Investigation of Imidazolinone Herbicide Resistance Gene with KASP Markers for Japonica/Geng Rice Varieties in the Huanghuaihai Region of China
Abstract
1. Introduction
2. Results
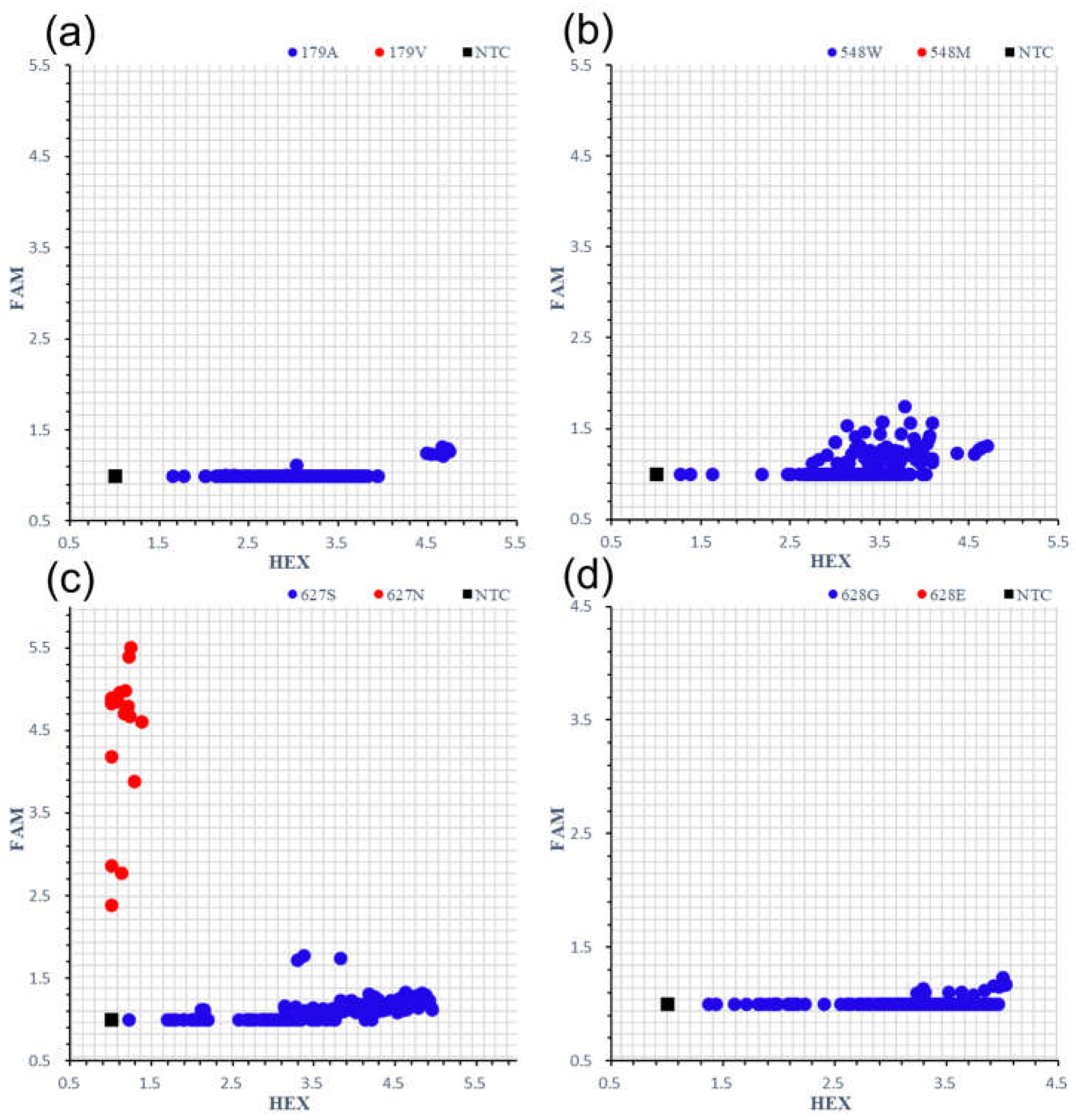
2.1. Development of Specific KASP Markers to Screen for Each Variant of ALS1179V/548M/627N/628E from Collected Japonica/Geng Rice Varieties or Lines
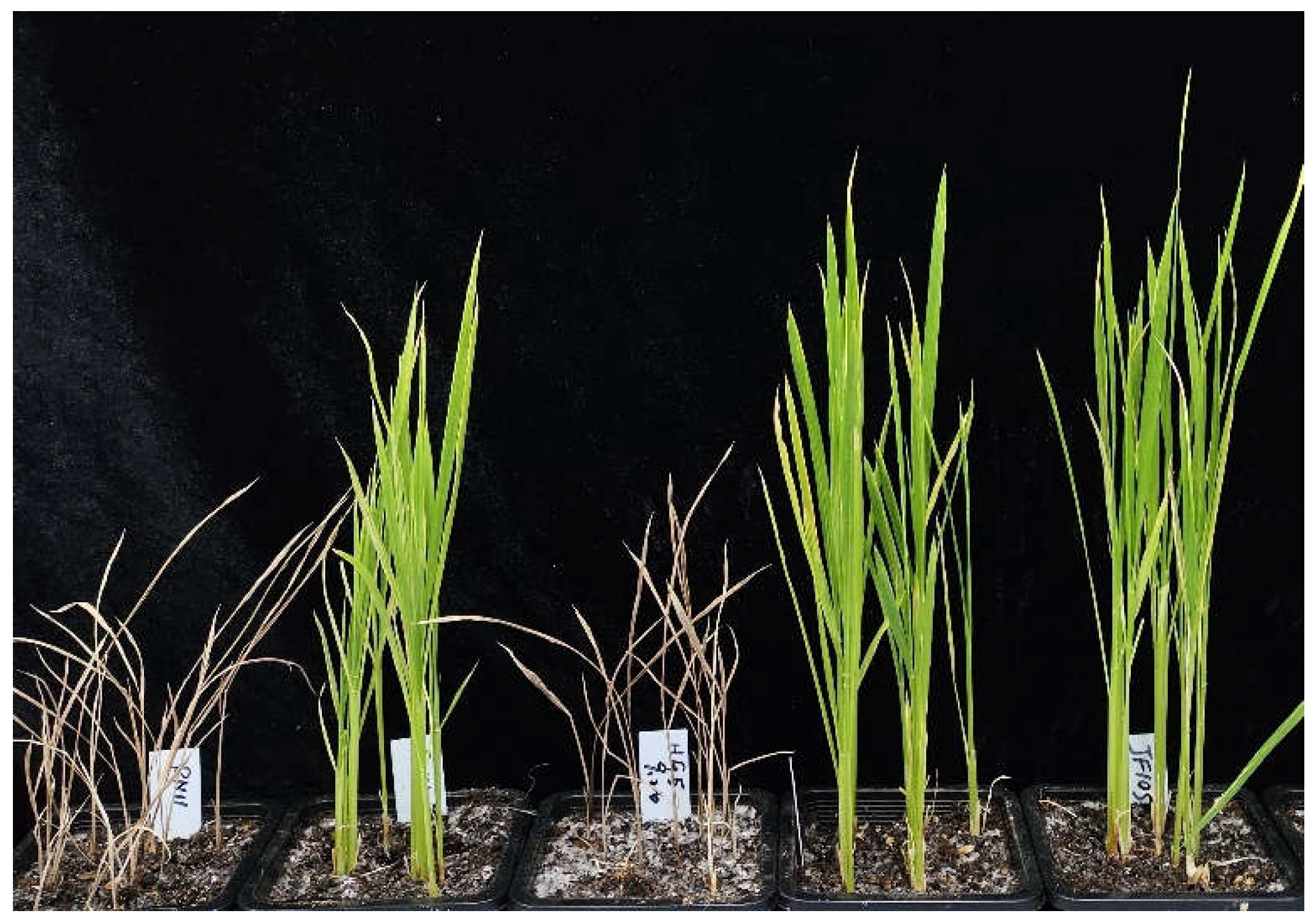
2.2. Imidazolinone Herbicide Resistance Assay for OsALS1627N Rice Varieties
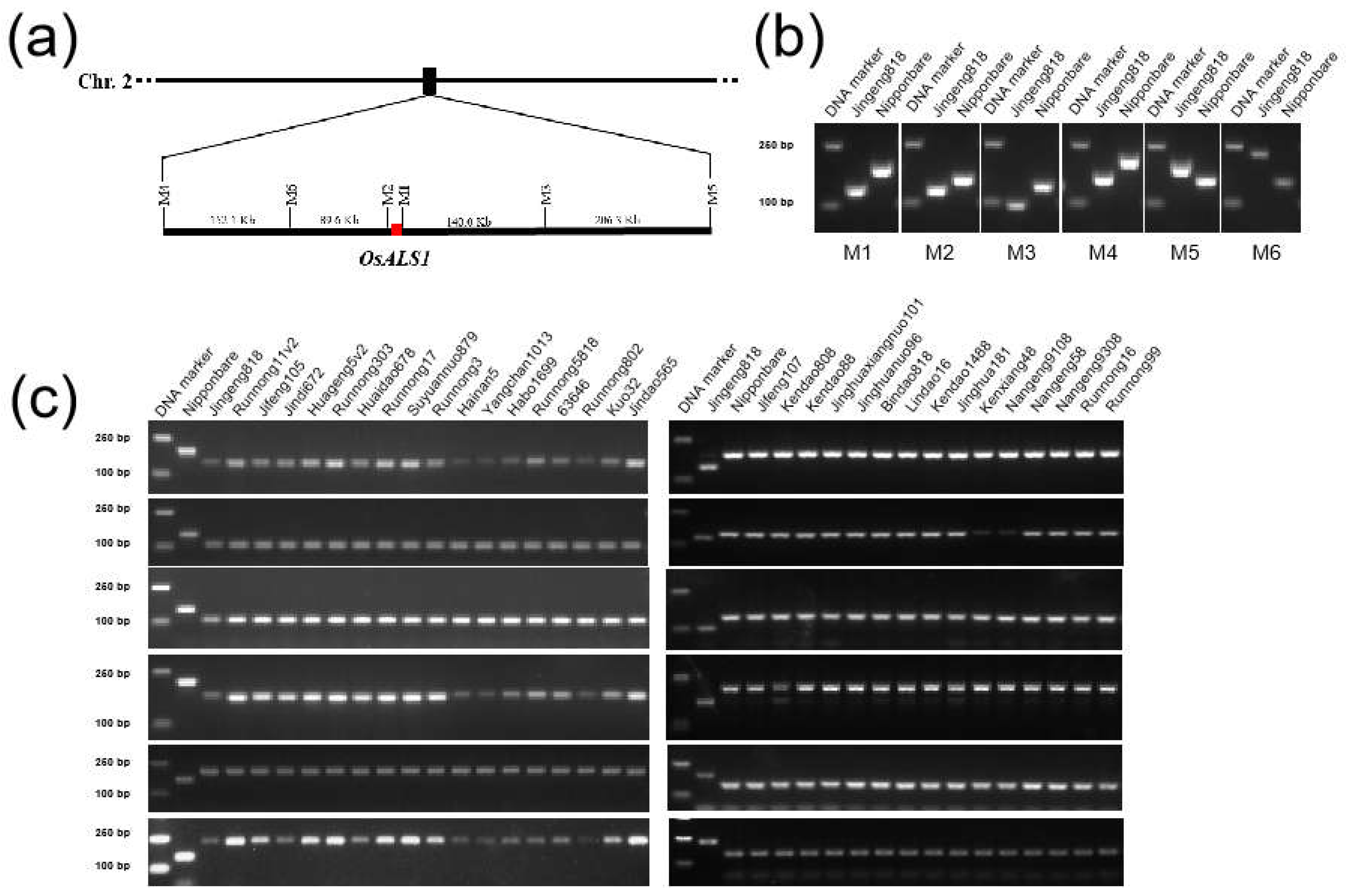
2.3. Development of Molecular Markers Tightly Linked to OsALS1627N from Sequenced JG818
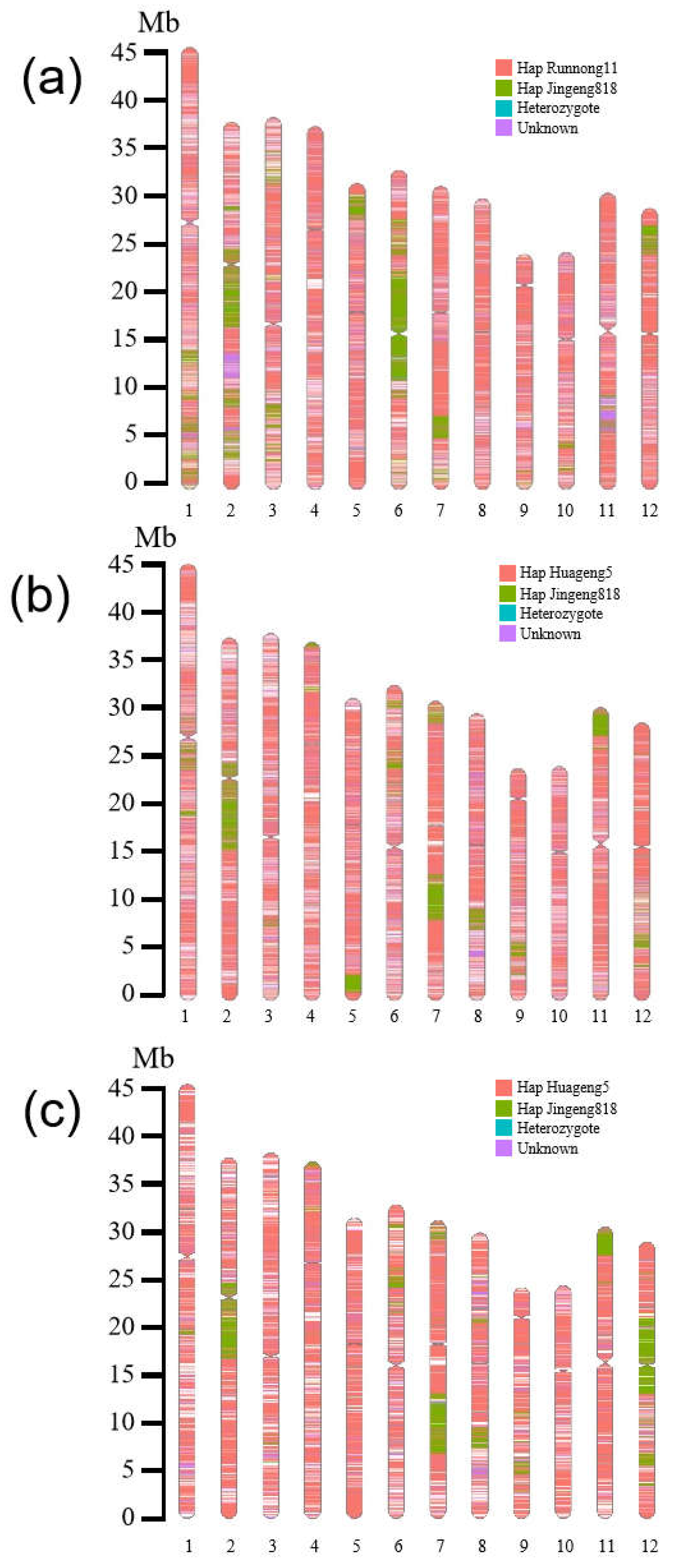
2.4. Comparative Analysis of the Genome Sequence of Herbicide-Resistant Rice
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Phenotyping of Imidazolinone Herbicide Resistance
4.3. DNA Extraction, InDel Marker Development and PCR
4.4. Whole-Genomic Sequencing and Data Analysis
4.5. KASP Marker Development and Genotyping Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Vanga, S.K.; Saxena, R.; Orsat, V.; Raghavan, V. Effect of Climate Change on the Yield of Cereal Crops: A Review. Climate 2018, 6, 41. [Google Scholar] [CrossRef]
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. In Technical Considerations for Rice Fortification in Public Health; DeRegil, L.M., Laillou, A., MoenchPfanner, R., PenaRosas, J.P., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2014; Volume 1324, pp. 7–14. [Google Scholar]
- Springmann, M.; Clark, M.; Mason-D’Croz, D.; Wiebe, K.; Bodirsky, B.L.; Lassaletta, L.; de Vries, W.; Vermeulen, S.J.; Herrero, M.; Carlson, K.M.; et al. Options for Keeping the Food System Within Environmental Limits. Nature 2018, 562, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Akbar, N.; Ehsanullah; Jabran, K.; Ali, M.A. Weed Management Improves Yield and Quality of Direct Seeded Rice. Aust. J. Crop. Sci. 2011, 5, 688–694. [Google Scholar]
- Fartyal, D.; Agarwal, A.; James, D.; Borphukan, B.; Ram, B.; Sheri, V.; Agrawal, P.K.; Achary, V.M.M.; Reddy, M.K. Developing Dual Herbicide Tolerant Transgenic Rice Plants for Sustainable Weed Management. Sci. Rep. 2018, 8, 11598. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Chen, L.; Deng, X.W.; Tang, X. Development of Herbicide Resistance Genes and Their Application in Rice. Crop. J. 2022, 10, 26–35. [Google Scholar] [CrossRef]
- Delye, C.; Jasieniuk, M.; Le Corre, V. Deciphering the Evolution of Herbicide Resistance in Weeds. Trends. Genet. 2013, 29, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Chen, S.; Meng, X.; Chai, Z.; Wang, D.; Yuan, Y.; Chen, K.; Jiang, L.; Li, J.; Gao, C. Generating Broad-Spectrum Tolerance to ALS-Inhibiting Herbicides in Rice by Base Editing. Sci. China-Life Sci. 2021, 64, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; He, D.; Liang, Q.; Zhang, Z.; Wu, J.; Zhang, Z.; Zhang, J.; Wang, L.; Zhang, C.; Zhao, P.; et al. Loss of OsARF18 Function Confers Glufosinate Resistance in Rice. Mol. Plant 2023, 16, 1355–1358. [Google Scholar] [CrossRef]
- Li, Y.; Liang, J.; Deng, B.; Jiang, Y.; Zhu, J.; Chen, L.; Li, M.; Li, J. Applications and Prospects of CRISPR/Cas9-Mediated Base Editing in Plant Breeding. Curr. Issues Mol. Biol. 2023, 45, 918–935. [Google Scholar] [CrossRef]
- Sudianto, E.; Beng-Kah, S.; Ting-Xiang, N.; Saldain, N.E.; Scott, R.C.; Burgos, N.R. Clearfield® Rice: Its Development, Success, and Key Challenges on a Global Perspective. Crop. Prot. 2013, 49, 40–51. [Google Scholar] [CrossRef]
- Tan, S.Y.; Evans, R.R.; Dahmer, M.L.; Singh, B.K.; Shaner, D.L. Imidazolinone-Tolerant Crops: History, Current Status and Future. Pest Manag. Sci. 2005, 61, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.D.; Nouwens, A.; Lonhienne, T.G.; Guddat, L.W. Comprehensive Understanding of Acetohydroxyacid Synthase Inhibition by Different Herbicide Families. Proc. Natl. Acad. Sci. USA 2017, 114, E1091–E1100. [Google Scholar] [CrossRef] [PubMed]
- Pozniak, C.J.; Birk, I.T.; O’Donoughue, L.S.; Ménard, C.; Hucl, P.J.; Singh, B.K. Physiological and Molecular Characterization of Mutation-Derived Imidazolinone Resistance in Spring Wheat. Crop. Sci. 2004, 44, 1434–1443. [Google Scholar] [CrossRef]
- Wright, T.R.; Bascomb, N.F.; Sturner, S.F.; Penner, D. Biochemical Mechanism and Molecular Basis for ALS-Inhibiting Herbicide Resistance in Sugarbeet (Beta vulgaris) Somatic Cell Selections. Weed Sci. 1998, 46, 13–23. [Google Scholar] [CrossRef]
- Thompson, C.; Tar’an, B. Genetic Characterization of the Acetohydroxyacid Synthase (AHAS) Gene Responsible for Resistance to Imidazolinone in Chickpea (Cicer arietinum L.). Theor Appl Genet. 2014, 127, 1583–1591. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.V.; Mendes, R.; Suzukawa, A.; Adegas, F.; Marcelino-Guimaraes, F.; Oliveira, R. Target-site Mutation Confers Cross-Resistance to ALS-Inhibiting Herbicides in Erigeron sumatrensis from Brazil. Plants 2022, 11, 467. [Google Scholar] [CrossRef] [PubMed]
- Tranel, P.J.; Wright, T.R. Resistance of Weeds to ALS-Inhibiting Herbicides: What Have We Learned? Weed Sci. 2002, 50, 700–712. [Google Scholar] [CrossRef]
- An, C.; Shao, Y.; Peng, Y.; Mao, B.; Zhao, B. Ala179Val Mutation in Rice Acetolactate Synthase Confers Broad-Spectrum Resistance to ALS Inhibitor Herbicides. J. Plant Genet. Res. 2024, 25, 259–269. (In Chinese) [Google Scholar]
- Chen, L.; Gu, G.; Wang, C.; Chen, Z.; Yan, W.; Jin, M.; Xie, G.; Zhou, J.; Deng, X.W.; Tang, X. Trp548Met Mutation of Acetolactate Synthase in Rice Confers Resistance to a Broad Spectrum of ALS-Inhibiting Herbicides. Crop. J. 2021, 9, 750–758. [Google Scholar] [CrossRef]
- Piao, Z.; Wang, W.; Wei, Y.; Zonta, F.; Wan, C.; Bai, J.; Wu, S.; Wang, X.; Fang, J. Characterization of an Acetohydroxy Acid Synthase Mutant Conferring Tolerance to Imidazolinone Herbicides in Rice (Oryza sativa). Planta 2018, 247, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, A.; Hao, Y.; Xia, X.; Khan, A.; Xu, Y.; Varshney, R.K.; He, Z. Crop Breeding Chips and Genotyping Platforms: Progress, Challenges, and Perspectives. Mol. Plant 2017, 10, 1047–1064. [Google Scholar] [CrossRef] [PubMed]
- Semagn, K.; Babu, R.; Hearne, S.; Olsen, M. Single Nucleotide Polymorphism Genotyping using Kompetitive Allele Specific PCR (KASP): Overview of the Technology and its Application in Crop Improvement. Mol. Breed. 2014, 33, 1–14. [Google Scholar] [CrossRef]
- Shavrukov, Y. Comparison of SNP and CAPS Markers Application in Genetic Research in Wheat and Barley. BMC Plant Biol. 2016, 16, 11. [Google Scholar] [CrossRef] [PubMed]
- Babu, P.; Baranwal, D.K.; Harikrishna; Pal, D.; Bharti, H.; Joshi, P.; Thiyagarajan, B.; Gaikwad, K.B.; Bhardwaj, S.C.; Singh, G.P.; et al. Application of Genomics Tools in Wheat Breeding to Attain Durable Rust Resistance. Front. Plant Sci. 2020, 11, 567147. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, B.; Yang, B.; Chen, T.; Xing, Y.; Sun, Z.; Xu, B.; Chi, M.; Lu, B.; Li, J.; et al. Genetic Analysis of Rice Neck Blast Resistance in Huang-Huai-Hai Region. Chin. J. Rice Sci. 2019, 33, 377–382. [Google Scholar]
- Gao, X.; Li, M.; Fang, F.; Li, J. Species Composition and Characterization of Weed Communities in Rice-Wheat Rotation Area in Huang-Huai-Hai Region. Acta Phytop. Sin. 2019, 46, 472–478. (In Chinese) [Google Scholar]
- Wang, F.; Yang, J.; Fan, F.; Li, W.; Wang, J.; Xu, Y.; Zhu, J.; Fei, Y.; Zhong, W. Development and Application of the Functional Marker for Imidazolinone Herbicides Resistant ALS Gene in Rice. Acta Agron Sin. 2018, 44, 324–331. [Google Scholar] [CrossRef]
- Li, M.; Feng, Z.; Cui, A.; Wang, G.; Xu, Z.; Huang, T.; Ding, W.; Hu, K.; Chen, Z.; Zuo, S. Improvement of Japonica Rice Resistance to Herbicides Through Introducing Imidazolinone Herbicide-Resistant Gene ALS627N. Jiangsu Agric. Sci. 2020, 50, 263–269. (In Chinese) [Google Scholar]
- Zhang, M.; Li, P.; Liu, X.; Zhang, H.; Li, Z.; Liu, F.; Yao, F. Breeding of Novel Rice Lines with Dual Resistance to Herbicide and Insect. Shangdong Agric. Sci. 2022, 54, 24–30. (In Chinese) [Google Scholar]
- Yu, Q.; Powles, S.B. Resistance to AHAS Inhibitor Herbicides: Current Understanding. Pest Manag. Sci. 2014, 70, 1340–1350. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Yu, Q.; Purba, E.; Li, M.; Walsh, M.; Friesen, S.; Powles, S.B. A Novel Amino Acid Substitution Ala-122-Tyr in ALS Confers High-Level and Broad Resistance Across ALS-Inhibiting Herbicides. Pest Manag. Sci. 2012, 68, 1164–1170. [Google Scholar] [CrossRef] [PubMed]
- Bernasconi, P.; Woodworth, A.R.; Rosen, B.A.; Subramanian, M.V.; Siehl, D.L. A Naturally Occurring Point Mutation Confers Broad Range Tolerance to Herbicides that Target Acetolactate Synthase. J. Biol. Chem. 1995, 270, 17381–17385. [Google Scholar] [CrossRef] [PubMed]
- Milliman, L.D.; Riechers, D.E.; Wax, L.M.; Simmons, F.W. Characterization of Two Biotypes of Imidazolinone-Resistant Eastern Black Nightshade (Solanum ptycanthum). Weed Sci. 2003, 51, 139–144. [Google Scholar] [CrossRef]
- Trucco, F.; Hager, A.G.; Tranel, P.J. Acetolactate Synthase Mutation Conferring Imidazolinone-Specific Herbicide Resistance in Amaranthus hybridus. J. Plant Physiol. 2006, 163, 475–479. [Google Scholar] [CrossRef]
- Whaley, C.A.; Wilson, H.P.; Westwood, J.H. ALS Resistance in Several Smooth Pigweed (Amaranthus hybridus) Biotypes. Weed Sci. 2006, 54, 828–832. [Google Scholar] [CrossRef]
- Webster, E.P.; Masson, J.A. Acetolactate Synthase-Inhibiting Herbicides on Imidazolinone-Tolerant Rice. Weed Sci. 2001, 49, 652–657. [Google Scholar] [CrossRef]
- Wang, F.; Li, Z.; Wang, Y.; Fu, J.; Yang, W.; Yin, H.; Wang, S.; Wang, Y.; Bai, T.; Zhang, Z. New Herbicide-Resistant Japonica Rice Germplasms Created by EMS Mutagenesis. J. Henan Agric. Sci. 2021, 50, 8–16. (In Chinese) [Google Scholar]
- Wu, Y.; Xiao, N.; Yu, L.; Cai, Y.; Pan, C.; Li, Y.; Zhang, X.; Huang, N.; Zhou, C.; Ji, H.; et al. Research Progress in Herbicide-Resistant Rice Germplasm Innovation in China. J. Plant Genet. Res. 2021, 22, 890–899. (In Chinese) [Google Scholar]
- Ren, Y.; Liu, B.; Jiang, H.; Cheng, W.; Tao, L.; Wu, K.; Wang, H.; Shen, G.; Fang, Y.; Zhang, C.; et al. Precision Editing of GLR1 Confers Glufosinate Resistance without Yield Penalty in Rice. Plant Biotechnol. J. 2023, 21, 2417–2419. [Google Scholar] [CrossRef]
- Qun, J.; Ling, X.; Tang, Z.; Zhou, Z.; Zhang, B. EMS Mutagenesis to Create Rice Anti-Acetyl-CoA Carboxylase Inhibitor-Herbicide Germplasm. Jiangsu J. Agric. Sci. 2023, 39, 305–312. (In Chinese) [Google Scholar]
- He, C.; Holme, J.; Anthony, J. SNP Genotyping: The KASP Assay. Methods Mol. Biol. 2014, 1145, 75–86. [Google Scholar] [PubMed]
- Addison, C.K.; Angira, B.; Kongchum, M.; Harrell, D.L.; Baisakh, N.; Linscombe, S.D.; Famoso, A.N. Characterization of Haplotype Diversity in the BADH2 Aroma Gene and Development of a KASP SNP Assay for Predicting Aroma in U.S. Rice. Rice 2020, 13, 47. [Google Scholar] [CrossRef] [PubMed]
- Cheon, K.; Jeong, Y.; Oh, H.; Oh, J.; Kang, D.; Kim, N.; Lee, E.; Baek, J.; Kim, S.L.; Choi, I.; et al. Development of 454 New Kompetitive Allele-Specific PCR (KASP) Markers for Temperate japonica Rice Varieties. Plants 2020, 9, 1531. [Google Scholar] [CrossRef] [PubMed]
- Grover, N.; Kumar, A.; Yadav, A.K.; Krishan, S.G.; Ellur, R.K.; Bhowmick, P.K.; Vinod, K.K.; Bollinedi, H.; Nagarajan, M.; Viswanathan, C.; et al. Marker Assisted Development and Characterization of Herbicide Tolerant Near Isogenic Lines of a Mega Basmati Rice Variety, “Pusa Basmati 1121”. Rice 2020, 13, 68. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-In-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]

| Marker | Forward Primer Sequence (5′-3′) | Reverse Primer Sequence (5′-3′) | Product Length (bp) |
|---|---|---|---|
| M1 | CCAAAGGTTCCCTAATCAAGTT | CGGGTTTAGATGTGGGTTTC | 142 1/191 2 |
| M2 | GGGAGTGATTCTGAATGTTA | AGATCCCAGATCATCCTTTC | 134 1/158 2 |
| M3 | ACCATCGTCATCGCTGTC | CGCTCAGGAGATCGAAGAT | 97 1/138 2 |
| M4 | TGTATGATAGTCAGCCTTGTG | GAGGTGGTAATAACGTGTGA | 170 1/219 2 |
| M5 | ACACAGTGTGAACTAACCTA | CTAAACAGGGCCGTAGTATA | 108 1/154 2 |
| M6 * | ATCAGCATCCTCAGTTCCATAA | GCACAAAGTGAAAGACACATTC | 220 1/146 2 |
| Varieties/Lines | Clean Reads (Gb) | Mapped Reads (Gb) | Coverage | Phenotype * | Number of SNPs | ||||
|---|---|---|---|---|---|---|---|---|---|
| JG818 | Sensitive Parent | Heterozygous | Unknown | Total | |||||
| Jingeng818 | 16.51 | 10.18 | 97.55% | T | - | - | - | - | - |
| Runnong11 | 6.39 | 6.34 | 97.41% | S | - | - | - | - | - |
| Runnong11v2 | 4.12 | 4.09 | 96.85% | T | 59,163 | 258,674 | 1330 | 25,384 | 344,551 |
| Huageng5 | 5.72 | 5.68 | 97.53% | S | - | - | - | - | - |
| Huageng5v2 | 5.25 | 5.23 | 97.33% | T | 44,277 | 280,909 | 1494 | 20,696 | 347,376 |
| Jifeng105 | 6.35 | 6.32 | 97.22% | S | 79,139 | 289,763 | 2066 | 20,510 | 391,478 |
| Marker | Primer Sequence (5′-3′) |
|---|---|
| ALS1179V | F1: GAAGGTGACCAAGTTCATGCTCTATGGGCGTCTCCTGGAAGA F2: GAAGGTCGGAGTCAACGGATTCTATGGGCGTCTCCTGGAAGG R: CCATCACGGGCCAGGTCCC |
| ALS1548M | F1: GAAGGTGACCAAGTTCATGCTCAACATTTGGGTATGGTGGTGCAAAT F2: GAAGGTCGGAGTCAACGGATTCAACATTTGGGTATGGTGGTGCAATG R: TCGCTCTCACATTCCGGGTTG |
| ALS1627N | F1: GAAGGTGACCAAGTTCATGCTATCATGTCCTTGAATGCGCCCCCAT F2: GAAGGTCGGAGTCAACGGATTATCATGTCCTTGAATGCGCCCCCAC R: CAGTCCGTGTAACAAAGAAGAGTGAAGTCC |
| ALS1628E | F1: GAAGGTGACCAAGTTCATGCTGTGCTGCCTATGATCCCAAGTGA F2: GAAGGTCGGAGTCAACGGATTGTGCTGCCTATGATCCCAAGTGG R: ACAGTCCTGCCATCACCATCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Feng, W.; Wang, T.; Zhang, H.; Mao, S.; Zhang, H.; Huang, W.; Liu, H.; Feng, S.; Chu, Z. Investigation of Imidazolinone Herbicide Resistance Gene with KASP Markers for Japonica/Geng Rice Varieties in the Huanghuaihai Region of China. Plants 2024, 13, 1097. https://doi.org/10.3390/plants13081097
Liu P, Feng W, Wang T, Zhang H, Mao S, Zhang H, Huang W, Liu H, Feng S, Chu Z. Investigation of Imidazolinone Herbicide Resistance Gene with KASP Markers for Japonica/Geng Rice Varieties in the Huanghuaihai Region of China. Plants. 2024; 13(8):1097. https://doi.org/10.3390/plants13081097
Chicago/Turabian StyleLiu, Peng, Wenjie Feng, Tao Wang, Huadong Zhang, Shuaige Mao, Hua Zhang, Wenchao Huang, Haifeng Liu, Shangzong Feng, and Zhaohui Chu. 2024. "Investigation of Imidazolinone Herbicide Resistance Gene with KASP Markers for Japonica/Geng Rice Varieties in the Huanghuaihai Region of China" Plants 13, no. 8: 1097. https://doi.org/10.3390/plants13081097
APA StyleLiu, P., Feng, W., Wang, T., Zhang, H., Mao, S., Zhang, H., Huang, W., Liu, H., Feng, S., & Chu, Z. (2024). Investigation of Imidazolinone Herbicide Resistance Gene with KASP Markers for Japonica/Geng Rice Varieties in the Huanghuaihai Region of China. Plants, 13(8), 1097. https://doi.org/10.3390/plants13081097






