyledon explants of tomato (CV. Micro-Tom). Int. J. Mol. Sci. 2020, 21, 5309. [Google Scholar] [CrossRef] [PubMed]
- Stoynova-Bakalova, E.; Karanov, E.; Petrov, P.; Hall, M.A. Cell division and cell expansion in cotyledons of Arabidopsis seedlings. New Phytol. 2004, 162, 471–479. [Google Scholar] [CrossRef]
- Jones, B.; Ljung, K.; Gunnerås, S.A.; Petersson, S.V.; Tarkowski, P.; Graham, N.; May, S.; Dolezal, K.; Sandberg, G. Cytokinin regulation of auxin synthesis in arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell 2010, 22, 2956–2969. [Google Scholar] [CrossRef]
- Růzǐčka, K.; Šimášková, M.; Duclercq, J.; Petrášek, J.; Zažímalová, E.; Simon, S.; Friml, J.; Van Montagu, M.C.E.; Benková, E. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl. Acad. Sci. USA 2009, 106, 4284–4289. [Google Scholar] [CrossRef] [PubMed]
- Dar, S.A.; Nawchoo, I.A.; Tyub, S.; Kamili, A.N. Effect of plant growth regulators on in vitro induction and maintenance of callus from leaf and root explants of Atropa acuminata Royle Ex Lindl. Biotechnol. Rep. 2021, 32, e00688. [Google Scholar] [CrossRef]
- Kinoshita, A.; Vayssières, A.; Richter, R.; Sang, Q.; Roggen, A.; Van Driel, A.D.; Smith, R.S.; Coupland, G. Regulation of shoot meristem shape by photoperiodic signaling and phytohormones during floral induction of Arabidopsis. elife 2020, 9, e60661. [Google Scholar] [CrossRef] [PubMed]
- Sandhya, D.; Jogam, P.; Venkatapuram, A.K.; Savitikadi, P.; Peddaboina, V.; Allini, V.R.; Abbagani, S. Highly efficient Agrobacterium-mediated transformation and plant regeneration system for genome engineering in tomato. Saudi J. Biol. Sci. 2022, 29, 103292. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, P.; Ashwath, N.; Senaratna, T.; Midmore, D. Tissue culture studies of tomato (Lycopersicon esculentum). Plant Cell Tissue Organ Cult. 2004, 78, 1–21. [Google Scholar]
- Raspor, M.; Motyka, V.; Kaleri, A.R.; Ninković, S.; Tubić, L.; Cingel, A.; Ćosić, T. Integrating the roles for cytokinin and auxin in de novo shoot organogenesis: From hormone uptake to signaling outputs. Int. J. Mol. Sci. 2021, 22, 8554. [Google Scholar] [CrossRef]
- Garcia, C.; de Almeida, A.-A.F.; Costa, M.; Britto, D.; Valle, R.; Royaert, S.; Marelli, J.P. Abnormalities in somatic embryogenesis caused by 2,4-D: An overview. Plant Cell Tissue Organ Cult. 2019, 137, 193–212. [Google Scholar] [CrossRef]
- Larriba, E.; Sánchez-García, A.B.; Martínez-Andújar, C.; Albacete, A.; Pérez-Pérez, J.M. Tissue-specific metabolic reprogramming during wound-induced organ formation in tomato hypocotyl explants. Int. J. Mol. Sci. 2021, 22, 10112. [Google Scholar] [CrossRef]
- Larriba, E.; Belén Sánchez-García, A.; Salud Justamante, M.; Martínez-Andújar, C.; Albacete, A.; Pérez-Pérez, J.M. Dynamic hormone gradients regulate wound-induced de novo organ formation in tomato hypocotyl explants. Int. J. Mol. Sci. 2021, 22, 11843. [Google Scholar] [PubMed]
- Chaudhry, Z.; Abbas, S.; Yasmin, A.; Rashid, H.; Ahmed, H.A.B.I.B.; Anjum, M.A. Tissue culture studies in tomato (Lycopersicon esculentum) var. Moneymaker. Pak. J. Bot. 2010, 42, 155–163. [Google Scholar]
- Yu, H.; Zhang, L.; Wang, W.; Tian, P.; Wang, W.; Wang, K.; Gao, Z.; Liu, S.; Zhang, Y.; Irish, V.F.; et al. TCP5 controls leaf margin development by regulating KNOX and BEL-like transcription factors in Arabidopsis. J. Exp. Bot. 2021, 72, 1809–1821. [Google Scholar] [PubMed]
- Navarro-Cartagena, S.; Micol, J.L. Is auxin enough? Cytokinins and margin patterning in simple leaves. Trends Plant Sci. 2023, 28, 54–73. [Google Scholar] [CrossRef] [PubMed]
- Tsukaya, H. The leaf meristem enigma: The relationship between the plate meristem and the marginal meristem. Plant Cell 2021, 33, 3194–3206. [Google Scholar] [CrossRef]
- Schuetz, M.; Smith, R.; Ellis, B. Xylem tissue specification, patterning, and differentiation mechanisms. J. Exp. Bot. 2013, 64, 11–31. [Google Scholar] [CrossRef] [PubMed]
- Omary, M.; Gil-Yarom, N.; Yahav, C.; Steiner, E.; Hendelman, A.; Efroni, I. A conserved superlocus regulates above- and belowground root initiation. Science 2022, 375, eabf4368. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Jiao, Y.; Meyerowitz, E.M. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 2010, 18, 463–471. [Google Scholar] [PubMed]
- Meng, W.J.; Cheng, Z.J.; Sang, Y.L.; Zhang, M.M.; Rong, X.F.; Wang, Z.W.; Tang, Y.Y.; Zhang, X.S. Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 2017, 29, 1357–1372. [Google Scholar] [CrossRef]
- Alaguero-Cordovilla, A.; Sánchez-García, A.B.; Ibáñez, S.; Albacete, A.; Cano, A.; Acosta, M.; Pérez-Pérez, J.M. An auxin-mediated regulatory framework for wound-induced adventitious root formation in tomato shoot explants. Plant. Cell Environ. 2021, 44, 1642–1662. [Google Scholar] [CrossRef]
- Mamidala, P.; Nanna, R.S. Effect of genotype, explant source and medium on in vitro regeneration of tomato. Int. J. Genet. Mol. Biol. 2021, 3, 45–50. [Google Scholar]
- Gubis, J.; Lajchová, Z.; Faragó, J.; Jureková, Z. Effect of growth regulators on shoot induction and plant regeneration in tomato (Lycopersicon esculentum Mill.). Biol. Bratisl. 2004, 59, 405–408. [Google Scholar]
- Alaguero-Cordovilla, A.; Gran-Gómez, F.; Tormos-Moltó, S.; Pérez-Pérez, J. Morphological characterization of root system architecture in diverse tomato genotypes during early growth. Int. J. Mol. Sci. 2018, 19, 3888. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Aguinis, H.; Gottfredson, R.K.; Joo, H. Best-practice recommendations for defining, identifying, and handling outliers. Organ. Res. Methods 2013, 16, 270–301. [Google Scholar]
- Pasternak, T.; Tietz, O.; Rapp, K.; Begheldo, M.; Nitschke, R.; Ruperti, B.; Palme, K. Protocol: An improved and universal procedure for whole-mount immunolocalization in plants. Plant Methods 2015, 11, 50. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, T.; Kircher, S.; Pérez-Pérez, J.M.; Palme, K. A simple pipeline for cell cycle kinetic studies in the root apical meristem. J. Exp. Bot. 2022, 73, 4683–4695. [Google Scholar] [PubMed]




| Accession | Organism | Name | Genotype 1 | |
| LA3475 | S. lycopersicum var. lycopersicum | M82 | sp; u; obv; I; Ve | |
| LA3911 | S. lycopersicum var. lycopersicum | Micro-Tom | d; sp; ej-2w; u; I; Sm | |
| LA2706 | S. lycopersicum var. lycopersicum | Moneymaker | sp+; ej-2w; u; obv+ | |
| Accession | Organism | Country | Site | Latitude, Longitude |
| BGV007910 | S. lycopersicum var. cerasiforme | México | Palo de Arco; Ciudad Valles; San Luis de Potosí | 21.91, 99.16 |
| BGV007927 | S. lycopersicum var. cerasiforme | México | El Vergel: Culiacán. Sinaloa | 24.73, 107.79 |
| BGV016054 | S. lycopersicum var. cerasiforme | Perú | Rumizapa | 6.45, 76.47 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yaroshko, O.; Pasternak, T.; Larriba, E.; Pérez-Pérez, J.M. Optimization of Callus Induction and Shoot Regeneration from Tomato Cotyledon Explants. Plants 2023, 12, 2942. https://doi.org/10.3390/plants12162942
Yaroshko O, Pasternak T, Larriba E, Pérez-Pérez JM. Optimization of Callus Induction and Shoot Regeneration from Tomato Cotyledon Explants. Plants. 2023; 12(16):2942. https://doi.org/10.3390/plants12162942
Chicago/Turabian StyleYaroshko, Olha, Taras Pasternak, Eduardo Larriba, and José Manuel Pérez-Pérez. 2023. "Optimization of Callus Induction and Shoot Regeneration from Tomato Cotyledon Explants" Plants 12, no. 16: 2942. https://doi.org/10.3390/plants12162942
APA StyleYaroshko, O., Pasternak, T., Larriba, E., & Pérez-Pérez, J. M. (2023). Optimization of Callus Induction and Shoot Regeneration from Tomato Cotyledon Explants. Plants, 12(16), 2942. https://doi.org/10.3390/plants12162942








