Unveiling the Neem (Azadirachta indica) Effects on Biofilm Formation of Food-Borne Bacteria and the Potential Mechanism Using a Molecular Docking Approach
Abstract
1. Introduction
2. Results
2.1. Phytochemical Analysis of Neem (A. indica) Using Gas Chromatography–Mass Spectrometry Analysis
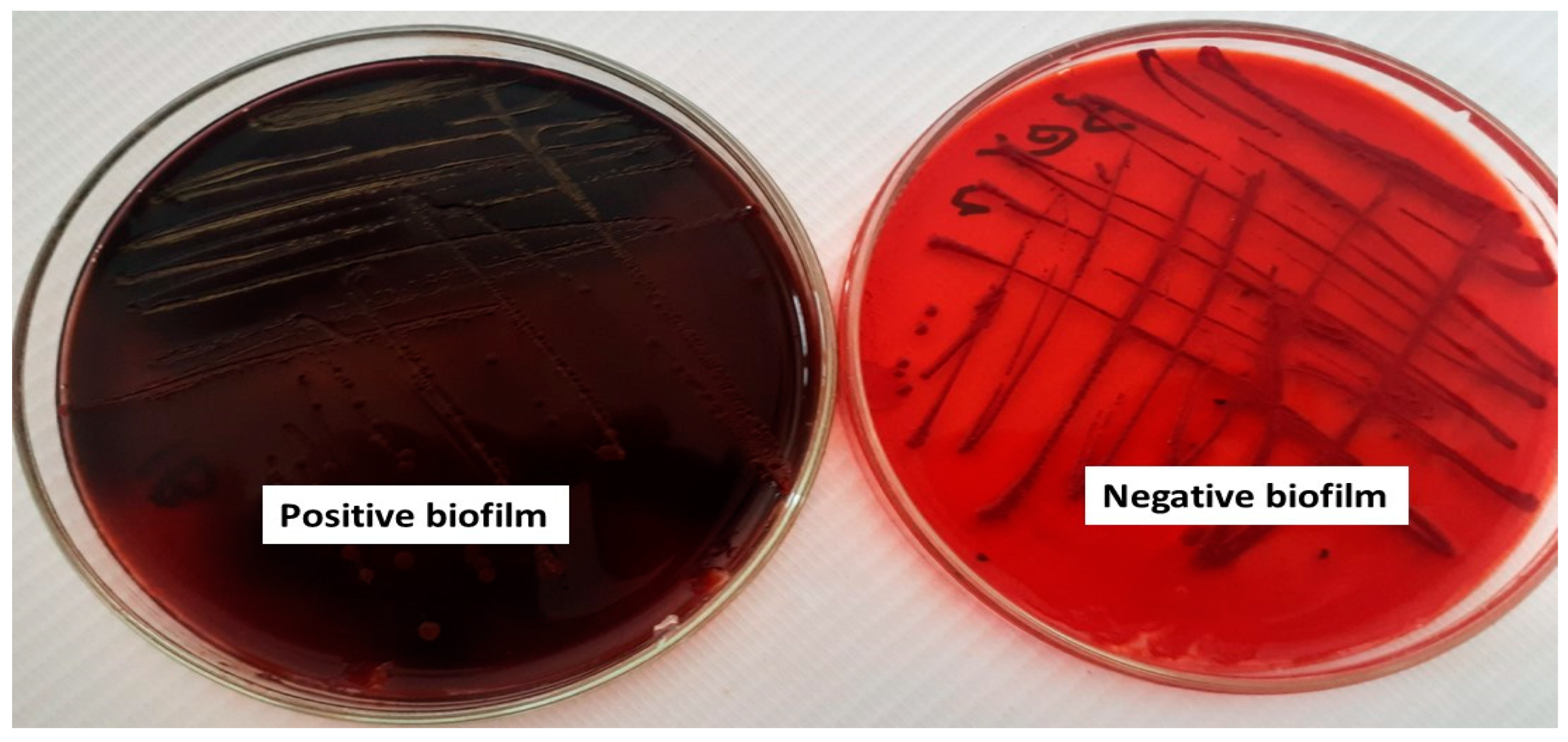
2.2. Qualitative and Quantitative Assessment of Biofilm Formation
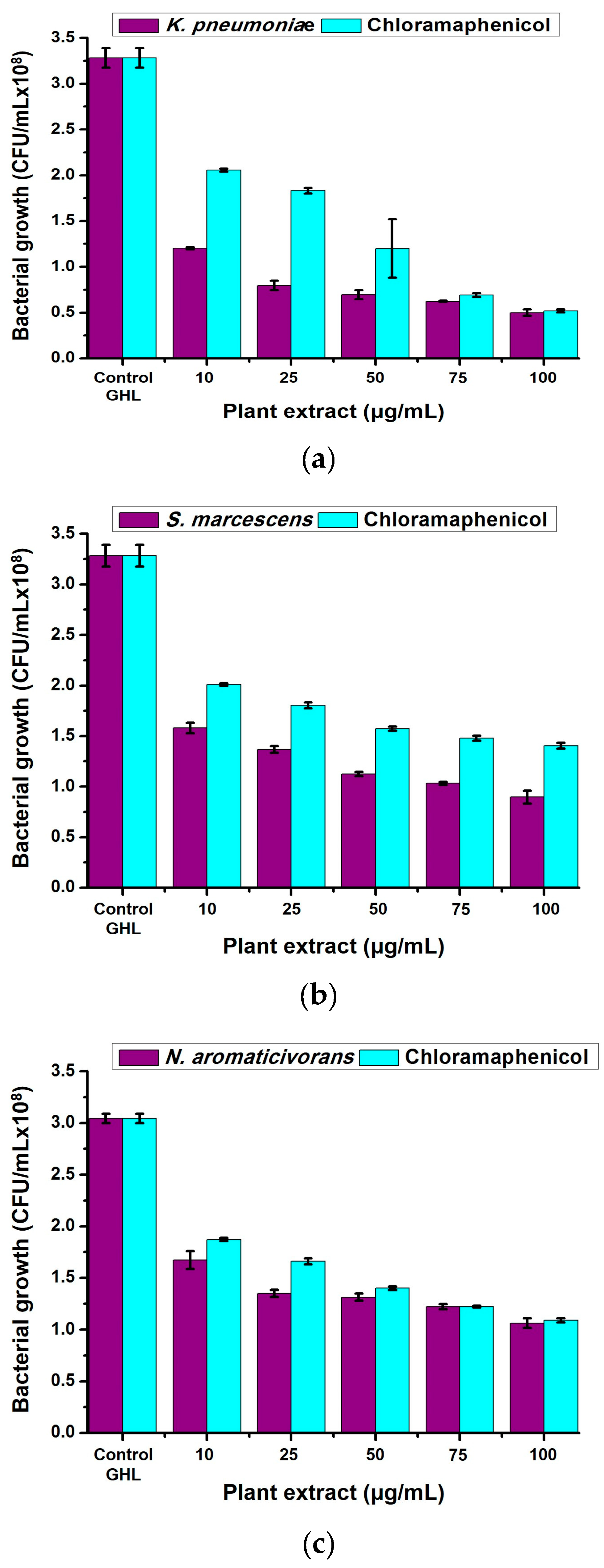
2.3. Antibacterial Properties of Neem (A. indica) Methanolic Extract
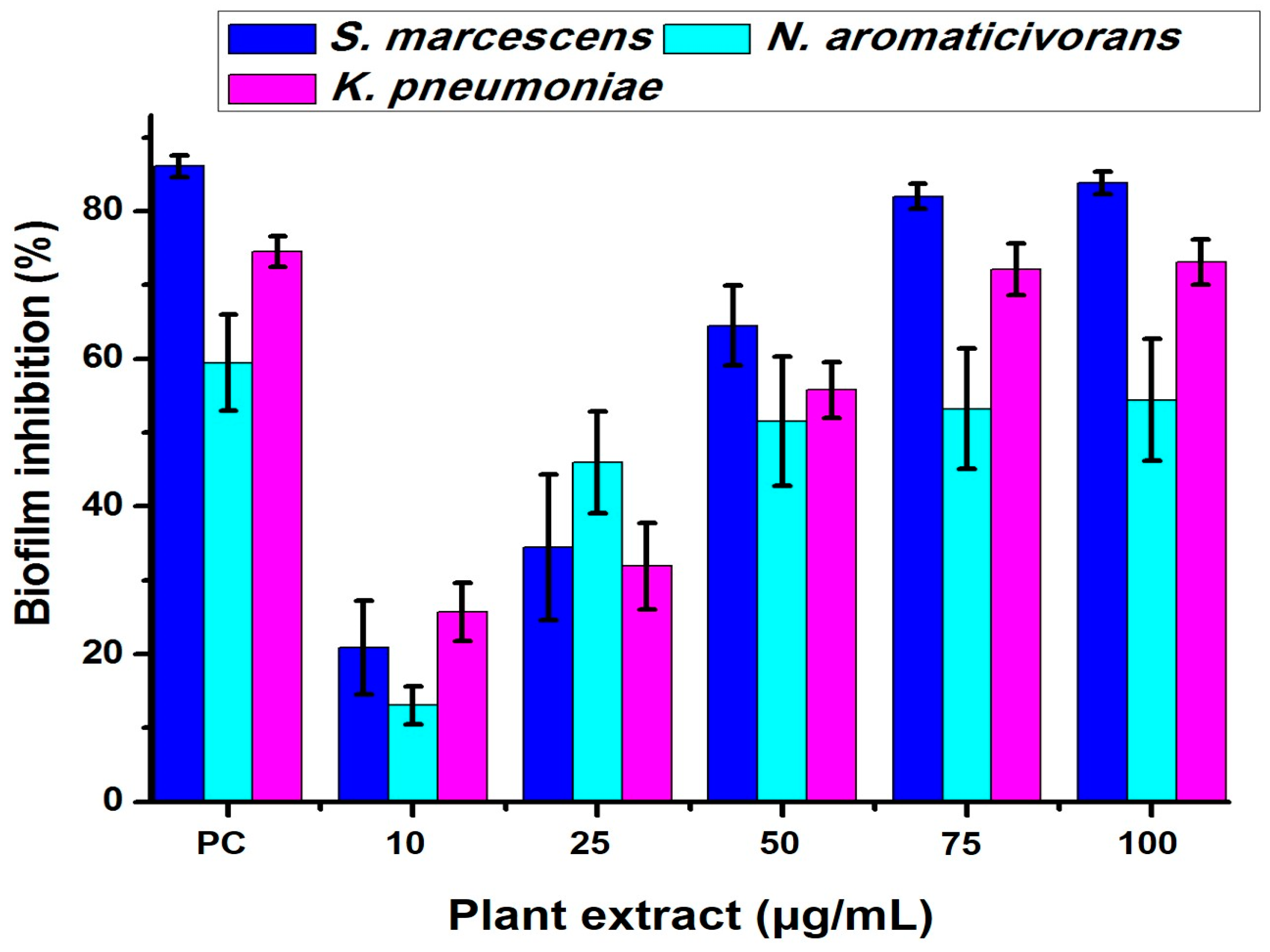
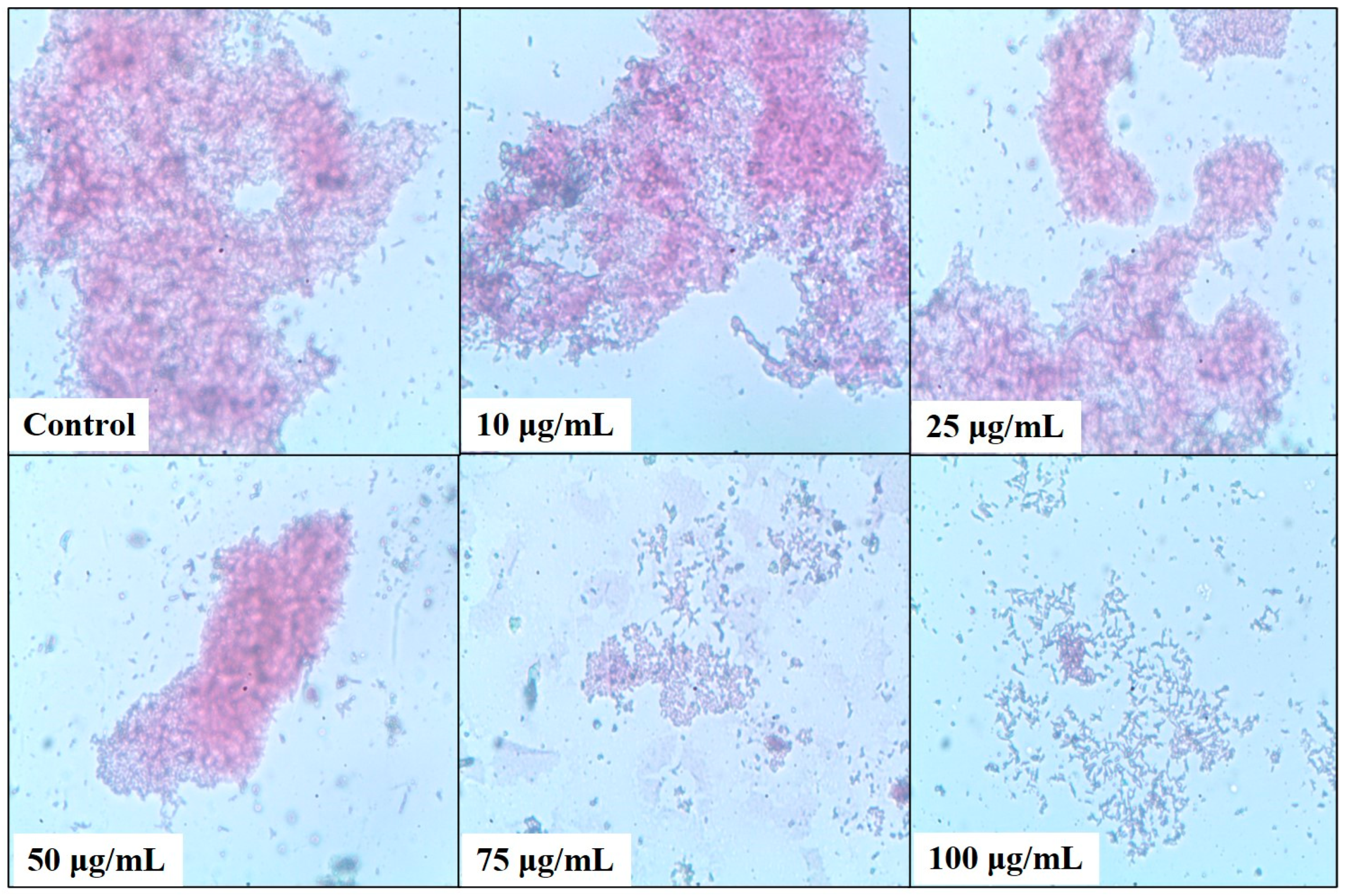
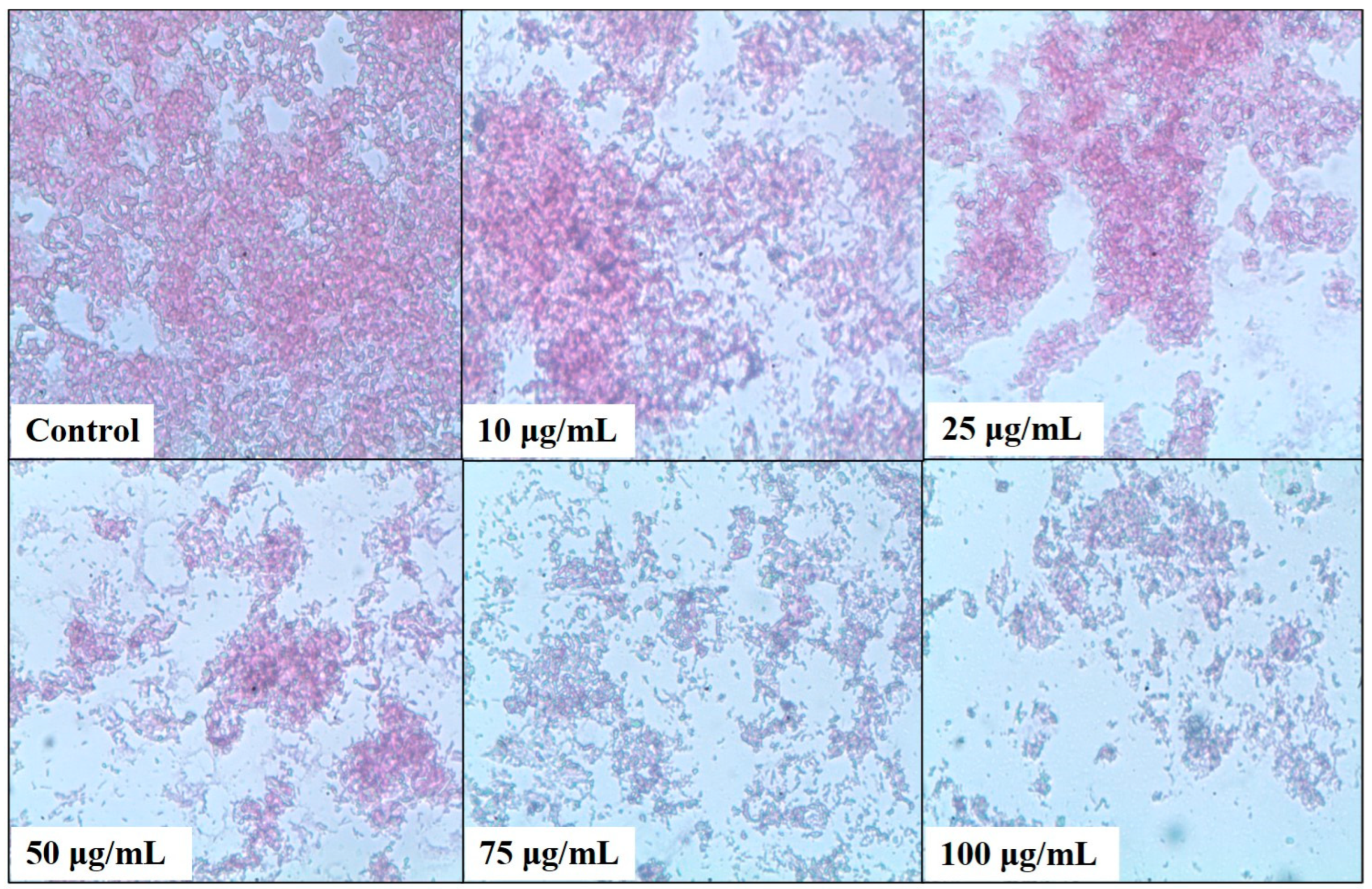
2.4. Bacterial Biofilm Inhibition Using Neem (A. indica) Extract
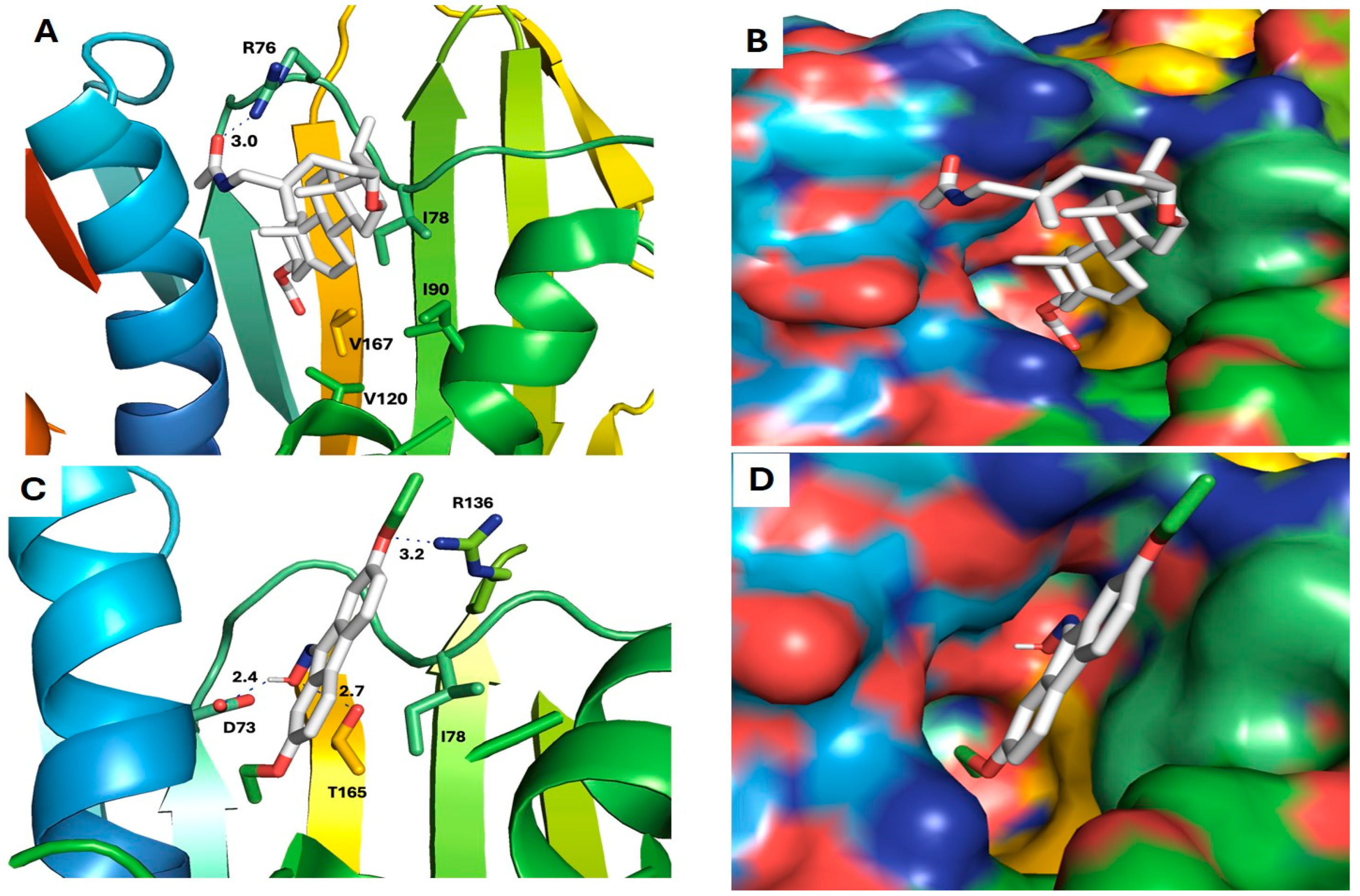
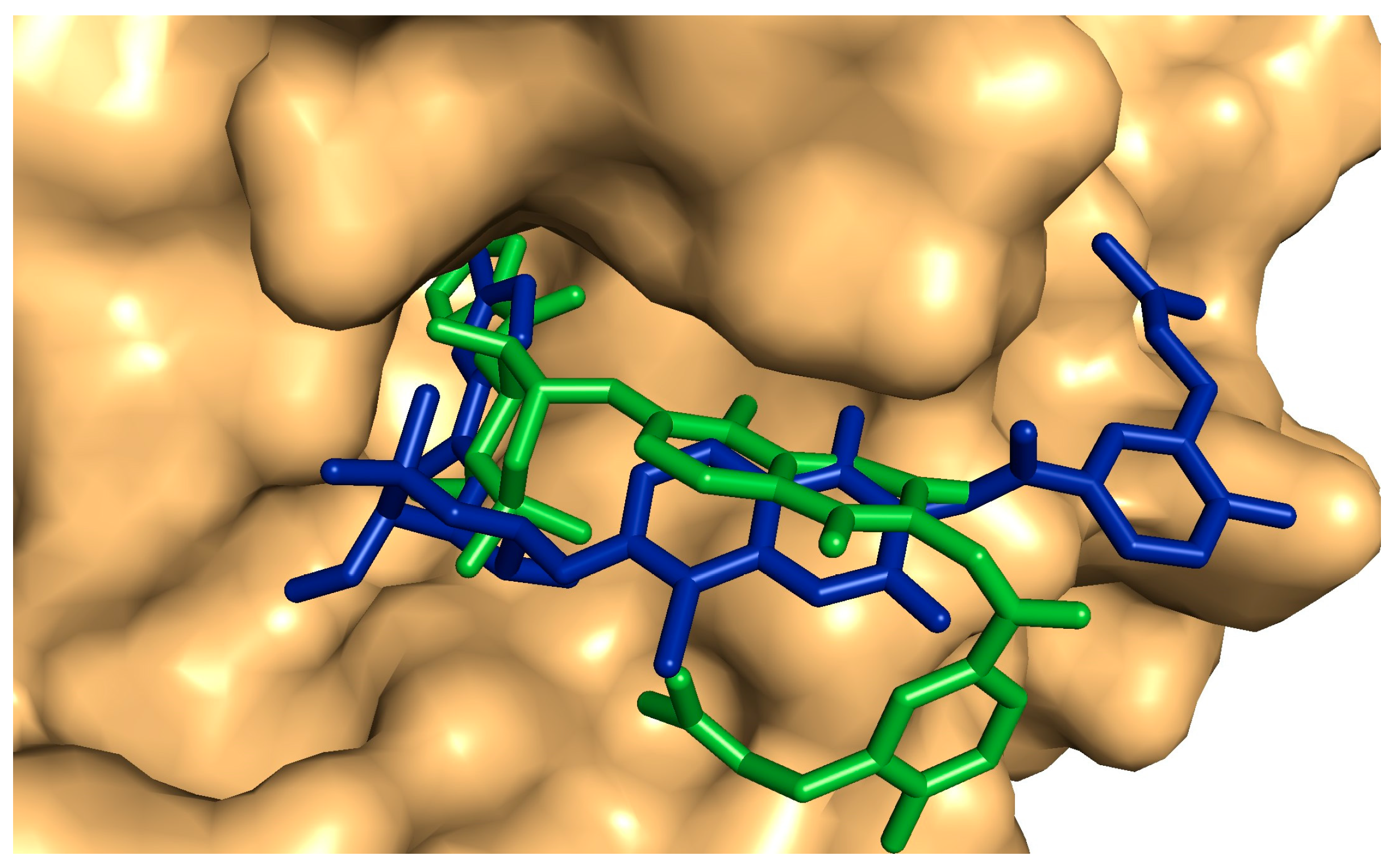
2.5. Antibiofilm and Antibacterial Mechanisms of Neem Extract Using Molecular Docking Approach
3. Discussion
4. Materials and Methods
4.1. Collection and Extraction of Neem (A. indica) Leaves
4.2. Phytochemical Analysis of Neem (A. indica) Extract
4.3. Bacterial Culture, Growth, and Maintenance
4.4. Assessment of the Biofilm Formation Capabilities
4.5. Quantitative Determination of Bacterial Biofilm
4.6. Testing the Antibacterial Properties of Neem (A. indica) Extract
4.7. Assessment of Bacterial Biofilm Inhibition Using Neem (A. indica) Extract
4.8. Light Microscopy Analysis of the Biofilm Formation
4.9. Molecular Docking
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Buscaroli, E.; Braschi, I.; Cirillo, C.; Fargue-Lelièvre, A.; Modarelli, G.C.; Pennisi, G.; Righini, I.; Specht, K.; Orsini, F. Reviewing chemical and biological risks in urban agriculture: A comprehensive framework for a food safety assessment of city region food systems. Food Control 2021, 126, 108085. [Google Scholar] [CrossRef]
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Doepfer, D.; Fazil, A.; Fischer-Walker, C.L.; Hald, T. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: A data synthesis. PLoS Med. 2015, 12, e1001921. [Google Scholar] [CrossRef]
- Kim, S.O.; Kim, S.S. Recent (2011–2017) food borne outbreak cases in the Republic of Korea compared to the United States: A review. Food Sci. Biotechnol. 2021, 30, 185–194. [Google Scholar] [CrossRef]
- Bai, X.; Nakatsu, C.H.; Bhunia, A.K. Bacterial Biofilms and Their Implications in Pathogenesis and Food Safety. Foods 2021, 10, 2117. [Google Scholar] [CrossRef]
- Rather, M.A.; Gupta, K.; Bardhan, P.; Borah, M.; Sarkar, A.; Eldiehy, K.S.; Bhuyan, S.; Mandal, M. Microbial biofilm: A matter of grave concern for human health and food industry. J. Basic Microbiol. 2021, 61, 380–395. [Google Scholar] [CrossRef]
- Kalia, V.C.; Patel, S.K.S.; Lee, J.K. Bacterial biofilm inhibitors: An overview. Ecotoxicol. Environ. Saf. 2023, 264, 115389. [Google Scholar] [CrossRef]
- Liu, S.S.; Brul, S.A.; Zaat, J. Bacterial persister-cells and spores in the food chain: Their potential inactivation by antimicrobial peptides (AMPs). Int. J. Mol. Sci. 2020, 21, 8967. [Google Scholar] [CrossRef]
- Sharma, S.; Mohler, J.; Mahajan, S.D.; Schwartz, S.A.; Bruggemann, L.; Aalinkeel, R. Microbial Biofilm: A Review on Formation, Infection, Antibiotic Resistance, Control Measures, and Innovative Treatment. Microorganisms 2023, 11, 1614. [Google Scholar] [CrossRef]
- Gado, D.A.; Abdalla, M.A.; Erhabor, J.O.; Ehlers, M.M.; McGaw, L.J. In vitro anti-biofilm effects of Loxostylis alata extracts and isolated 5-demethyl sinensetin on selected foodborne bacteria. S. Afr. J. Bot. 2023, 156, 29–34. [Google Scholar] [CrossRef]
- Rupani, R.; Chavez, A. Medicinal plants with traditional use: Ethnobotany in the Indian subcontinent. Clin. Dermatol. 2018, 36, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Panda, A.K.; De Mandal, S.; Shakeel, M.; Bisht, S.S.; Khan, J. Natural anti-biofilm agents: Strategies to control biofilm-forming pathogens. Front. Microbiol. 2020, 11, 566325. [Google Scholar] [CrossRef]
- Bodede, O.; Shaik, S.; Chenia, H.; Singh, P.; Moodley, R. Quorum sensing inhibitory potential and in silico molecular docking of flavonoids and novel terpenoids from Senegalia nigrescens. J. Ethnopharmacol. 2018, 216, 134–146. [Google Scholar] [CrossRef]
- Dogan, S.; Gokalsin, B.; Senkardes, I.; Dogan, A.; Sesal, N.C. Anti-quorum sensing and anti-biofilm activities of Hypericum perforatum extracts against Pseudomonas aeruginosa. J. Ethnopharmacol. 2019, 235, 293–300. [Google Scholar] [CrossRef]
- Famuyide, I.M.; Aro, A.O.; Fasina, F.O. Antibacterial and antibiofilm activity of acetone leaf extracts of nine under-investigated south African Eugenia and Syzygium (Myrtaceae) species and their selectivity indices. BMC Complement. Altern. Med. 2019, 19, 141. [Google Scholar] [CrossRef]
- Alam, K.; Al Farraj, D.A.; Mah-e-Fatima, S.; Yameen, M.A.; Elshikh, M.S.; Alkufeidy, R.M.; Mustafa, A.M.A.; Bhasme, P.; Alshammari, M.K.; Alkubaisi, N.A.; et al. Anti-biofilm activity of plant derived extracts against infectious pathogen-Pseudomonas aeruginosa PAO1. J. Infect. Public Health 2020, 13, 1734–1741. [Google Scholar] [CrossRef]
- Saeloh, D.; Visutthi, M. Efficacy of Thai Plant Extracts for Antibacterial and Anti-Biofilm Activities against Pathogenic Bacteria. Antibiotics 2021, 10, 1470. [Google Scholar] [CrossRef]
- Olawuwo, O.S.; Famuyide, I.M.; McGaw, L.J. Antibacterial and Antibiofilm Activity of Selected Medicinal Plant Leaf Extracts Against Pathogens Implicated in Poultry Diseases. Front. Vet. Sci. 2022, 9, 820304. [Google Scholar] [CrossRef]
- Adeyemo, R.O.; Famuyide, I.M.; Dzoyem, J.P.; Lyndy, J.M. Anti-Biofilm, Antibacterial, and Anti-Quorum Sensing Activities of Selected South African Plants Traditionally Used to Treat Diarrhoea. Evid. Based Complement. Alternat Med. 2022, 2022, 1307801. [Google Scholar] [CrossRef]
- Wylie, M.R.; Merrell, D.S. The Antimicrobial Potential of the Neem Tree Azadirachta indica. Front. Pharmacol. 2022, 30, 891535. [Google Scholar] [CrossRef]
- Islas, J.F.; Acosta, E.; Buentello, Z.G.; Delgado-Gallegos, J.L.; Moreno-Treviño, M.G.; Escalante, B.; Moreno-Cuevas, J.E. An overview of Neem (Azadirachta indica) and its potential impact on health. J. Funct. Food 2020, 74, 104171. [Google Scholar] [CrossRef]
- Hernández-Aquino, E.; Muriel, P. Beneficial effects of naringenin in liver diseases: Molecular mechanisms. World J. Gastrol. 2018, 24, 1679–1707. [Google Scholar] [CrossRef]
- Saleem, S.; Muhammad, G.; Hussain, M.A.; Bukhari, S.N.A. A comprehensive review of phytochemical profile, bioactives for pharmaceuticals, and pharmacological attributes of Azadirachta indica. Phytothe. Res. 2018, 32, 1241–1272. [Google Scholar] [CrossRef]
- Altayb, H.N.; Yassin, N.F.; Hosawi, S.; Kazmi, I. In-vitro and in-silico antibacterial activity of Azadirachta indica (Neem), methanolic extract, and identification of Beta.d-Mannofuranoside as a promising antibacterial agent. BMC Plant Biol. 2022, 25, 262. [Google Scholar] [CrossRef]
- Thamer, F.H.T.; Thamer, N. Gas chromatography—Mass spectrometry (GC-MS) profiling reveals newly described bioactive compounds in Citrullus colocynthis (L.) seeds oil extracts. Heliyon 2023, 9, e16861. [Google Scholar] [CrossRef]
- Ali, E.; Islam, M.S.; Hossen, M.I.; Khatun, M.M.; Islam, M.A. Extract of neem (Azadirachta indica) leaf exhibits bactericidal effect against multidrug resistant pathogenic bacteria of poultry. Vet. Med. Sci. 2021, 7, 1921–1927. [Google Scholar] [CrossRef]
- Mehrishi, P.; Agarwal, P.; Broor, S.; Sharma, A. Antibacterial and Antibiofilm Properties of Azadirachta indica (Neem), Aloe vera (Aloe vera), and Mentha piperita (Peppermint) against Multidrug-Resistant Clinical Isolates. Biomed. Biotechnol. Res. J. 2022, 6, 98–104. [Google Scholar] [CrossRef]
- Agu, P.C.; Afiukwa, C.A.; Orji, O.U. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci. Rep. 2023, 13, 13398. [Google Scholar] [CrossRef]
- de Melo, A.L.F.; Rossato, L.; Barbosa, M.D.S.; Palozi, R.A.C.; Alfredo, T.M.; Antunes, K.A.; Eduvirgem, J.; Ribeiro, S.M.; Simionatto, S. From the environment to the hospital: How plants can help to fight bacteria biofilm. Microbiol. Res. 2022, 261, 127074. [Google Scholar] [CrossRef]
- Asghari, M.; Khodadadi, E.; Pakdel, F.; Kamounah, F.S. Baicalin, a natural antimicrobial and anti-biofilm agent. J. Herb. Med. 2021, 27, 100432. [Google Scholar] [CrossRef]
- Ye, C.; Xu, S.; Wu, Z.; Wang, M. Cu3(PO4)2/BiVO4 photoelectrochemical sensor for sensitive and selective determination of synthetic antioxidant propyl gallate. Anal. Bioanal. Chem. 2022, 414, 4139–4147. [Google Scholar] [CrossRef]
- Ribeiro, S.M.; Felício, M.R.; Boas, E.V.; Gonçalves, S.; Costa, F.F.; Samy, R.P.; Santos, N.C.; Franco, O.L. New frontiers for anti-biofilm drug development. Pharmacol. Ther. 2016, 160, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Hu, W.; Tian, Z.; Yuan, D.; Yi, G.; Zhou, Y.; Cheng, Q.; Zhu, J.; Li, M. Developing natural products as potential anti-biofilm agents. Chin. Med. 2019, 14, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Huang, J.; Yi, Y.; Liu, S.; Liu, R.; Xiao, Z.; Li, C. Effects of Rhapontigenin as a Novel Quorum-Sensing Inhibitor on Exoenzymes and Biofilm Formation of Pectobacterium carotovorum subsp. carotovorum and Its Application in Vegetables. Molecules 2022, 27, 8878. [Google Scholar] [CrossRef]
- Prabu, S.L.; Umamaheswari, A.; Felciya, S.J.G. Investigation on the bio film eradication potential of selected medicinal plants against methicillin-resistant Staphylococcus aureus. Biotechnol. Rep. 2020, 28, e00523. [Google Scholar] [CrossRef]
- Ray, R.R.; Pattnaik, S. Contribution of phytoextracts in challenging the biofilms of pathogenic bacteria. Biocatal. Agric. Biotechnol. 2023, 48, 102642. [Google Scholar] [CrossRef]
- Kumar, S.; Das, R.; Lavigne, R.; Luyten, W. Indian medicinal plant extracts to control multidrug-resistant S. aureus, including in biofilms. S. Afr. J. Bot. 2020, 128, 283–291. [Google Scholar] [CrossRef]
- Al Bratty, M.; Makeen, H.A.; Alhazmi, H.A. Phytochemical, cytotoxic, and antimicrobial evaluation of the fruits of miswak plant, Salvadora persica L. J. Chem. 2020, 3, 4521951. [Google Scholar] [CrossRef]
- Levent, G.; Anderson, R.C.; Petrujkić, B. Evaluation of Thymol-β-d-Glucopyranoside as a Potential Prebiotic Intervention to Reduce Carriage of Zoonotic Pathogens in Weaned and Feeder Pigs. Microorganisms 2021, 9, 860. [Google Scholar] [CrossRef]
- Kumar, P.; Sati, S.C. Chemical composition, antioxidant and antimicrobial activities of Himalayan Fraxinus micrantha Lingelsh leaf extract. Nat. Prod. Res. 2021, 35, 3519–3523. [Google Scholar] [CrossRef]
- Eltabey, S.M.; Ibrahim, A.H.; Zaky, M.M.; Ibrahim, A.E.; Alrashdi, Y.B.A.; El Deeb, S.; Saleh, M.M. The Promising Effect of Ascorbic Acid and Paracetamol as Anti-Biofilm and Anti-Virulence Agents against Resistant Escherichia coli. Curr. Issues Mol. Biol. 2024, 46, 6805–6819. [Google Scholar] [CrossRef]
- Sun, C.Q.; Johnson, K.D.; Wong, H.; Foo, L.Y. Biotransformation of Flavonoid Conjugates with Fatty Acids and Evaluations of Their Functionalities. Front. Pharmacol. 2017, 8, 759. [Google Scholar] [CrossRef]
- Youssef, A.M.M.; Maaty, D.A.M.; Al-Saraireh, Y.M. Phytochemical Analysis and Profiling of Antioxidants and Anticancer Compounds from Tephrosia purpurea (L.) subsp. apollinea Family Fabaceae. Molecules 2023, 28, 3939. [Google Scholar] [CrossRef]
- Sun, M.; Zhou, Z.; Dong, J.; Zhang, J.; Xia, Y.; Shu, R. Antibacterial and antibiofilm activities of docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) against periodontopathic bacteria. Microb. Pathog. 2016, 99, 196–203. [Google Scholar] [CrossRef]
- Ghorab, M.M.M.; Soliman, A.; El-Sayyad, G.S.; Abdel-Kader, M.S.; El-Batal, A.I. Synthesis, Antimicrobial, and Antibiofilm Activities of Some Novel 7-Methoxyquinoline Derivatives Bearing Sulfonamide Moiety against Urinary Tract Infection-Causing Pathogenic Microbes. Int. J. Mol. Sci. 2023, 24, 8933. [Google Scholar] [CrossRef]
- Hrichi, S.; Chaâbane-Banaoues, R.; Alibrando, F. Chemical Composition, Antifungal and Anti-Biofilm Activities of Volatile Fractions of Convolvulus althaeoides L. Roots from Tunisia. Molecules 2022, 27, 6834. [Google Scholar] [CrossRef]
- Hikaambo, C.; Kaacha, L.; Mudenda, S.; Nyambe, M.; Chabalenge, B.; Phiri, M.; Biete, L.; Akapelwa, T.; Mufwambi, W.; Chulu, M.; et al. Phytochemical Analysis and Antibacterial Activity of Azadirachta indica Leaf Extracts against Escherichia coli. Pharmacol. Pharm. 2022, 13, 1–10. [Google Scholar] [CrossRef]
- Ibrahim, N.; Kebede, A. In Vitro antibacterial Activities of Methanol and Aqueous Leave Extracts of Selected Medicinal Plants against Human Pathogenic Bacteria. Saudi J. Biol. Sci. 2020, 27, 2261–2268. [Google Scholar] [CrossRef]
- Kaur, R.; Tiwari, A.; Manish, M.; Maurya, I.K.; Bhatnagar, R.; Singh, S. Common Garlic (Allium sativum L.) Has Potent Anti-Bacillus Anthracis Activity. J. Ethnopharmacol. 2021, 264, 113230. [Google Scholar] [CrossRef]
- Morales-Covarrubias, M.S.; García-Aguilar, N.; Bolan-Mejía, M.D.; Puello-Cruz, A.C. Evaluation of Medicinal Plants and Colloidal Silver Efficiency against Vibrio parahaemolyticus Infection in Litopenaeus vannamei Cultured at Low Salinity. Dis. Aquat. Org. 2016, 122, 57–65. [Google Scholar] [CrossRef]
- Ravva, S.V.; Korn, A. Effect of Neem (Azadirachta indica) on the Survival of Escherichia coli O157:H7 in Dairy Manure. Int. J. Environ. Res. Public Health 2015, 12, 7794–7803. [Google Scholar] [CrossRef]
- Bharitkar, Y.P.; Bathini, S.; Ojha, D.; Ghosh, S.; Mukherjee, H.; Kuotsu, K.; Chattopadhyay, D.; Mondal, N.B. Antibacterial and antiviral evaluation of sulfonoquinovosyldiacylglyceride: A glycolipid isolated from Azadirachta indica leaves. Lett. Appl. Microbiol. 2014, 58, 184–189. [Google Scholar] [CrossRef]
- Haris, Z.; Ahmad, I. Evaluation of antioxidant-rich bioactive plant extracts with special reference to Moringa oleifera Lam. for their interference to bacterial quorum sensing and biofilm, Phytomed. Plus 2024, 4, 100511. [Google Scholar] [CrossRef]
- Mazaheri, T.; Cervantes-Huamán, B.R.H.; Bermúdez-Capdevila, M.; Ripolles-Avila, C.; Rodríguez-Jerez, J.J. Listeria monocytogenes biofilms in the food industry: Is the current hygiene program sufficient to combat the persistence of the pathogen? Microorganisms 2021, 9, 181. [Google Scholar] [CrossRef]
- Kakoullis, L.; Papachristodoulou, E.; Chra, P.; Panos, G. Mechanisms of antibiotic resistance in important Gram-positive and Gram-negative pathogens and novel antibiotic solutions. Antibiotics 2021, 10, 415. [Google Scholar] [CrossRef]
- Zhou, J.W.; Ji, P.C.; Wang, C.Y.; Yang, Y.J.; Zhao, X.Y.; Tang, H.Z.; Tang, S.R. Synergistic effect of propyl gallate and antibiotics against biofilms of Serratia marcescens and Erwinia carotovora in vitro. LWT Food Sci. Technol. 2023, 173, 114258. [Google Scholar] [CrossRef]
- Géry, A.; Gravey, F.; Fines-Guyon, M.; Guerin, F.; Ethuin, F.; Borgey, F.; Lubrano, J.; Le Hello, S. Investigation of Serratia marcescens surgical site infection outbreak associated with peroperative ultrasonography probe. J. Hosp. Infect. 2021, 111, 184–188. [Google Scholar] [CrossRef]
- Francés-Cuesta, C.; Sánchez-Hellín, V.; Gomila, B.; González-Candelas, F. Is there a widespread clone of Serratia marcescens producing outbreaks worldwide? J. Hosp. Infect. 2021, 108, 7–14. [Google Scholar] [CrossRef]
- Guerra, M.E.S.; Destro, G.; Vieira, B.; Lima, A.S.; Ferraz, L.F.C.; Hakansson, A.P.; Darrieux, M.; Converso, T.R. Klebsiella pneumoniae biofilms and their role in disease pathogenesis. Front. Cell. Infect. Microbiol. 2022, 12, 877995. [Google Scholar] [CrossRef]
- Saifi, S.; Ashraf, A.; Hasan, G.M.; Shamsi, A.; Hassan, I. Insights into the preventive actions of natural compounds against Klebsiella pneumoniae infections and drug resistance. Fitoterapia 2024, 173, 105811. [Google Scholar] [CrossRef]
- Etikala, A.; Thamburaj, S.; Johnson, A.M.; Sarma, C.; Mummaleti, G.; Kalakandan, S.K. Incidence, toxin gene profile, antibiotic resistance and antibacterial activity of Allium parvum and Allium cepa extracts on Bacillus cereus isolated from fermented millet-based food. LWT 2022, 160, 113314. [Google Scholar] [CrossRef]
- Rouzeau-Szynalski, K.; Stollewerk, K.; Messelh¨ausser, U.; Ehling-Schulz, M. Why be serious about emetic Bacillus cereus: Cereulide production and industrial challenges. Food Microbiol. 2020, 85, 103279. [Google Scholar] [CrossRef] [PubMed]
- Min Park, K.; Jeong, M.; Park, K.J.; Koo, M. Prevalence, enterotoxin genes, and antibiotic resistance of Bacillus cereus isolated from raw vegetables in Korea. J. Food Prot. 2018, 81, 1590–1597. [Google Scholar] [CrossRef]
- Dietrich, R.; Jessberger, N.; Ehling-Schulz, M.; M¨artlbauer, E.; Granum, P.E. The food poisoning toxins of Bacillus cereus. Toxins 2021, 13, 98. [Google Scholar] [CrossRef]
- Ranasinghe, S.; Fhogartaigh, C.N. Bacterial gastroenteritis. Medicine 2021, 49, 687–693. [Google Scholar] [CrossRef]
- Sah, S.K.; Rasool, U.; Hemalatha, S. Andrographis paniculata extract inhibit growth, biofilm formation in multidrug resistant strains of Klebsiella pneumoniae. J. Tradit. Complement. Med. 2019, 10, 599–604. [Google Scholar] [CrossRef]
- Lahiri, D.; Nag, M.; Dutta, B.; Mukherjee, I.; Ghosh, S.; Dey, A. Catechin as the Most Efficient Bioactive Compound from Azadirachta indica with Antibiofilm and Anti-Quorum Sensing Activities against Dental Biofilm: An In Vitro and In Silico Study. Appl. Biochem. Biotechnol. 2021, 193, 1617–1630. [Google Scholar] [CrossRef]
- Guchhait, K.C.; Manna, T.; Barai, M.; Karmakar, M.; Nandi, S.K.; Jana, D. Antibiofilm and Anticancer Activities of Unripe and Ripe Azadirachta indica (Neem) Seed Extracts. BMC Complement. Med. Ther. 2022, 22, 42. [Google Scholar] [CrossRef]
- Mistry, K.S.; Sanghvi, Z.; Parmar, G.; Shah, S.; Pushpalatha, K. Antibacterial Efficacy of Azadirachta indica, Mimusops elengi and 2% CHX on Multispecies Dentinal Biofilm. J. Conserv. Dent. 2015, 18, 461–466. [Google Scholar] [CrossRef]
- Quelemes, P.V.; Perfeito, M.L.; Guimarães, M.A.; dos Santos, R.C.; Lima, D.F.; Nascimento, C. Effect of Neem (Azadirachta indica A. Juss) Leaf Extract on Resistant Staphylococcus aureus Biofilm Formation and Schistosoma mansoni Worms. J. Ethnopharmacol. 2015, 175, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, K.; Bharath, N.; Natarajan, R.; Dinesh, S.; Murugesan, S.; Selvaraj, S. Comparative Evaluation of Antimicrobial Efficacy of Toothpastes Containing Probiotic and Neem as Primary Ingredient on Salivary Streptococcus mutans in Melmaruvathur Population: An In Vivo Study. J. Pharm. Bioallied Sci. 2020, 12, S595–S600. [Google Scholar] [CrossRef]
- Tasanarong, T.; Patntirapong, S.; Aupaphong, V. The Inhibitory Effect of a Novel Neem Paste against Cariogenic Bacteria. J. Clin. Exp. Dent. 2021, 13, e1083–e1088. [Google Scholar] [CrossRef]
- Wylie, M.R.; Windham, I.H.; Blum, F.C.; Wu, H.; Merrell, D.S. In Vitro antibacterial Activity of Nimbolide against Helicobacter pylori. J. Ethnopharmacol. 2021, 285, 114828. [Google Scholar] [CrossRef] [PubMed]
- Reece, R.J.; Maxwell, A. DNA gyrase: Structure and function. Crit. Rev. Biochem. Mol. Biol. 1991, 26, 335–375. [Google Scholar] [CrossRef] [PubMed]
- Lafitte, D.; Lamour, V.; Tsvetkov, B.O.; Makarov, A.A.; Klich, M.; Deprez, P.; Moras, D.; Briand, C.; Gilli, R. DNA gyrase interaction with coumarin-based inhibitors: The role of the hydroxybenzoate isopentenyl moiety and the 5′-methyl group of the noviose. Biochemistry 2002, 41, 7217–7223. [Google Scholar] [CrossRef] [PubMed]
- Gennaro, L. Extraction Technology for Medicinal and Aromatic Plant; United Nations Industrial Development Organization and the International Centre for Science and High Technology: Trieste, Italy, 2008; p. 23. [Google Scholar]
- Abdel-Hakeem, S.S.; Mahmoud, G.A.-E.; Abdel-Hafeez, H.H. Evaluation and Microanalysis of Parasitic and Bacterial Agents of Egyptian Fresh Sushi, Salmo salar. Microsc. Microanal. 2019, 25, 1498–1508. [Google Scholar] [CrossRef]
- Mahmoud, G.A.-E.; Osman, Y.A.; Abdel-Hakeem, S.S. Hydrolytic bacteria associated with natural helminth infection in the midgut of Red Sea marbled spinefoot rabbit fish Siganus rivulatus. Microb. Pathog. 2020, 147, 104404. [Google Scholar] [CrossRef]
- Elshahawy, A.M.; Shalaby, M.S.; Rashad, M.; Taha, E.O.; Mahmoud, G.A.-E. Multi-characterization and antipathogenic bacterial activities of cerium doped manganese nano ferrites: Structural, and magnetic insights. Inorg. Chem. Commun. 2024, 163, 112283. [Google Scholar] [CrossRef]
- de Castro Melo, P.; Ferreira, L.M.; Filho, A.N.; Zafalon, L.F.; Vicente, H.I.; de Souza, V. Comparison of methods for the detection of biofilm formation by Staphylococcus aureus isolated from bovine subclinical mastitis. Braz. J. Microbiol. 2013, 44, 119–124. [Google Scholar] [CrossRef]
- Cruz, C.D.; Shah, S.; Tammela, P. Defining conditions for biofilm inhibition and eradication assays for Gram-positive clinical reference strains. BMC Microbiol. 2018, 18, 173. [Google Scholar] [CrossRef]
- Yimgang, L.V.; Kouipou Toghueo, R.M.; Mbekou, I.M.; Dize, D.; Boyom, F.F. Crude metabolites from endophytic fungi inhabiting Cameroonian Annona muricata inhibit the causative agents of urinary tract infections. PLoS ONE 2022, 17, e0267246. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J.; Vina, A.D. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]






| No. | Compounds | Calculated Binding Energies (kcal/mol) | Peak Area (%) |
|---|---|---|---|
| 1 | (2-Methyl-[1,3]dioxolan-2-yl)-acetic acid, phenyl ester | −5.68 ± 0.50 | 1.42 |
| 2 | 2-Acetonyl-9-[3-deoxy-a-d-ribouranosyl]hypoxanthine | −6.34 ± 0.44 | 0.21 |
| 3 | Ethyl iso-allocholate | −6.67 ± 0.38 | 0.85 |
| 4 | Pseudosolasodine diacetate | −7.07 ± 0.45 | 3.32 |
| 5 | 4-Methoxy-6-methyl-5-nitroisobenzofuran-1,3-dione | −5.82 ± 0.33 | 0.12 |
| 6 | Benzofuran, 7-(2,4-dinitrophenoxy)-3-ethoxy-2,3-dihydro-2,2-dimethyl- | −6.2 ± 0.33 | 0.03 |
| 7 | Methyl 4-O-acetyl-2,3,6-tri-O-ethyl-a-d-mannopyranoside | −4.54 ± 0.23 | 0.58 |
| 8 | 2-[4-methyl-6-(2,6,6-trimethylcyclohex-1-enyl)hexa-1,3,5-trienyl]cyclohex-1-en 1 carboxaldehyde | −6.6 ± 0.35 | 0.80 |
| 9 | Preg-4-en-3-one, 17a-hydroxy-17a-cyano- | −6.35 ± 0.34 | 0.06 |
| 10 | 9-Oximino-2,7-diethoxyfluorene | −7.12 ± 0.61 | 0.06 |
| 11 | 17a-Allyl-3a-acetoxy-17a-aza-D-homoandrost-5-ene-17-one | −6.95 ± 0.79 | 0.06 |
| 12 | N-[3-Diethylaminopropyl]-4-oxo-1,2,3,4,5,6,7,8-octohydroquinoline | −4.8 ± 0.44 | 0.21 |
| 13 | Carbonic acid, (ethyl)(1,2,4-triazol-1-ylmethyl) diester | −4.37 ± 0.32 | 0.16 |
| 14 | Sarreroside | −7.53 ± 0.29 | 0.18 |
| 15 | 9,10-Secocholesta-5,7,10(19)-triene-3,24,25-triol, (3a,5Z,7E)- | −6.67 ± 0.22 | 0.24 |
| 16 | Pyrimidine-2,4(1H,3H)-dione,1,3-dimethyl-6-[2-(4-morpholyl)ethenyl]-5-nitro- | −5.74 ± 0.36 | 0.11 |
| 17 | Ergosta-5,22-dien-3-ol, acetate, (3a,22E)- | −7.1 ± 0.60 | 0.65 |
| 18 | Cholestan−3-ol, 2-methylene-, (3a,5a)- | −7.35 ± 0.50 | 0.29 |
| 19 | Pregn-4-ene-3,20-dione, 17,21-dihydroxy-, bis(O-methyloxime) | −6.11 ± 0.35 | 0.39 |
| 20 | Estra-1,3,5(10)-trien-17a-ol | −7.00 ± 0.92 | 2.17 |
| 21 | Clorobiocin (Standard) | −8.35 ± 0.68 | |
| 1KZN: The complex crystal structure of DNA gyrase with its inhibitor (Clorobiocin) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahmoud, G.A.-E.; Rashed, N.M.; El-Ganainy, S.M.; Salem, S.H. Unveiling the Neem (Azadirachta indica) Effects on Biofilm Formation of Food-Borne Bacteria and the Potential Mechanism Using a Molecular Docking Approach. Plants 2024, 13, 2669. https://doi.org/10.3390/plants13182669
Mahmoud GA-E, Rashed NM, El-Ganainy SM, Salem SH. Unveiling the Neem (Azadirachta indica) Effects on Biofilm Formation of Food-Borne Bacteria and the Potential Mechanism Using a Molecular Docking Approach. Plants. 2024; 13(18):2669. https://doi.org/10.3390/plants13182669
Chicago/Turabian StyleMahmoud, Ghada Abd-Elmonsef, Nahed M. Rashed, Sherif M. El-Ganainy, and Shimaa H. Salem. 2024. "Unveiling the Neem (Azadirachta indica) Effects on Biofilm Formation of Food-Borne Bacteria and the Potential Mechanism Using a Molecular Docking Approach" Plants 13, no. 18: 2669. https://doi.org/10.3390/plants13182669
APA StyleMahmoud, G. A.-E., Rashed, N. M., El-Ganainy, S. M., & Salem, S. H. (2024). Unveiling the Neem (Azadirachta indica) Effects on Biofilm Formation of Food-Borne Bacteria and the Potential Mechanism Using a Molecular Docking Approach. Plants, 13(18), 2669. https://doi.org/10.3390/plants13182669








