Analysis of the CHS Gene Family Reveals Its Functional Responses to Hormones, Salinity, and Drought Stress in Moso Bamboo (Phyllostachys edulis)
Abstract
:1. Introduction
2. Results
2.1. Identification and Physicochemical Property Analysis of the PeCHS Gene Family Members
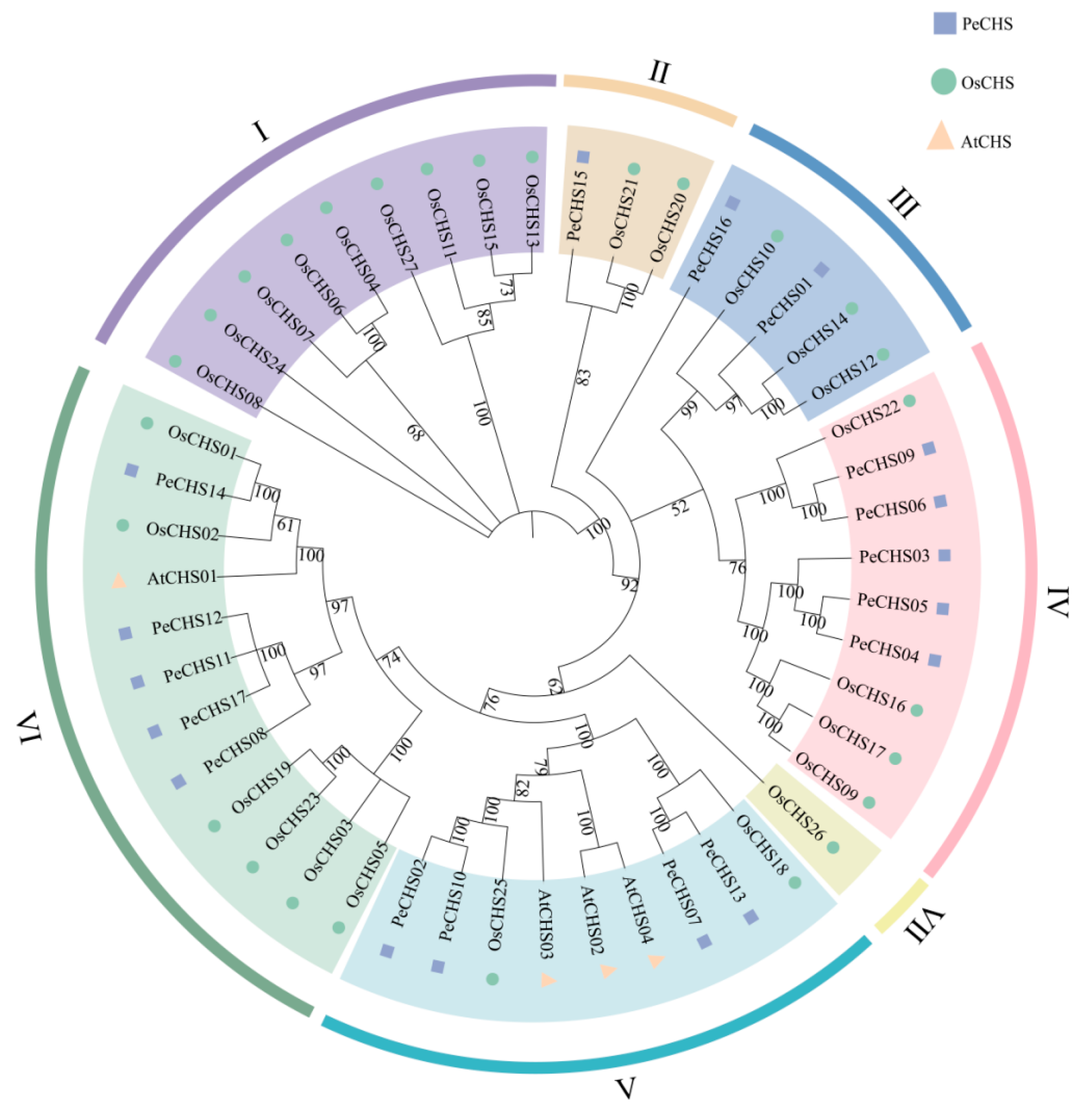
2.2. Classification and Phylogenetic Analysis of the PeCHS Gene Family
2.3. Chromosomal Localization and Collinearity Analysis of PeCHS Genes
2.4. Motif and Gene Structure Analysis of the PeCHS Gene Family
2.5. Conserved Domain and Tertiary Structure Prediction of PeCHS Proteins
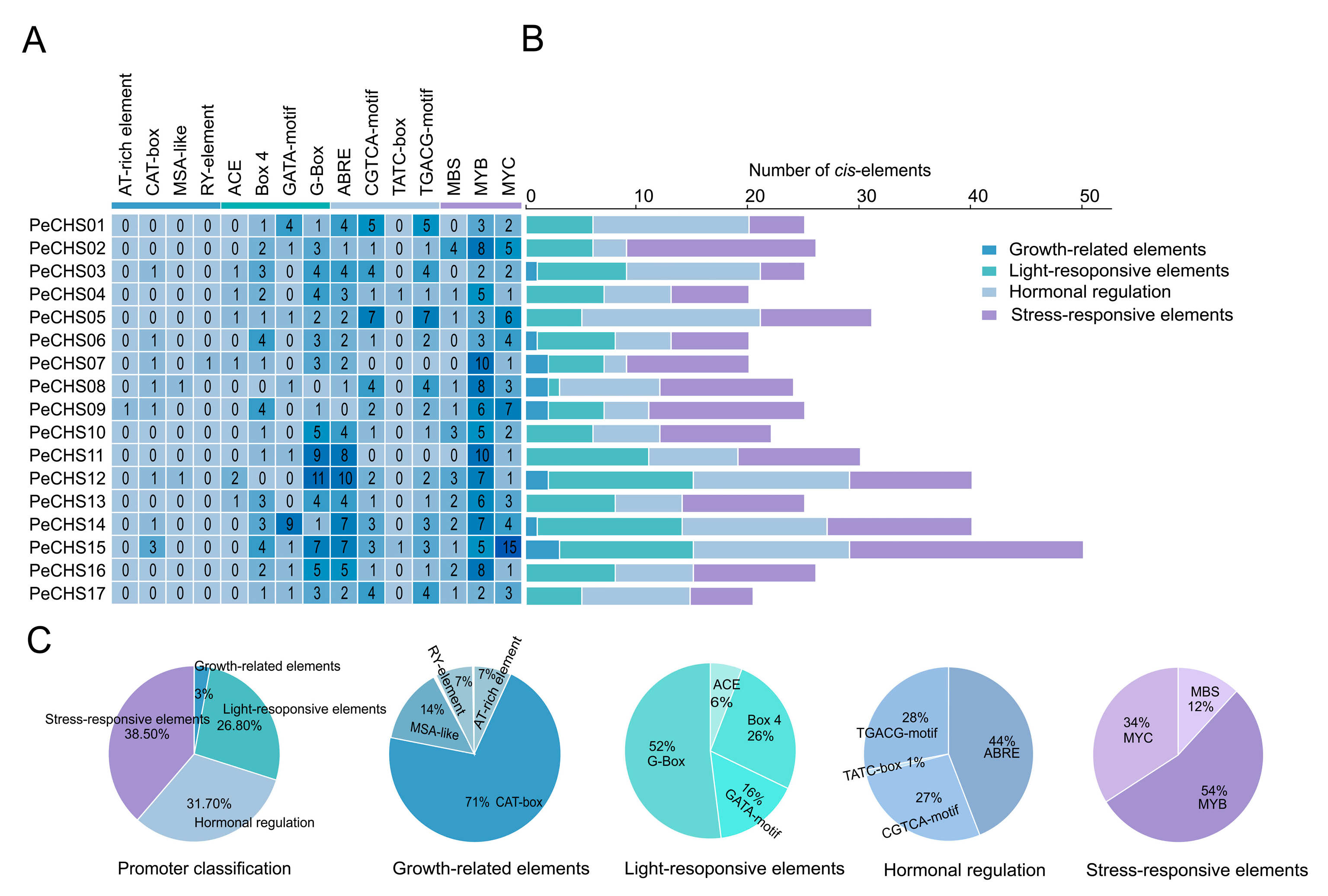
2.6. Cis-Acting Element Analysis of the PeCHS Gene Family
2.7. RNA-Seq Expression Profile Analysis of the PeCHS Gene Family
2.8. GO Functional Enrichment Analysis of the PeCHS Gene Family
2.9. Correlation Analysis of the Regulatory Network Between PeCHS Genes and Transcription Factors
2.10. Subcellular Localization of PeCHS14 and PeCHS15
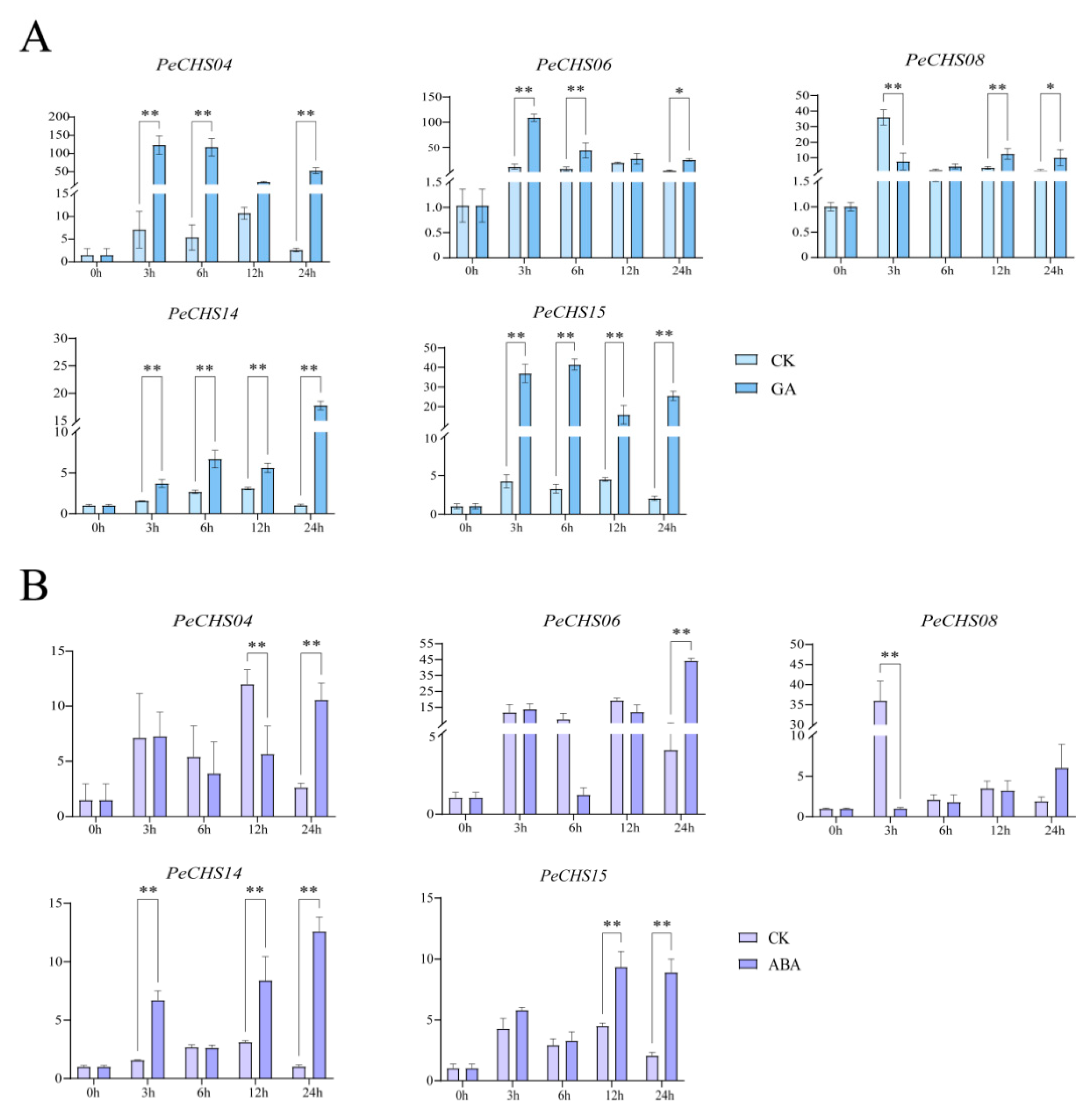
2.11. QRT-PCR Analysis of the PeCHS Gene Family
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of the PeCHS Gene Family
4.3. Analysis of Physicochemical Properties
4.4. Phylogenetic Tree Construction
4.5. Collinearity Analysis
4.6. Gene Structure, Motif Composition, and Promoter Element Analysis
4.7. Prediction of Protein Tertiary Structure
4.8. Gene Expression Analysis Based on RNA-Seq Data
4.9. GO Enrichment Analysis
4.10. Construction of the PeCHS Gene Regulatory Network
4.11. Subcellular Localization of PeCHS14 and PeCHS15
4.12. QRT-PCR Experiment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Guo, D.; Wang, H.; Zhang, S.; Lan, T. The type III polyketide synthase supergene family in plants: Complex evolutionary history and functional divergence. Plant J. 2022, 112, 414–428. [Google Scholar] [CrossRef] [PubMed]
- Brasileiro, A.C.M.; Gimenes, M.A.; Pereira, B.M.; Mota, A.P.Z.; Aguiar, M.N.; Martins, A.C.Q.; Passos, M.A.S.; Guimaraes, P.M. The Stilbene Synthase Family in Arachis: A Genome-Wide Study and Functional Characterization in Response to Stress. Genes 2023, 14, 2181. [Google Scholar] [CrossRef] [PubMed]
- Yoshihiko, K.; Miku, M.; Nobutaka, F.; Sueharu, H. In Vitro Synthesis of Curcuminoids by Type III Polyketide Synthase from Oryza sativa. J. Biol. Chem. 2007, 282, 37702–37709. [Google Scholar]
- Miku, M.; Yoshihiko, K.; Nobutaka, F.; Sueharu, H. Alkylresorcylic acid synthesis by type III polyketide synthases from rice Oryza sativa. Phytochemistry 2010, 71, 1059–1067. [Google Scholar]
- Mengqi, L.; Fengwang, M.; Fengying, W.; Cuiling, J.; Yuejin, W. Expression of stilbene synthase VqSTS6 from wild Chinese Vitis quinquangularis in grapevine enhances resveratrol production and powdery mildew resistance. Planta 2019, 250, 1997–2007. [Google Scholar]
- Zhang, J.; Yan, X.; Huang, T.; Liu, H.; Liu, F.; Yang, M.; Yang, M.; Ma, L. Overexpressing 4-coumaroyl-CoA ligase and stilbene synthase fusion genes in red raspberry plants leads to resveratrol accumulation and improved resistance against Botrytis cinerea. Plant Biochem. Biotechnol. 2022, 32, 85–91. [Google Scholar] [CrossRef]
- He, X.; Xue, F.; Zhang, L.; Guo, H.; Ma, L.; Yang, M. Overexpressing fusion proteins of 4-coumaroyl-CoA ligase (4CL) and stilbene synthase (STS) in tobacco plants leading to resveratrol accumulation and improved stress tolerance. Plant Biotechnol. Rep. 2018, 12, 295–302. [Google Scholar] [CrossRef]
- Lanqing, M.; Xiaobin, P.; Haiyan, S.; Gaobin, P.; Huahong, W.; Caiyan, L.; Wei, H.; Guofeng, L.; Benye, L.; Hechun, Y. A novel type III polyketide synthase encoded by a three-intron gene from Polygonum cuspidatum. Planta 2008, 229, 457–469. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, X.; Feng, D.; Liang, Y.; Wu, Z.; Du, S.; Zhou, Y.; Geng, C.; Men, P.; Fu, C.; et al. Discovery of a Unique Flavonoid Biosynthesis Mechanism in Fungi by Genome Mining. Angew. Chem. Int. Ed. Engl. 2023, 62, e202215529. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, R.; Li, D.; Wang, F. Transcriptomic and Coexpression Network Analyses Revealed Pine Chalcone Synthase Genes Associated with Pine Wood Nematode Infection. Int. J. Mol. Sci. 2021, 22, 11195. [Google Scholar] [CrossRef]
- Schüz, R.; Heller, W.; Hahlbrock, K. Substrate specificity of chalcone synthase from Petroselinum hortense. Formation of phloroglucinol derivatives from aliphatic substrates. J. Biol. Chem. 1983, 258, 6730–6734. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Jeridi, M.; Siddiqui, S.; Shah, A.Z.; Ali, S. Genome-wide identification, characterization, and expression analysis of the Chalcone Synthase gene family in Oryza sativa under Abiotic Stresses. Plant Stress 2023, 9, 100201–100215. [Google Scholar] [CrossRef]
- Gu, L.; Cao, Y.; Chen, X.; Wang, H.; Zhu, B.; Du, X.; Sun, Y. The Genome-Wide Identification, Characterization, and Expression Analysis of the Strictosidine Synthase-like Family in Maize (Zea mays L.). Int. J. Mol. Sci. 2023, 24, 14733. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhi, S.; Liu, C.; Xu, F.; Zhao, A.; Wang, X.; Tang, X.; Li, Z.; Huang, P.; Yu, M. Isolation and characterization of a novel chalcone synthase gene family from mulberry. Plant Physiol. Biochem. 2017, 115, 107–118. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, M.; Huang, Z. Genomic and Expression Analysis of Cassava (Manihot esculenta Crantz) Chalcone Synthase Genes in Defense against Tetranychus cinnabarinus Infestation. Genes 2024, 15, 336. [Google Scholar] [CrossRef]
- Pettinga, D.J.; Ou, J.; Patterson, E.L.; Jugulam, M.; Westra, P.; Gaines, T.A. Increased chalcone synthase (CHS) expression is associated with dicamba resistance in Kochia scoparia. Pest Manag. Sci. 2017, 74, 2306–2315. [Google Scholar] [CrossRef]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- Furumura, S.; Ozaki, T.; Sugawara, A.; Morishita, Y.; Tsukada, K.; Ikuta, T.; Inoue, A.; Asai, T. Identification and Functional Characterization of Fungal Chalcone Synthase and Chalcone Isomerase. J. Nat. Prod. 2023, 86, 398–405. [Google Scholar] [CrossRef]
- Zhang, Y.; Shu, H.; Mumtaz, M.A.; Hao, Y.; Li, L.; He, Y.; Jin, W.; Li, C.; Zhou, Y.; Lu, X.; et al. Transcriptome and Metabolome Analysis of Color Changes during Fruit Development of Pepper (Capsicum baccatum). Int. J. Mol. Sci. 2022, 23, 12524. [Google Scholar] [CrossRef]
- Hwang, H.G.; Milito, A.; Yang, J.; Jang, S.B.; Jung, G.Y. Riboswitch-guided chalcone synthase engineering and metabolic flux optimization for enhanced production of flavonoids. Metab. Eng. 2023, 75, 143–152. [Google Scholar] [CrossRef]
- Noelia, C.; Lorena, M.Z.; Francisco, A.H. Postharvest UV radiation enhanced biosynthesis of flavonoids and carotenes in bell peppers. Postharvest Biol. Technol. 2022, 184, 111774. [Google Scholar] [CrossRef]
- Rahmatullah, J.; Muhammad Aaqil, K.; Sajjad, A.; Lubna, L.; Muhammad, W.; Jae-Ryoung, P.; Saleem, A.; Nari, K.; In-Jung, L.; Kyung Min, K. Drought and UV Radiation Stress Tolerance in Rice Is Improved by Overaccumulation of Non-Enzymatic Antioxidant Flavonoids. Antioxidants 2022, 11, 917. [Google Scholar] [CrossRef] [PubMed]
- Göran, S. Effects of Flavonoids on the Polar Transport of Auxins. Physiol. Plant. 1976, 38, 262–266. [Google Scholar] [CrossRef]
- Hussain, S. Bioinformatics-based characterization of the chalcone synthase (CHS) family genes in flowering plants. Sabrao J. Breed. Genet. 2023, 55, 1950–1962. [Google Scholar] [CrossRef]
- Brigitte, U.; Bruns, K.H.; Schäfer, E. Fluence dependence of the ultraviolet-light-induced accumulation of chalcone synthase mRNA and effects of blue and far-red light in cultured parsley cells. Planta 1986, 169, 393–398. [Google Scholar] [CrossRef]
- Sebastian, R.; Janet, F.B.; Gottfried, W. Phenylpropanoid compounds in primary leaf tissues of rye (Secale cereale). Light response of their metabolism and the possible role in UV-B protection. Physiol. Plant. 1996, 97, 160–168. [Google Scholar] [CrossRef]
- Zhou, B.; Wang, Y.; Zhan, Y.; Li, Y.; Kawabata, S. Chalcone synthase family genes have redundant roles in anthocyanin biosynthesis and in response to blue/UV-A light in turnip (Brassica rapa; Brassicaceae). Am. J. Bot. 2013, 100, 2458–2467. [Google Scholar] [CrossRef]
- Bai, Y.; Gu, Y.; Liu, S.; Jiang, L.; Han, M.; Geng, D. Flavonoids metabolism and physiological response to ultraviolet treatments in Tetrastigma hemsleyanum Diels et Gilg. Front. Plant Sci. 2022, 13, 926197–926219. [Google Scholar] [CrossRef]
- Thi Thanh Hien, D.; Huub, J.M.L.; Robert, V. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011, 10, 397–412. [Google Scholar] [CrossRef]
- Wang, F.; Ren, G.; Li, F.; Qi, S.; Xu, Y.; Wang, B.; Yang, Y.; Ye, Y.; Zhou, Q.; Chen, X. A chalcone synthase gene AeCHS from Abelmoschus esculentus regulates flavonoid accumulation and abiotic stress tolerance in transgenic Arabidopsis. Acta Physiol. Plant. 2018, 40, 97–110. [Google Scholar] [CrossRef]
- Ahmad, S.; Ali, S.; Shah, A.Z.; Khan, A.; Faria, S. Chalcone synthase (CHS) family genes regulate the growth and response of cucumber (Cucumis sativus L.) to Botrytis cinerea and abiotic stresses. Plant Stress 2023, 8, 100159–100171. [Google Scholar] [CrossRef]
- Bai, Y.; Shi, K.; Shan, D.; Wang, C.; Yan, T.; Hu, Z.; Zheng, X.; Zhang, T.; Song, H.; Li, R.; et al. The WRKY17-WRKY50 complex modulates anthocyanin biosynthesis to improve drought tolerance in apple. Plant Sci. 2024, 340, 111965–111977. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Vidal, J.; Cobo, A.; Ortega-Morente, E.; Gálvez, A.; Alejo-Armijo, A.; Salido, S.; Altarejos, J. Antimicrobial and antioxidant activities of flavonoids isolated from wood of sweet cherry tree (Prunus avium L.). J. Wood Chem. Technol. 2021, 41, 104–117. [Google Scholar] [CrossRef]
- Chen, L.; Guo, H.; Lin, Y.; Cheng, H. Chalcone synthase EaCHS1 from Eupatorium adenophorum functions in salt stress tolerance in tobacco. Plant Cell Rep. 2015, 34, 885–894. [Google Scholar] [CrossRef]
- Wang, X.; Qiao, Q.; Zhao, K.; Zhai, W.; Zhang, F.; Dong, H.; Lin, L.; Xing, C.; Su, Z.; Pan, Z.; et al. PbWRKY18 promotes resistance against black spot disease by activation of the chalcone synthase gene PbCHS3 in pear. Plant Sci. 2024, 341, 112015–112025. [Google Scholar] [CrossRef]
- Bai, Y.; Cai, M.; Mu, C.; Cheng, W.; Zheng, H.; Cheng, Z.; Li, J.; Mu, S.; Gao, J. New Insights Into the Local Auxin Biosynthesis and Its Effects on the Rapid Growth of Moso Bamboo (Phyllostachys edulis). Front. Plant Sci. 2022, 13, 858686–858697. [Google Scholar] [CrossRef]
- Xiao, X.; Zhu, C.; Yang, K.; Liu, Y.; Gao, Z. Identification of Late Flavonoid Biosynthesis Genes of Moso Bamboo Reveals the Potential Function of PeANR4 Involved in Osmotic and Salt Stress. Forests 2023, 14, 1399. [Google Scholar] [CrossRef]
- Shao, S.-Y.; Ting, Y.; Wang, J.; Sun, J.; Guo, X.-F. Characterization and identification of the major flavonoids in Phyllostachys edulis leaf extract by UPLC–QTOF–MS/MS. Acta Chromatogr. 2020, 32, 228–237. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Cheng, J.; Zhao, J.; Shi, R.; He, L.; Li, Q.; Chen, Y. Efficient purification of flavonoids from bamboo shoot residues of Phyllostachys edulis by macroporous resin and their hypoglycemic activity. Food Chem. X 2022, 16, 100505–100515. [Google Scholar] [CrossRef]
- Yang, J.-P.; He, H.; Lu, Y.-H. Four Flavonoid Compounds from Phyllostachys edulis Leaf Extract Retard the Digestion of Starch and Its Working Mechanisms. J. Agric. Food Chem. 2014, 62, 7760–7770. [Google Scholar] [CrossRef]
- Muthusamy, R.; Kim, Y.; Vinod, K.K.; Anket, S.; Jungnam, C.; Viswanathan, S.; Mingbing, Z. Genetics and genomics of moso bamboo (Phyllostachys edulis): Current status, future challenges, and biotechnological opportunities toward a sustainable bamboo industry. Food Energy Secur. 2020, 9, e229. [Google Scholar] [CrossRef]
- Liu, H.; Lyu, H.; Zhu, K.; Van de Peer, Y.; Cheng, Z. The emergence and evolution of intron-poor and intronless genes in intron-rich plant gene families. Plant J. 2021, 105, 1072–1082. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Lim, C.W.; Lee, S.C. Role of pepper MYB transcription factor CaDIM1 in regulation of the drought response. Front. Plant Sci. 2022, 13, 1028392–1028404. [Google Scholar] [CrossRef] [PubMed]
- Sonia, S.; Deena Nath, G.; Ankita Singh, K.; Ashwani, S.; Ramasare, P. Identification and characterization of the Cyamopsis tetragonoloba transcription factor MYC (CtMYC) under drought stress. Gene 2023, 882, 147654. [Google Scholar] [CrossRef]
- Shan, W.; Yongbin, W.; Rui, Y.; Wei, C.; Yaning, L.; Duanrong, Z.; Meng, L.; Ping, W.; Bin, H. Genome-Wide Identification and Analysis Uncovers the Potential Role of JAZ and MYC Families in Potato under Abiotic Stress. Int. J. Mol. Sci. 2023, 24, 6706. [Google Scholar] [CrossRef]
- Masayuki, O.; Hiroyuki, T.; Toshio, K.; Masakazu, S.; Hidenari, T. Molecular cloning and characterization of a novel salt-inducible gene encoding an acidic isoform of PR-5 protein in soybean (Glycinemax [L.] Merr.). Plant Physiol. Biochem. 2006, 44, 574–580. [Google Scholar] [CrossRef]
- Fumiyuki, S.; Fumiyuki, T.; Kazuko, Y.S.; Kazuo, S. Cellular Phosphorylation Signaling and Gene Expression in Drought Stress Responses: ABA-Dependent and ABA-Independent Regulatory Systems. Plants 2021, 10, 756. [Google Scholar] [CrossRef]
- Qi, S.; Tuan-Hua David, H. Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell 1995, 7, 295–307. [Google Scholar] [CrossRef]
- Lo, L.; Yu, M.; Yao, W.; Ding, Y.; Lin, S. Research advance in growth and development of bamboo organs. Ind. Crop. Prod. 2023, 205, 117428–117441. [Google Scholar] [CrossRef]
- Sikdar, S.; Datta, S. A novel statistical approach for identification of the master regulator transcription factor. BMC Bioinform. 2017, 18, 79–90. [Google Scholar] [CrossRef]
- Sharma, N.; Bhalla, P.L.; Singh, M.B. Transcriptome-wide profiling and expression analysis of transcription factor families in a liverwort, Marchantia polymorpha. BMC Genom. 2013, 14, 915. [Google Scholar] [CrossRef] [PubMed]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription Factors Associated with Abiotic and Biotic Stress Tolerance and Their Potential for Crops Improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef]
- Cheng, Z.; Luan, Y.; Meng, J.; Sun, J.; Tao, J.; Zhao, D. WRKY Transcription Factor Response to High-Temperature Stress. Plants 2021, 10, 2211. [Google Scholar] [CrossRef]
- Gong, Y.; Liu, X.; Chen, S.; Li, H.; Duanmu, H. Genome-Wide Identification and Salt Stress Response Analysis of the bZIP Transcription Factor Family in Sugar Beet. Int. J. Mol. Sci. 2022, 23, 11573. [Google Scholar] [CrossRef]
- Zepeda, B.; Marcelis, L.F.M.; Kaiser, E.; Verdonk, J.C. Petunia as a model for MYB transcription factor action under salt stress. Front. Plant Sci. 2023, 14, 1286547. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, Z.; Wang, P.; Qin, C.; He, L.; Kong, L.; Ren, W.; Liu, X.; Ma, W. Genome-wide identification of the NAC transcription factors family and regulation of metabolites under salt stress in Isatis indigotica. Int. J. Biol. Macromol. 2023, 240, 124436. [Google Scholar] [CrossRef]
- Danial, H.; Xueqing, F.; Qian, S.; Muhammad, K.; Jocelyn, K.C.R.; Kexuan, T. Parallel Transcriptional Regulation of Artemisinin and Flavonoid Biosynthesis. Trends Plant Sci. 2020, 25, 466–476. [Google Scholar] [CrossRef]
- Tong, Y.; Lyu, Y.; Xu, S.; Zhang, L.; Zhou, J. Optimum chalcone synthase for flavonoid biosynthesis in microorganisms. Crit. Rev. Biotechnol. 2021, 41, 1194–1208. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, Z.; Li, J.; Zhang, H.; Peng, Y.; Li, Z. Uncovering Hierarchical Regulation among MYB-bHLH-WD40 Proteins and Manipulating Anthocyanin Pigmentation in Rice. Int. J. Mol. Sci. 2022, 23, 8203. [Google Scholar] [CrossRef]
- Ajay Kumar, M.; Tomáš, K.; Vishnu Sukumari, N.; Ahamed, K.; Matoušek, J.; Khaled, M.H.; Naganeeswaran, S.; Karel, K.; Jutta, L.M.; Khaled, M.A.A. The multifaceted roles of R2R3 transcription factor HlMYB7 in the regulation of flavonoid and bitter acids biosynthesis, development and biotic stress tolerance in hop (Humulus lupulus L.). Plant Physiol. Biochem. 2023, 197, 107636–107650. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.; Li, J.; Liu, B.; Wang, D.; Gao, C. The R2R3-MYB transcription factor ThRAX2 recognized a new element MYB-T (CTTCCA) to enhance cadmium tolerance in Tamarix hispida. Plant Sci. 2023, 329, 111574. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Gao, Y.; Sui, Y.; Dang, Y.; Wu, F.; Wang, X.; Zhang, F.; Du, X.; Sui, N. R2R3 MYB transcription factor SbMYBHv33 negatively regulates sorghum biomass accumulation and salt tolerance. Theor. Appl. Genet. 2023, 136, 5. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, P.A.; Gurav, T.P.; Kondhare, K.R.; Giri, A.P. MYB proteins: Versatile regulators of plant development, stress responses, and secondary metabolite biosynthetic pathways. Int. J. Biol. Macromol. 2024, 138588. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Duan, J.; Wang, Q.; Zhang, M.; Zhi, H.; Bai, Z.; Zhang, Y.; Luo, J. The Paeonia qiui R2R3-MYB Transcription Factor PqMYBF1 Positively Regulates Flavonol Accumulation. Plants 2023, 12, 1427. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, C.; Xu, X.; Su, Y.; Gao, Y.; Yang, J.; Xie, C.; Ma, J. A MYB family transcription factor TdRCA1 from wild emmer wheat regulates anthocyanin biosynthesis in coleoptile. Theor. Appl. Genet. 2024, 137, 208–217. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, T.; Yuan, L.; Qiu, F.; Tang, Y.; Li, D.; Zhang, F.; Zeng, L.; Yang, C.; Nagdy, M.M.; et al. A Fruit-Expressed MYB Transcription Factor Regulates Anthocyanin Biosynthesis in Atropa belladonna. Int. J. Mol. Sci. 2024, 25, 4963. [Google Scholar] [CrossRef]
- Deng, X.; Bashandy, H.; Ainasoja, M.; Kontturi, J.; Pietiäinen, M.; Laitinen, R.A.E.; Albert, V.A.; Valkonen, J.P.T.; Elomaa, P.; Teeri, T.H. Functional diversification of duplicated chalcone synthase genes in anthocyanin biosynthesis of Gerbera hybrida. New Phytol. 2013, 201, 1469–1483. [Google Scholar] [CrossRef]
- Su, L.; Lv, A.; Wen, W.; Fan, N.; Li, J.; Gao, L.; Zhou, P.; An, Y. MsMYB741 is involved in alfalfa resistance to aluminum stress by regulating flavonoid biosynthesis. Plant J. 2022, 112, 756–771. [Google Scholar] [CrossRef]
- Han, T.; Wu, W.; Li, W. Transcriptome Analysis Revealed the Mechanism by Which Exogenous ABA Increases Anthocyanins in Blueberry Fruit During Veraison. Front. Plant Sci. 2021, 12, 758215–758228. [Google Scholar] [CrossRef]
- Bai, M.Y.; Zhang, L.Y.; Gampala, S.S.; Zhu, S.W.; Song, W.Y.; Chong, K.; Wang, Z.Y. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 13839–13844. [Google Scholar] [CrossRef]
- Cui, X.-Y.; Gao, Y.; Guo, J.; Yu, T.-F.; Zheng, W.-J.; Liu, Y.-W.; Chen, J.; Xu, Z.-S.; Ma, Y.-Z. BES/BZR Transcription Factor TaBZR2 Positively Regulates Drought Responses by Activation of TaGST1. Plant Physiol. 2019, 180, 605–620. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Zhao, S.; Bao, T.; Zhao, P.; Peng, K.; Guo, Q.; Gao, X.; Qin, J. Tomato BZR/BES transcription factor SlBZR1 positively regulates BR signaling and salt stress tolerance in tomato and Arabidopsis. Plant Sci. 2021, 302, 110719. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Kim, Y.; Shim, Y.; Cho, S.; Yang, T.; Song, Y.H.; Kang, K.; Paek, N. Rice OsGATA16 is a positive regulator for chlorophyll biosynthesis and chloroplast development. Plant J. 2023, 117, 599–615. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Yang, Y.; Lou, S.; Wei, W.; Zhao, Z.; Ren, Y.; Lin, C.; Ma, L. Genome-Wide Characterization and Gene Expression Analyses of GATA Transcription Factors in Moso Bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 2019, 21, 14. [Google Scholar] [CrossRef] [PubMed]
- Jaina, M.; Sara, C.; Lowri, W.; Matloob, Q.; Gustavo, S.; Erik, L.L.S.; Silvio, C.E.T.; Lisanna, P.; Shriya, R.; Lorna, R.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Procter, J.B.; Carstairs, G.M.; Soares, B.; Mourão, K.; Ofoegbu, T.C.; Barton, D.; Lui, L.; Menard, A.; Sherstnev, N.; Roldan-Martinez, D.; et al. Alignment of Biological Sequences with Jalview. Methods Mol. Biol. 2021, 2231, 203–224. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Timothy, L.B.; James, E.J.; Charles, E.G.; William Stafford, N. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yu, P.; Huang, B.; Ma, R.; Vinod, K.K.; Muthusamy, R. Genome-wide identification and expression characterization of the DoG gene family of moso bamboo (Phyllostachys edulis). BMC Genom. 2022, 23, 357. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Chen, J.; Huang, B.; Huang, Z.; Zhang, Z. The BBX gene family in Moso bamboo (Phyllostachys edulis): Identification, characterization and expression profiles. BMC Genom. 2021, 22, 533. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Jiaqi, T.; Hui, G.; Bin, H.; Yibin, Y.; Muthusamy, R.; Zhijun, Z. Genome-wide analysis of the KNOX gene family in Moso bamboo: Insights into their role in promoting the rapid shoot growth. Res. Sq. 2023, 24, 213–232. [Google Scholar] [CrossRef]
- Jaime, A.C.-M.; Rafael, R.-P.; Ieva, R.; Roza Berhanu, L.; Laura, T.; Romain, B.-M.; Jérémy, L.; Paul, B.; Aziz, K.; Nicolás Manosalva, P.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2021, 50, D165–D173. [Google Scholar] [CrossRef]
- Tan, J.; Xuan, X.; Su, S.; Jiao, Y.; Guo, H.; Zhang, Z. Comprehensive analysis of the CPP gene family in Moso bamboo: Insights into their role in rapid shoot growth. BMC Genom. 2024, 25, 1173. [Google Scholar] [CrossRef]
- Xuan, X.; Su, S.; Tan, J.; Guo, H.; Jiao, Y.; Zhang, Z. Genome-wide identification, characterization, and expression pattern analysis of the JAZ gene family in Moso bamboo during rapid shoot development. Adv. Bamboo Sci. 2024, 7, 100083–100097. [Google Scholar] [CrossRef]
- Guo, H.; Tan, J.; Jiao, Y.; Huang, B.; Ma, R.; Ramakrishnan, M.; Qi, G.; Zhang, Z. Genome-wide identification and expression analysis of the HAK/KUP/KT gene family in Moso bamboo. Front. Plant Sci. 2024, 15, 1331710–1331730. [Google Scholar] [CrossRef]
- Huang, B.; Huang, Z.; Ma, R.; Ramakrishnan, M.; Chen, J.; Zhang, Z.; Yrjälä, K. Genome-wide identification and expression analysis of LBD transcription factor genes in Moso bamboo (Phyllostachys edulis). BMC Plant Biol. 2021, 21, 296. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]







| Name | Gene ID | Number of Amino Acids | MW (KDa) | PI | Instability Index | GRAVY |
|---|---|---|---|---|---|---|
| PeCHS01 | PH02Gene38478.t1 | 405 | 42.724 | 6.32 | 35.55 | 0.052 |
| PeCHS02 | PH02Gene37360.t1 | 390 | 42.303 | 7.08 | 43.45 | −0.068 |
| PeCHS03 | PH02Gene38613.t1 | 391 | 42.172 | 5.7 | 34.46 | 0.001 |
| PeCHS04 | PH02Gene38615.t1 | 394 | 42.538 | 5.93 | 32.1 | −0.004 |
| PeCHS05 | PH02Gene49028.t1 | 394 | 42.538 | 5.93 | 32.1 | −0.004 |
| PeCHS06 | PH02Gene50391.t1 | 391 | 42.317 | 5.96 | 40.66 | −0.017 |
| PeCHS07 | PH02Gene30447.t1 | 390 | 42.314 | 6.08 | 33.3 | −0.066 |
| PeCHS08 | PH02Gene43021.t1 | 388 | 42.208 | 5.72 | 36.63 | −0.038 |
| PeCHS09 | PH02Gene13137.t1 | 402 | 43.789 | 5.66 | 39.11 | −0.055 |
| PeCHS10 | PH02Gene24703.t1 | 354 | 38.067 | 6.41 | 43.08 | −0.096 |
| PeCHS11 | PH02Gene35700.t1 | 393 | 42.014 | 5.89 | 34.69 | 0.036 |
| PeCHS12 | PH02Gene35703.t1 | 309 | 32.996 | 6.83 | 32.85 | 0.079 |
| PeCHS13 | PH02Gene11618.t1 | 390 | 42.066 | 5.7 | 35.84 | −0.031 |
| PeCHS14 | PH02Gene44458.t1 | 368 | 39.940 | 6.1 | 43.11 | −0.036 |
| PeCHS15 | PH02Gene01692.t1 | 396 | 42.945 | 5.54 | 35.49 | −0.096 |
| PeCHS16 | PH02Gene43281.t1 | 402 | 43.266 | 6.38 | 40.58 | −0.069 |
| PeCHS17 | PH02Gene48527.t1 | 393 | 41.954 | 6.02 | 33.84 | 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, S.; Xuan, X.; Tan, J.; Yu, Z.; Jiao, Y.; Zhang, Z.; Ramakrishnan, M. Analysis of the CHS Gene Family Reveals Its Functional Responses to Hormones, Salinity, and Drought Stress in Moso Bamboo (Phyllostachys edulis). Plants 2025, 14, 161. https://doi.org/10.3390/plants14020161
Su S, Xuan X, Tan J, Yu Z, Jiao Y, Zhang Z, Ramakrishnan M. Analysis of the CHS Gene Family Reveals Its Functional Responses to Hormones, Salinity, and Drought Stress in Moso Bamboo (Phyllostachys edulis). Plants. 2025; 14(2):161. https://doi.org/10.3390/plants14020161
Chicago/Turabian StyleSu, Shiying, Xueyun Xuan, Jiaqi Tan, Zhen Yu, Yang Jiao, Zhijun Zhang, and Muthusamy Ramakrishnan. 2025. "Analysis of the CHS Gene Family Reveals Its Functional Responses to Hormones, Salinity, and Drought Stress in Moso Bamboo (Phyllostachys edulis)" Plants 14, no. 2: 161. https://doi.org/10.3390/plants14020161
APA StyleSu, S., Xuan, X., Tan, J., Yu, Z., Jiao, Y., Zhang, Z., & Ramakrishnan, M. (2025). Analysis of the CHS Gene Family Reveals Its Functional Responses to Hormones, Salinity, and Drought Stress in Moso Bamboo (Phyllostachys edulis). Plants, 14(2), 161. https://doi.org/10.3390/plants14020161






