Unveiling a Meaningful Form of Cypripedium × ventricosum Sw. (Cypripedioideae, Orchidaceae) from Changbai Mountain, China: Insights from Morphological, Molecular, and Plastome Analyses
Abstract
1. Introduction
2. Material and Methods
2.1. Plant Materials
2.2. Morphological Analysis
2.3. Extraction and Sequencing of Chloroplast Genomic DNA
2.4. Assembly and Annotation of Chloroplast Genome Sequence
2.5. Codon Usage Bias
2.6. Analysis of Repeat Sequences
2.7. Comparative Genome Analysis
2.8. Phylogenetic Analysis
3. Results
3.1. Morphological Characteristics
3.2. Characteristics of Plastome of W1
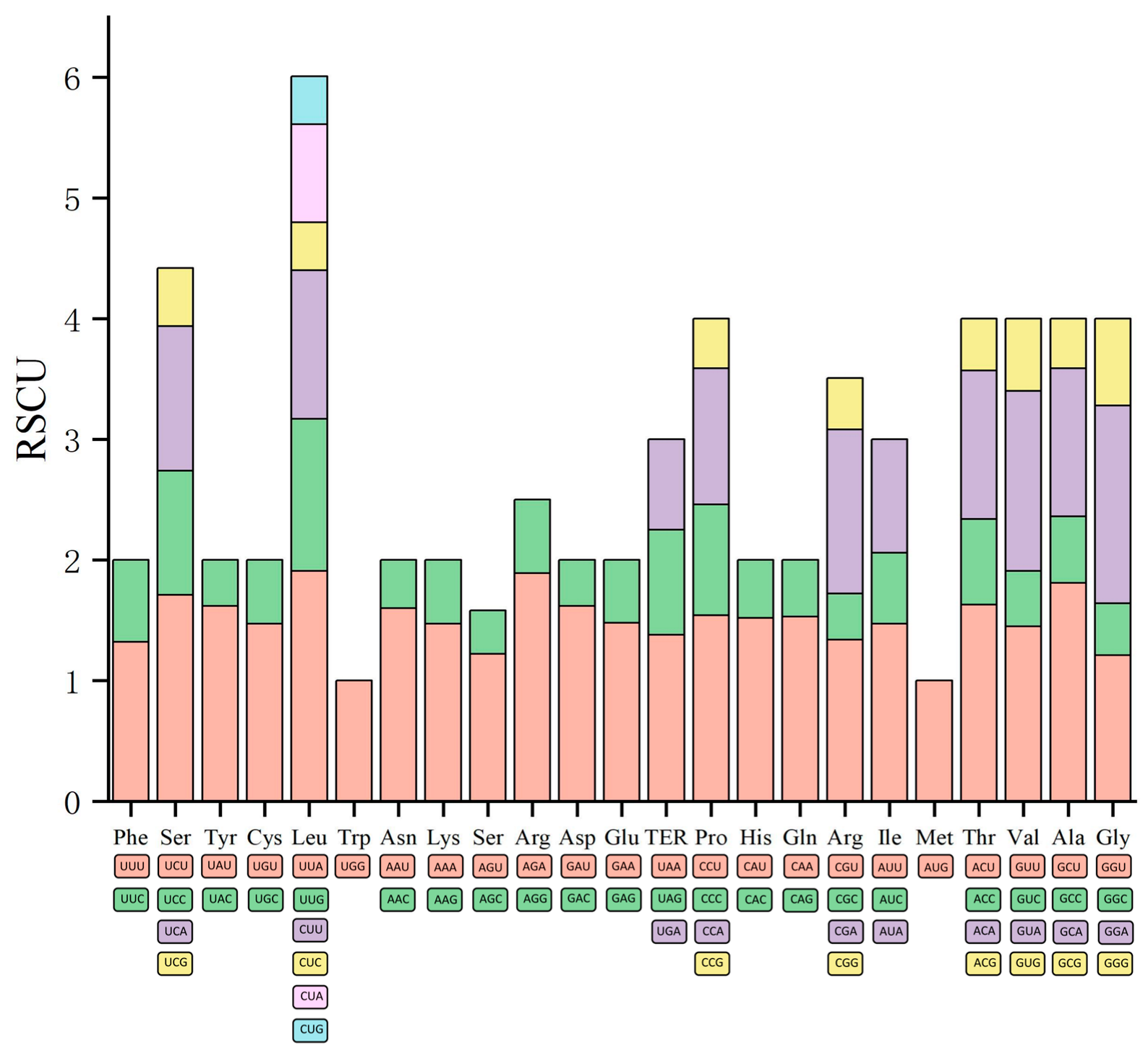
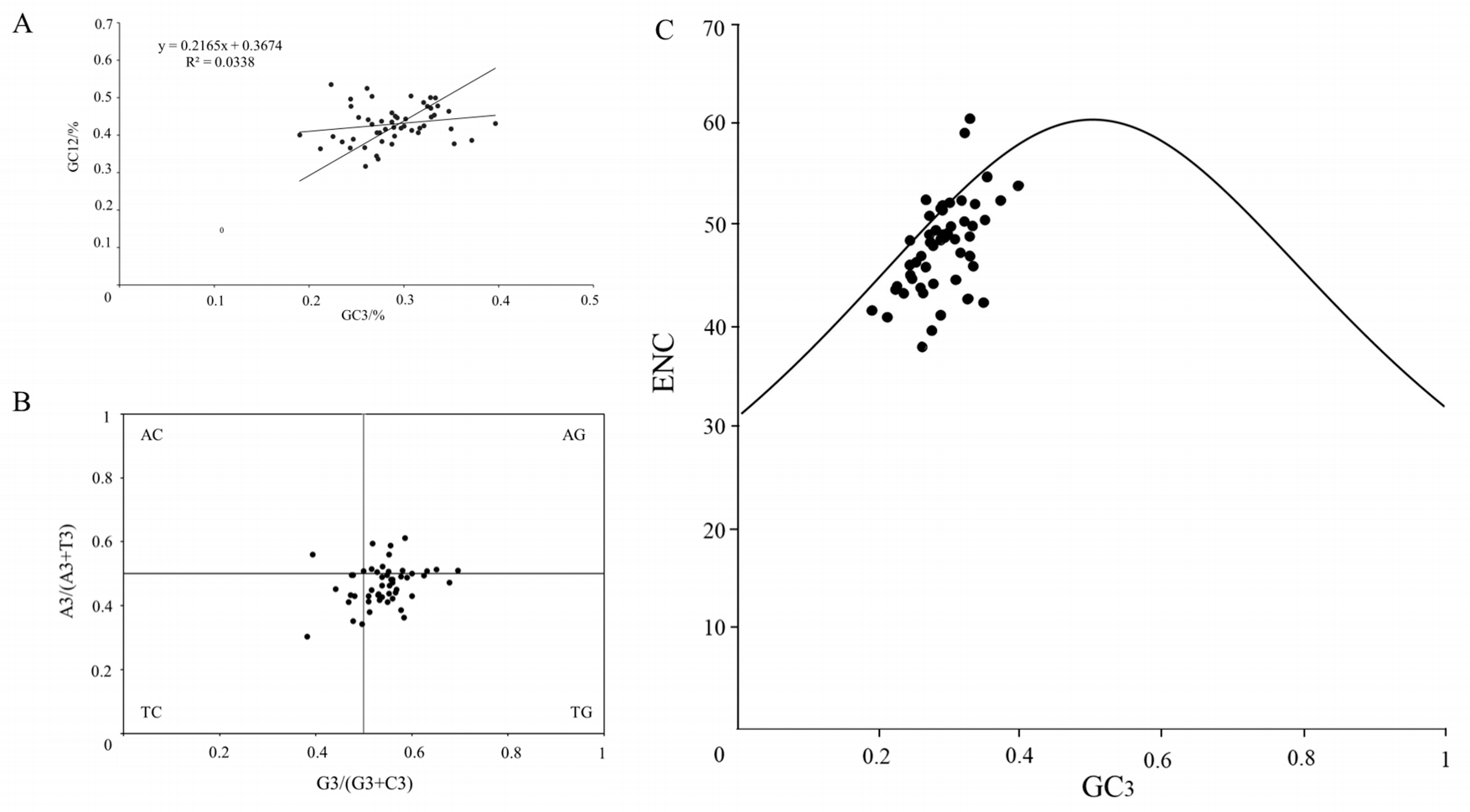
3.3. Codon Usage Bias
3.4. Repeat Sequence Analysis
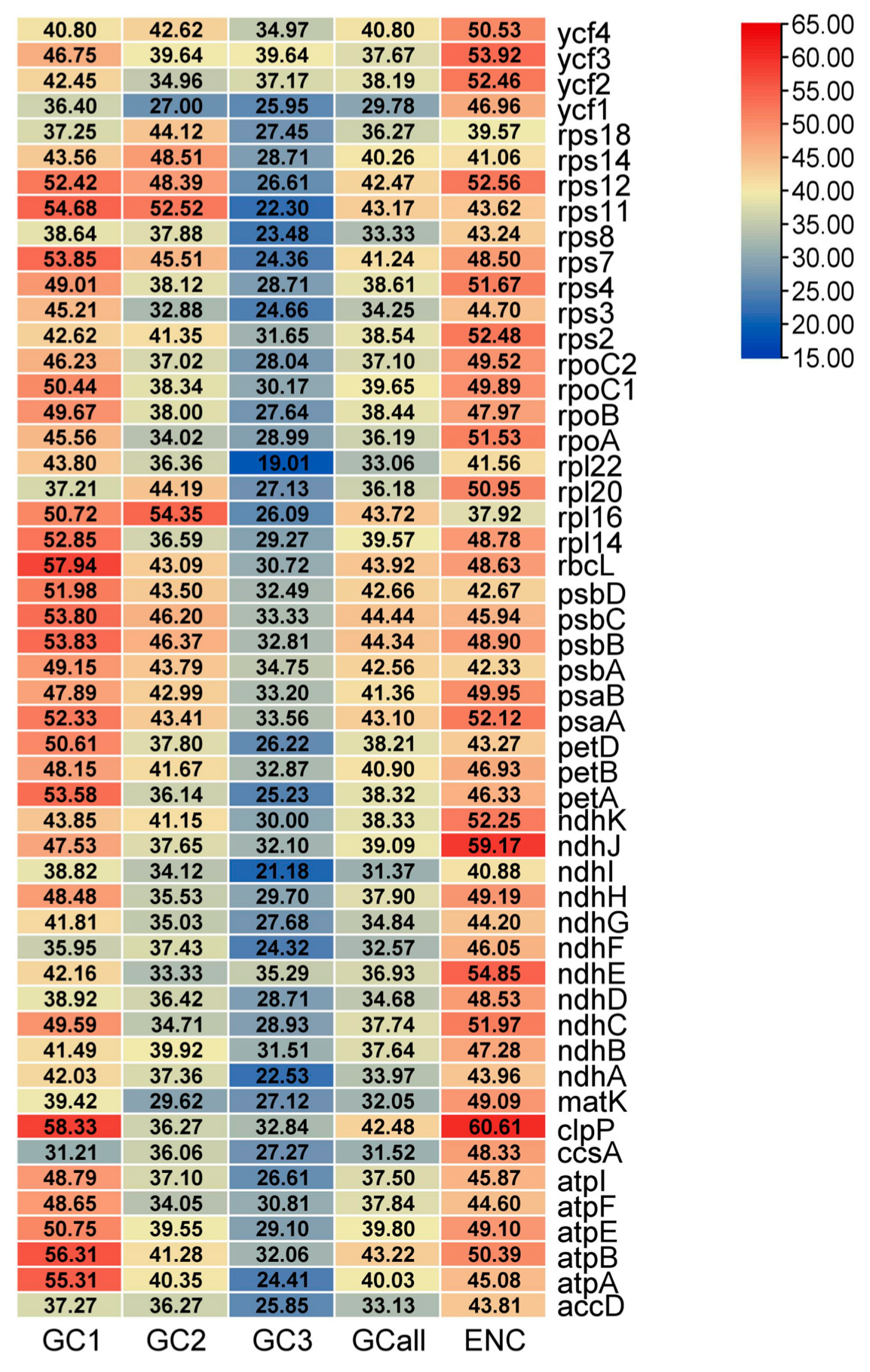
3.5. Comparative Genome Analysis
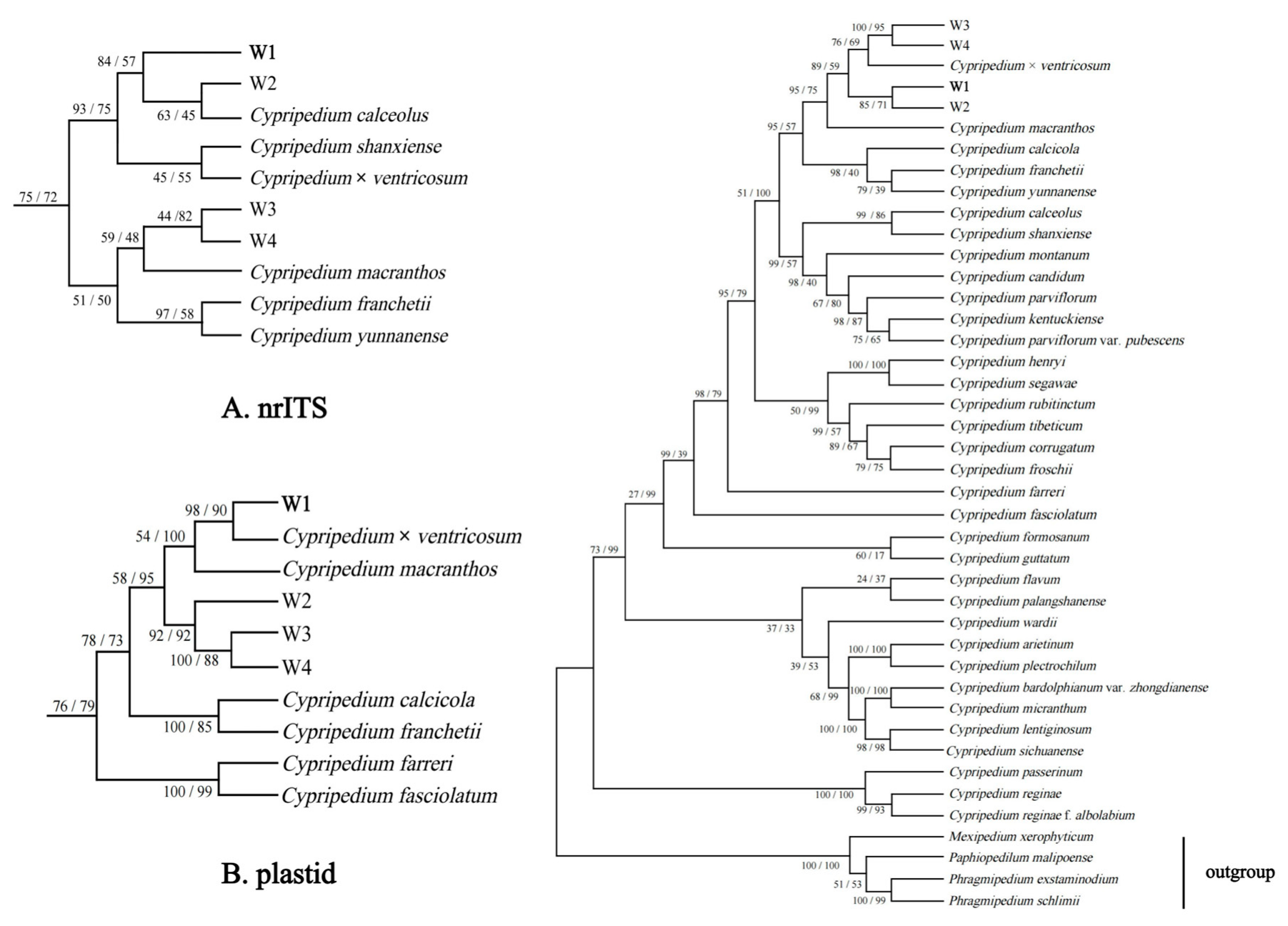
3.6. Phylogenetic Analysis
4. Discussion
4.1. Morphological and Molecular Evidence of W1
4.2. Chloroplast Genome Characterization of W1
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cribb, P.; Sandison, M.S. A preliminary assessment of the conservation status of Cypripedium species in the wild. Bot. J. Linn. Soc. 1998, 126, 183–190. [Google Scholar] [CrossRef][Green Version]
- Jiang, H.; Chen, M.C.; Lee, Y.I. In vitro germination and low-temperature seed storage of Cypripedium lentiginosum P.J.Cribb S.C.Chen, a rare and endangered lady’s slipper orchid. Sci. Hortic. 2017, 225, 471–479. [Google Scholar] [CrossRef]
- Li, H.; Liu, Z.J.; Salazar, G.A.; Bernhardt, P.; Perner, H.; Tomohisa, Y.; Jin, X.H.; Chung, S.W.; Luo, Y.B. Molecular phylogeny of Cypripedium (Orchidaceae: Cypripedioideae) inferred from multiple nuclear and chloroplast regions. Mol. Phylogenetics Evol. 2011, 61, 308–320. [Google Scholar] [CrossRef]
- Cribb, P. The Genus Cypripedium; Timber Press: Portland, OR, USA, 1997. [Google Scholar]
- Atwood, J. The relationships of the slipper orchids (subfamily Cypripedioideae, Orchidaceae). Selbyana 1984, 7, 129–247. [Google Scholar]
- Hennessy, E.F.; Hedge, T.A. The Slipper Orchids, Selenipedium, Phragmipedium, Criosanthes, Cypripedium, Paphiopedilum; Acorn: Randburg, South Africa, 1989. [Google Scholar]
- Lindley, J. The Genera and Species of Orchidaceous Plants; Ridgways: London, UK, 1840. [Google Scholar]
- Pfitzer, E.H.H. Orchidaceae-Pleonandrae; Verlag von Wilhelm Engelmann: Leipzig, Germany, 1903. [Google Scholar]
- Eccarius, W. Die Orchideengattung Cypripedium; Echinomedia Verlag: Albersdorf, Germany, 2009. [Google Scholar]
- Givnish, T.J.; Spalink, D.; Ames, M.; Lyon, S.P.; Hunter, S.J.; Zuluaga, A.; Iles, W.J.D.; Clements, M.A.; Arroyo, M.T.K.; Leebens-Mack, J.; et al. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. R. Soc. B Biol. Sci. 2015, 282, 20151553. [Google Scholar] [CrossRef] [PubMed]
- Li, M.H.; Zhang, G.Q.; Lan, S.R.; Liu, Z.J.; Consortium, C.P. A molecular phylogeny of Chinese orchids. J. Syst. Evol. 2016, 54, 349–362. [Google Scholar] [CrossRef]
- Kim, Y.K.; Jo, S.J.; Cheon, S.H.; Joo, M.J.; Hong, J.R.; Kwak, M.H.; Kim, K.J. Plastome Evolution and Phylogeny of Orchidaceae, With 24 New Sequences. Front. Plant Sci. 2020, 11, 22. [Google Scholar] [CrossRef]
- Pérez-Escobar, O.A.; Dodsworth, S.; Bogarín, D.; Bellot, S.; Balbuena, J.A.; Schley, R.J.; Kikuchi, I.A.; Morris, S.K.; Epitawalage, N.; Cowan, R.; et al. Hundreds of nuclear and plastid loci yield novel insights into orchid relationships. Am. J. Bot. 2021, 108, 1166–1180. [Google Scholar] [CrossRef]
- Serna-Sánchez, M.A.; Pérez-Escobar, O.A.; Bogarín, D.; TorresJimenez, M.F.; Alvarez-Yela, A.C.; Arcila-Galvis, J.E.; Hall, C.F.; De Barros, F.; Pinheiro, F.; Dodsworth, S.; et al. Plastid phylogenomics resolves ambiguous relationships within the orchid family and provides a solid timeframe for biogeography and macroevolution. Sci. Rep. 2021, 11, 6858. [Google Scholar]
- Chen, S.C.; Liu, Z.J.; Chen, L.J.; Li, L.Q. The Genus Cypripedium in China; Science Press: Beijing, China, 2013. [Google Scholar]
- Szlachetko, D.L.; Gorniak, M.; Kowalkowska, A.K.; Kolanowska, M.; Jurczak-Kurek, A.; Morales, F.A. The natural history of the genus Cypripedium (Orchidaceae). Plant Biosyst. 2021, 155, 772–796. [Google Scholar] [CrossRef]
- Liu, H.C.; Jacquemyn, H.; Chen, W.; Janssens, S.B.; He, X.Y.; Yu, S.; Huang, Y.Q. Niche evolution and historical biogeography of lady slipper orchids in North America and Eurasia. J. Biogeogr. 2021, 48, 2727–2741. [Google Scholar] [CrossRef]
- Pérez-Escobar, O.A.; Bogarín, D.; Przelomska, N.A.S.; Ackerman, J.D.; Balbuena, J.A.; Bellot, S.; Bühlmann, R.P.; Cabrera, B.; Cano, J.A.; Charitonidou, M.; et al. The origin and speciation of Orchids. New Phytol. 2024, 242, 700–716. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Liao, M.; Cheng, Y.H.; Feng, Y.; Ju, W.B.; Deng, H.N.; Li, X.; Plenković-Moraj, A.; Xu, B. Comparative chloroplast genomics of seven endangered Cypripedium species and phylogenetic relationships of Orchidaceae. Front. Plant Sci. 2022, 13, 911702. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Zhang, J.Y.; Feng, Y.; Ren, Z.X.; Deng, H.N.; Xu, B. Phylogenomic insights into the historical biogeography, character-state evolution, and species diversifcation rates of Cypripedioideae (Orchidaceae). Mol. Phylogenetics Evol. 2024, 199, 108138. [Google Scholar] [CrossRef]
- Harland, S. Introgressive Hybridization. Nature 1950, 166, 243–244. [Google Scholar] [CrossRef]
- Stebbins, G. The role of hybridization in evolution. Proc. Am. Philos. Soc. 1959, 103, 231–251. [Google Scholar]
- Andriamihaja, C.F.; Ramarosandratana, A.V.; Grisoni, M.; Jeannoda, V.; Besse, P. The Leafless Vanilla Species-Complex from the South-West Indian Ocean Region: A Taxonomic Puzzle and a Model for Orchid Evolution and Conservation Research. Diversity 2020, 12, 443. [Google Scholar] [CrossRef]
- Cozzolino, S.; D’Emerico, S.; Widmer, A. Evidence for reproductive isolate selection in Mediterranean Orchids: Karyotype differences compensate for the lack of pollinator specificity. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2004, 271, S259–S262. [Google Scholar] [CrossRef] [PubMed]
- Esposito, P.; Vereecken, N.J.; Gammella, M.; Rinaldi, R.; Laurent, P.; Tyteca, D. Characterization of sympatric Platanthera bifolia and Platanthera chlorantha (Orchidaceae) populations with intermediate plants. Peer J. 2018, 6, e4256. [Google Scholar] [CrossRef]
- van der Cingel, N. An Atlas of Orchid Pollination; Balkema Publishers: Rotterdam, The Netherlands, 1995. [Google Scholar]
- Jersáková, J.; Johnson, S.D.; Kindlmann, P. Mechanisms and evolution of deceptive pollination in orchids. Biol. Rev. Camb. Philos. Soc. 2006, 81, 219–235. [Google Scholar] [CrossRef]
- Knyasev, M.; Kulikov, P.; Knyaseva, O.; Semerikov, V. On the interspecific hybridization in Eurasian species of Cypripedium (Orchidaceae) and taxonomic status of C. ventricosum. Bot. Zhurnal 2020, 85, 94–102. [Google Scholar]
- Worley, A.; Sawich, L.S.; Ghazvini, H.; Ford, B. Hybridization and introgression between a rare and a common lady’s slipper orchid, Cypripedium candidum and C. parviflorum (Orchidaceae). Botany 2009, 87, 1054–1065. [Google Scholar] [CrossRef]
- Knyasev, M.S.; Pavel, V.; Kulikov, O.I.; Semerikov, V.L. Interspecific hybridization in northern Eurasian Cypripedium: Morphometric and genetic evidence of the hybrid origin of C. ventricosum. Biol. Environ. Sci. 2000, 15, 10–20. [Google Scholar]
- Bänziger, H.; Sun, H.Q.; Luo, Y.B. Pollination of wild lady slipper orchids Cypripedium yunnanense and C. flavum (Orchidaceae) in south-west China: Why are there no hybrids? Bot. J. Linn. Soc. 2008, 156, 51–64. [Google Scholar] [CrossRef]
- Hu, S.J.; Hu, H.; Yan, N.; Huang, J.L.; Li, S.Y. Hybridization and asymmetric introgression between Cypripedium tibeticum and C. yunnanense in Shangrila County, Yunnan Province, China. Nord. J. Botany. 2011, 29, 625–631. [Google Scholar] [CrossRef]
- Tonti-Filippini, J.; Nevill, P.G.; Dixon, K.; Small, I. What can we do with 1000 plastid genomes? Plant J. 2017, 90, 808–818. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Comes, H.P.; Qiu, Y.X. Phylogenomic insights into the temporal-spatial divergence history, evolution of leaf habit and hybridization in Stachyurus (Stachyuraceae). Mol. Phylogenetics Evol. 2020, 150, 106878. [Google Scholar] [CrossRef]
- Li, E.; Liu, K.J.; Deng, R.Y.; Gao, Y.W.; Liu, X.Y.; Dong, W.P.; Zhang, Z.X. Insights into the phylogeny and chloroplast genome evolution of Eriocaulon (Eriocaulaceae). BMC Plant Biol. 2023, 23, 32. [Google Scholar] [CrossRef]
- Yue, Y.L.; Li, J.W.; Sun, X.G.; Li, Z.; Jiang, B.J. Polymorphism analysis of the chloroplast and mitochondrial genomes in soybean. BMC Plant Biol. 2023, 23, 15. [Google Scholar] [CrossRef]
- Grevich, J.J.; Daniell, H. Chloroplast Genetic Engineering: Recent Advances and Future Perspectives. Crit. Rev. Plant Sci. 2005, 24, 83–107. [Google Scholar] [CrossRef]
- Wang, Y.; Zhan, D.F.; Jia, X.; Mei, W.L.; Dai, H.F.; Chen, X.T.; Peng, S.Q. Complete Chloroplast Genome Sequence of Aquilaria sinensis (Lour.) Gilg and Evolution Analysis within the Malvales Order. Front. Plant Sci. 2016, 7, 280. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.H.; Hao, Z.D.; Xu, H.B.; Yang, L.M.; Liu, G.; Sheng, Y.; Zheng, C.; Zheng, W.; Cheng, T.; Shi, J. The complete chloroplast genome sequence of the relict woody plant Metasequoia glyptostroboides Hu et Cheng. Front. Plant Sci. 2015, 6, 447. [Google Scholar] [CrossRef]
- Brownlee, A.G. A rapid DNA isolation procedure applicable to many refractory flamentous fungi. Fungal Genet. Rep. 1988, 35, 8. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate denovo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.I. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef]
- Matthew, K.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Chase, M.W.; Cameron, K.M.; Freudenstein, J.V.; Pridgeon, A.M.; André, S. An updated classification of Orchidaceae. Bot. J. Linn. Soc. 2015, 177, 151–174. [Google Scholar] [CrossRef]
- Chapter H Names of Hybrids. 2025. Volume 2, p. 17. Available online: https://www.iapt-taxon.org/nomen/pages/main/art_h4.html (accessed on 17 February 2025).
- Wissemann, V. Plant evolution by means of hybridization. Syst. Biodivers. 2007, 5, 243–253. [Google Scholar] [CrossRef]
- Abbott, R.J.; Hegarty, M.I.; Hiscock, S.J.; Brennan, A.C. Homoploid hybrid speciation in action. Taxon 2010, 59, 1375–1386. [Google Scholar] [CrossRef]
- Burgess, K.S.; Morgan, M.; Husband, B.C. Interspecific seed discounting and the fertility cost of hybridization in an endangered species. New Phytol. 2008, 177, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.Y.; Yang, J.X.; Li, H.K.; Zhao, H.S. Chloroplast genomes of two species of Cypripedium: Expanded genome size and proliferation of AT-biased repeat sequences. Front. Plant Sci. 2021, 12, 609729. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Zhang, M.F.; Xue, J.; Dong, R.; Zhang, X.H. Chloroplast genomic resources for phylogeny and DNA barcoding: A case study on Fritillaria. Sci. Rep. 2018, 8, 1184. [Google Scholar] [CrossRef]
- Meng, J.; Li, X.P.; Li, H.T.; Yang, J.B.; Wang, H.; He, J. Comparative Analysis of the Complete Chloroplast Genomes of Four Aconitum Medicinal Species. Molecules 2018, 23, 1015. [Google Scholar] [CrossRef]
- Hishamuddin, M.S.; Lee, S.Y.; Ng, W.L.; Ramlee, S.I.; Lamasudin, D.U.; Mohamed, R. Comparison of eight complete chloroplast genomes of the endangered Aquilaria tree species (Thymelaeaceae) and their phylogenetic relationships. Sci. Rep. 2020, 10, 13034. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Ying, Z.Q.; Yu, S.S.; Wang, Q.; Liao, G.; Ge, Y.; Cheng, R. Complete chloroplast genome of Stephania tetrandra (Menispermaceae) from Zhejiang Province: Insights into molecular structures, comparative genome analysis, mutational hotspots and phylogenetic relationships. Sci. Rep. 2021, 22, 880. [Google Scholar] [CrossRef]
- Somaratne, Y.; Guan, D.L.; Wang, W.Q.; Zhao, L.; Xu, S.Q. The Complete Chloroplast Genomes of Two Lespedeza Species: Insights into Codon Usage Bias, RNA Editing Sites, and Phylogenetic Relationships in Desmodieae (Fabaceae: Papilionoideae). Plants 2020, 9, 51. [Google Scholar] [CrossRef]
- Kim, J.S.; Kim, H.T.; Kim, J.H. The largest plastid genome of monocots: A novel genome type containing AT residue repeats in the slipper Orchid Cypripedium japonicum. Plant Mol. Biol. Report. 2015, 33, 1210–1220. [Google Scholar] [CrossRef]
- Singh, R.B.; Mahenderakar, M.D.; Jugran, A.K.; Singh, R.K.; Srivastava, R.K. Assessing genetic diversity and population structure of sugarcane cultivars, progenitor species and genera using microsatellite (SSR) markers. Gene 2020, 753, 144800. [Google Scholar] [CrossRef]
- Yu, J.Y.; Dossa, K.; Wang, L.H.; Zhang, Y.X.; Wei, X.; Liao, B.S.; Zhang, X.R. PMDBase: A database for studying microsatellite DNA and marker development in plants. Nucleic Acids Res. 2017, 45, 1046–1053. [Google Scholar] [CrossRef]
- Gong, L.; Ding, X.X.; Guan, W.; Zhang, D.C.; Zhang, J.; Bai, J.Q.; Xu, W.; Huang, J.; Qiu, X.H.; Zheng, X.S. Comparative chloroplast genome analyses of Amomum: Insights into evolutionary history and species identification. BMC Plant Biol. 2022, 22, 520. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhao, W.J.; Xu, J.; Li, M.F.; Zhang, Y.J. Chloroplast Genome Evolution and Species Identification of Styrax (Styracaceae). Biomed. Res. Int. 2022, 2022, 5364094. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.G.; Yao, H.; Chen, X.L.; Li, Y.; Song, J.Y. Molecular Structure and Phylogenetic Analyses of Complete Chloroplast Genomes of Two Aristolochia Medicinal Species. Int. J. Mol. Sci. 2017, 18, 1839. [Google Scholar] [CrossRef] [PubMed]

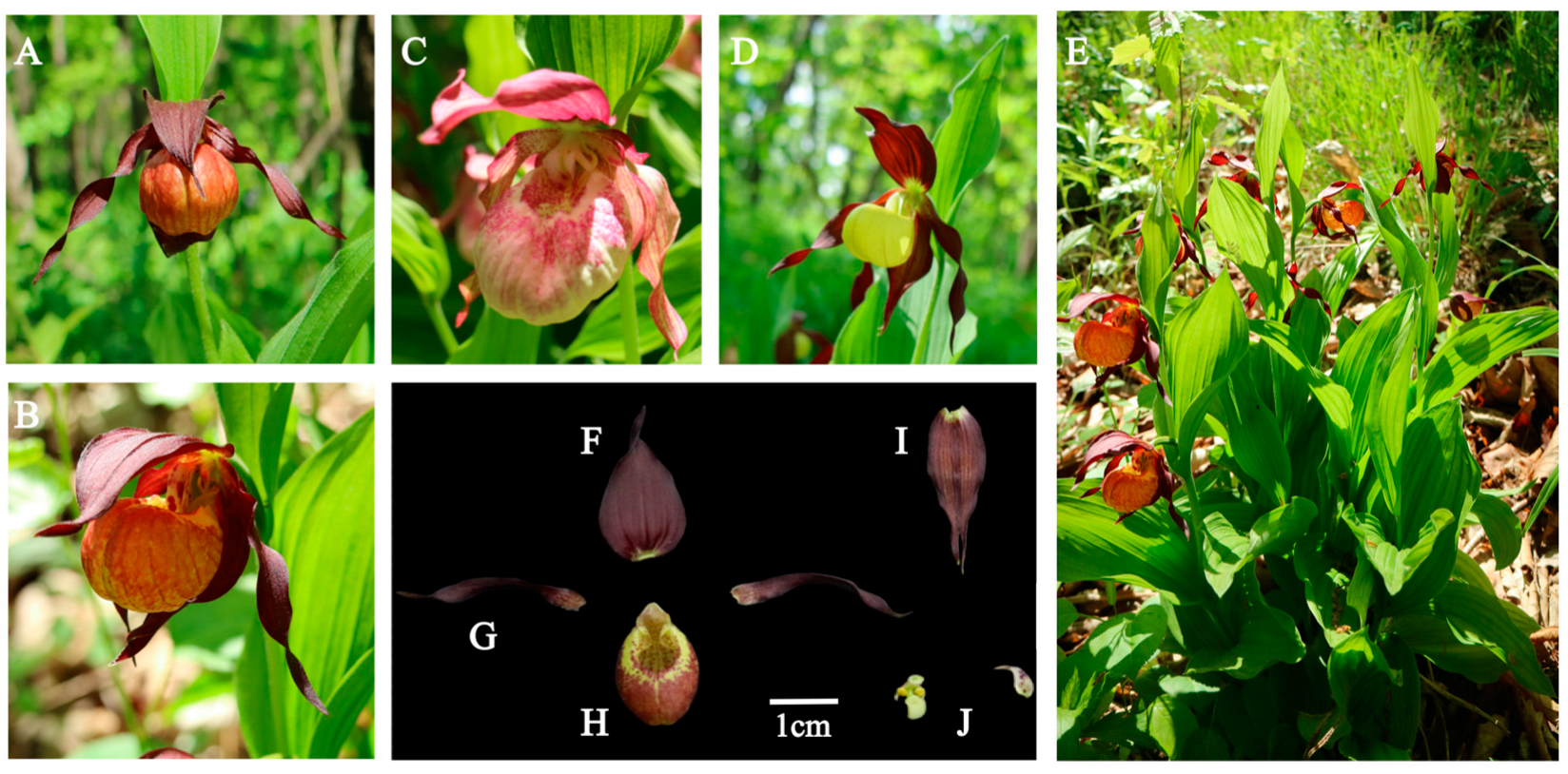





| Taxa | Voucher Specimen | ITS | trnH-psbA | trnS-trnfM | trnL Intron | trnL-F Spacer |
|---|---|---|---|---|---|---|
| C. × ventricosum | -(NOCC) | JF796923 | JF796979 | JF797097 | JF796882 | JF797031 |
| W1 | CLF02 | PQ619134 | * | * | * | * |
| C. bardolphianum | Li JH S6163 (NOCC) | JF796900 | JF796939 | JF797058 | - | JF797002 |
| C. calceolus | Zhang Y E8044a | Z78521 | JF796982 | JF797102 | JF796869 | AY557224 |
| C. calcicola | Li JH S6153 (NOCC) | JF796921 | JF796977 | JF797095 | JF796880 | JF797029 |
| C. candidum | Steele B S1299a | EF370092 | JF796942 | JF797061 | JF796864 | JF797014 |
| C. corrugatum | Gunnlaugson D S1149a | JF796901 | JF796943 | JF797062 | JF796871 | JF797020 |
| C. debile | Zhong SW S6527 (PE) | JF796929 | JF796987 | JF797107 | - | JF797048 |
| C. farreri | Li JH S6181 (NOCC) | JF796904 | JF796946 | JF797064 | JF796873 | JF797022 |
| C. fasciculatum | Kouzes R N6160 (PE) | JF796927 | JF796984 | JF797104 | - | JF797044 |
| C. fasciolatum | Li JH S6156 (NOCC) | Z78530 | JF796947 | JF797065 | JF796874 | JF797023 |
| C. flavum | Li JH S6161 (NOCC) | Z78517 | JF796948 | JF797066 | JF796857 | JF797005 |
| C. formosanum | Gunnlaugson D S1010a | Z78524 | JF796951 | JF797069 | JF796890 | JF797041 |
| C. franchetii | Li JH S6261 (NOCC) | JF796908 | JF796953 | JF797071 | JF796875 | JF797024 |
| C. froschii | Gunnlaugson D S1150a | JF796909 | JF796954 | JF797072 | JF796876 | JF797025 |
| C. guttatum | Li JH S6620 (NOCC) | Z78526 | JF796955 | JF797073 | JF796894 | JF797049 |
| C. henryi | Li JH S6157 (NOCC) | JF796910 | JF796956 | JF797074 | JF796861 | JF797011 |
| C. himalaicum | - | Z78523 | - | - | - | - |
| C. irapeanum | Salazar GA 7234 (MEXU) | FR720328 | FR851212 | FR851227 | FR851217 | - |
| C. kentuckiense | Gunnlaugson D S1146a | JF796911 | JF796958 | JF797076 | JF796870 | JF797019 |
| C. lentiginosum | Li JH S7704 (NOCC) | JF796912 | JF796959 | JF797077 | JF796855 | JF797000 |
| C. lichiangense | Li JH S6303 (NOCC) | Z78529 | JF796960 | JF797078 | - | JF797001 |
| C. macranthos | Ling CY S9032a (PE) | Z78522 | JF796962 | JF797080 | JF796877 | JF797026 |
| C. molle | Salazar GA 6883 (MEXU) | FR720327 | FR851211 | FR851226 | FR851216 | - |
| C. montanum | Frystak M S0613 (PE) | EF370091 | JF796965 | JF797083 | JF796867 | JF797017 |
| C. parviflorum | Taylor SL S0516 (PE) | EF370089 | JF796968 | JF797086 | JF796863 | JF797013 |
| C. plectrochilum | Li JH S6301 (NOCC) | Z78528 | JF796972 | JF797090 | JF796891 | JF797042 |
| C. reginae | Steele B S1159a | JF796917 | JF796971 | JF797089 | JF796860 | JF797010 |
| C. rubitinctum | Gunnlaugson D S1152a | JF796918 | JF796973 | JF797091 | JF796879 | JF797028 |
| C. segawae | Gunnlaugson D S1145a | Z78520 | JF796974 | JF797092 | JF796862 | JF797012 |
| C. shanxiense | Ling CY S0609 (PE) | JF796919 | JF796975 | JF797093 | JF796868 | JF797018 |
| C. sichuanense | Li JH S6154 (NOCC) | JF796920 | JF796976 | JF797094 | JF796854 | JF796999 |
| C. subtropicum | Jin XH N8003 (PE) | JF796896 | JF796934 | JF797053 | - | JF796994 |
| C. tibeticum | Li JH S6132 (NOCC) | JF796922 | JF796978 | JF797096 | JF796881 | JF797030 |
| C. wardii | Li JH S6101 (NOCC) | JF796924 | - | JF797098 | JF796853 | JF796996 |
| C. wumengense | Li JH S6102 (NOCC) | JF796913 | JF796961 | JF797079 | - | JF797033 |
| C. yunnanense | Li JH S6601 (NOCC) | JF796925 | - | JF797099 | JF796883 | JF797032 |
| Outgroup | ||||||
| Mexipedium xerophyticum | Salazar GA 5627 | FR720330 | FR851210 | FR851225 | FR851215 | - |
| Paphiopedilum malipoense | Liu ZJ E7030a | Z78498 | JF796981 | JF797101 | JF796885 | JF797035 |
| Phragmipedium exstaminodium | Salazar GA 7499 (MEXU) | FR720329 | FR851209 | FR851224 | FR851214 | - |
| Phragmipedium schlimii | Perner H S7032a | Z78514 | JF796980 | JF797100 | JF796884 | JF797034 |
| Characteristics | C. × ventricosum | W1 | C. calceolus | W2 | W3 | W4 |
|---|---|---|---|---|---|---|
| Stem | puberulous | puberulous | glandular-hairy | puberulous | puberulous | puberulous |
| Leaves | 5–6, elliptical or ovate-elliptical, glabrous or occasionally puberulent on the veins on both sides. | 5, broadly elliptic to ovate-elliptic, puberulous | 5, elliptical or ovate-elliptical, and sparsely pubescent on the back. | 5–6, elliptical, glabrous, or occasionally puberulent on the veins on both sides. | 5–6, ovate-elliptical, occasionally puberulent on the veins on both sides. | 5–6, elliptical or ovate-elliptical, glabrous or occasionally puberulent on the veins on both sides. |
| 14.0–23.5 × 6.5–11.1 cm | 14.0–16.3 × 4.7–7.5 cm | 13.2–14.2 × 5.8–6.2 cm | 18.0–19.0 × 8.0–9.2 cm | 13.0–15.5 × 4.7–7.0 cm | 14.2–17.8 × 6.0–7.7 cm | |
| Inflorescences | 1- or 2-flowered | 1- or 2-flowered | 1 | 1 | 1- or 2-flowered | 1- or 2-flowered |
| Flowers | purple-red to white | dark reddish-brown to yellow-green | maroon sepals and petals and yellow lip | yellow-green throughout | white to deep pink | light pink throughout |
| Dorsal sepal | ovate-lanceolate, apex twisted and acuminate | ovate, apex twisted, and acuminate | ovate or ovate-lanceolate, apex twisted and acuminate | ovate, apex twisted, and acuminate | ovate, apex twisted, and acuminate | ovate or ovate-lanceolate, apex twisted and acuminate |
| 4.4–5.9 × 2.1–2.8 cm | 3.9–4.8 × 2.0–2.3 cm | 4.4–4.6 × 1.6–1.7 cm | 6.5–6.6 × 2.7–3.3 cm | 5.4–5.5 × 2.3–2.4 cm | 5.0–5.2 × 2.6–3.2 cm | |
| Synsepal | small than dorsal sepal | similar to dorsal sepal, apex shallowly bilobed | similar to dorsal sepal, apex slightly bilobed | small than dorsal sepal | more elongated and thinner than dorsal sepal | small than dorsal sepal |
| Petals | liner-lanceolate or narrowly ovate-lanceolate, slightly twisted | liner-lanceolate, twisted | linear-tapering or linear-lanceolate, twisted | liner-lanceolate or narrowly ovate-lanceolate, slightly twisted | liner-lanceolate or narrowly ovate-lanceolate, slightly twisted | lanceolate with a relatively smooth margin |
| 4.3–5.7 × 0.7–1.9 cm | 3.6–4.6 × 0.6–0.8 cm | 3.7–4.5 × 0.5–0.6 cm | 6.3–6.7 × 1.1–1.3 cm | 5.8–7.1 × 1.0–1.4 cm | 4.3–5.6 × 1.1–1.8 cm | |
| Lip | deeply pouched, subellipsoid, or obvoid-globose, with a paler-rimmed mouth | deeply pouched, ellipsoid, or obovoid-globose, with a yellow-green rim to the mouth | deeply pouched, ellipsoid | deeply pouched, obovoid-globose | deeply pouched, ellipsoid, or obovoid-globose | deeply pouched, ellipsoid, or obovoid-globose |
| 3.4–3.9 × 2.5–2.9 cm | 2.8–3.3 × 1.7–2.7 cm | 3.2–3.4 × 2.1–2.2 cm | 3.4–3.6 × 2.1–2.3 cm | 3.1–3.3 × 2.2–2.3 cm | 3.5–3.7 × 2.4–2.7 cm | |
| Stamonode | subovate-oblong | subovate-oblong | suboblong-elliptic | subovate-oblong | subovate-oblong | subovate-oblong |
| 1.2–1.5 × 0.7–0.9 cm | 0.9–1.2 × 0.6–0.7 cm | 0.7–1.0 × 0.5–0.7 cm | 1.0–1.2 × 0.7–0.8 cm | 0.9–1.1 × 0.6–0.7 cm | 0.8–1.3 × 0.8–1.0 cm | |
| Flowering period | June | June | June | June | June | June |
| Composition | C. × ventricosum | W1 | C. calceolus | W2 | W3 | W4 |
|---|---|---|---|---|---|---|
| Lip | Pink White A | Brilliant Greenish Yellow A | Brilliant Greenish Yellow A | Greenish Yellow A | Strong Pink Red A | Pink White A |
| Strong Pink Red A | Greyish Brown A | - | - | White A | - | |
| Dorsal sepal | Pink White A | Dark Red A | Dark Red A | Greenish Yellow A | Strong Pink Red A | Pink White A |
| Strong Pink Red A | Brilliant Greenish Yellow A | Brilliant Greenish Yellow A | - | White A | - | |
| Synsepal | Pink White A | Dark Red A | Dark Red A | Greenish Yellow A | Strong Pink Red A | Pink White A |
| Strong Pink Red A | Brilliant Greenish Yellow A | Brilliant Greenish Yellow A | - | White A | - | |
| Left Petal | Strong Pink Red A | Dark Red A | Dark Red A | Greenish Yellow A | Pink White A to White A | Pink White A |
| Right Petal | Strong Pink Red A | Dark Red A | Dark Red A | Greenish Yellow A | Pink White A to White A | Pink White A |
| Category of Genes | Gene Grouping | Name of Genes |
|---|---|---|
| Photosynthesis | Subunits of ATP synthase | atpA, atpB, atpE, atpF *, atpH, atpI |
| Subunits of NADH dehydrogenase | ndhA *, ndhB * (×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Subunits of cytochrome | petA, petB *, petD *, petG, petL, petN | |
| Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunit of rubisco | rbcL | |
| Self-replication | Large subunit of ribosome | rpl14, rpl16 *, rpl2 * (×2), rpl20, rpl22, rpl23(×2), rpl32, rpl33, rpl36 |
| DNA-dependent RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | |
| Small subunit of ribosome | rps11, rps12 * (×2), rps14, rps15, rps18, rps19(×2), rps2, rps3, rps4, rps7(×2), rps8 | |
| rRNA | rrn16(×2), rrn23(×2), rrn4.5(×2), rrn5(×2) | |
| tRNA | trnA-UGC * (×2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnM-CAU trnH-GUG (×2), trnI-CAU (×2), trnL-CAA (×2), trnL-UAG, trnL-UAA *, trnV-GAC (×2), trnI-GAU * (×2), trnR-ACG (×2), trnR-UCU, trnN-GUU (×2), trnfM-CAU, trnG-GCC, trnG-UCC, trnY-GUA, trnT-GGU, trnT-UGU, trnS-UGA, trnS-GGA, trnS-GCU, trnV-UAC *, trnW-CCA trnP-UGG, trnQ-UUG | |
| Other genes | Subunitof Acetyl-CoA-carboxylase | accD |
| c-type cytochrome synthesis gene | ccsA | |
| Protease clpP | clpP ** | |
| Translational initiation factor | infA | |
| Maturase | matK | |
| Unknown | Conserved open reading frames | ycf1, ycf2(×2), ycf3 **, ycf4 |
| AminoAcid | Codon | Number | RSCU | Amino Acid | Codon | Number | RSCU |
|---|---|---|---|---|---|---|---|
| Phe | UUU | 814 | 1.32 | Pro | CCU | 331 | 1.54 |
| UUC | 422 | 0.68 | CCC | 197 | 0.92 | ||
| Ser | UCU | 470 | 1.71 | CCA | 244 | 1.13 | |
| UCC | 283 | 1.03 | CCG | 88 | 0.41 | ||
| UCA | 328 | 1.20 | His | CAU | 397 | 1.52 | |
| UCG | 132 | 0.48 | CAC | 125 | 0.48 | ||
| Tyr | UAU | 652 | 1.62 | Gln | CAA | 585 | 1.53 |
| UAC | 154 | 0.38 | CAG | 182 | 0.47 | ||
| Cys | UGU | 185 | 1.47 | Arg | CGU | 277 | 1.34 |
| UGC | 67 | 0.53 | CGC | 78 | 0.38 | ||
| Leu | UUA | 698 | 1.91 | CGA | 282 | 1.36 | |
| UUG | 462 | 1.26 | CGG | 88 | 0.43 | ||
| CUU | 449 | 1.23 | Ile | AUU | 881 | 1.47 | |
| CUC | 145 | 0.40 | AUC | 357 | 0.59 | ||
| CUA | 297 | 0.81 | AUA | 566 | 0.94 | ||
| CUG | 145 | 0.40 | Met | AUG | 512 | 1.00 | |
| Trp | UGG | 378 | 1.00 | Thr | ACU | 429 | 1.63 |
| Asn | AAU | 804 | 1.60 | ACC | 187 | 0.71 | |
| AAC | 203 | 0.40 | ACA | 325 | 1.23 | ||
| Lys | AAA | 836 | 1.47 | ACG | 112 | 0.43 | |
| AAG | 299 | 0.53 | Val | GUU | 417 | 1.45 | |
| Ser | AGU | 335 | 1.22 | GUC | 134 | 0.46 | |
| AGC | 98 | 0.36 | GUA | 430 | 1.49 | ||
| Arg | AGA | 390 | 1.89 | GUG | 172 | 0.60 | |
| AGG | 126 | 0.61 | Ala | GCU | 515 | 1.81 | |
| Asp | GAU | 712 | 1.62 | GCC | 156 | 0.55 | |
| GAC | 165 | 0.38 | GCA | 349 | 1.23 | ||
| Glu | GAA | 871 | 1.48 | GCG | 118 | 0.41 | |
| GAG | 309 | 0.52 | Gly | GGU | 438 | 1.21 | |
| TER | UAA | 24 | 1.38 | GGC | 156 | 0.43 | |
| UAG | 15 | 0.87 | GGA | 591 | 1.64 | ||
| UGA | 13 | 0.75 | GGG | 260 | 0.72 |
| Index | GC1 | GC2 | GC3 | GCall | ENC |
|---|---|---|---|---|---|
| GC1 | 1 | ||||
| GC2 | 0.397 ** | 1 | |||
| GC3 | 0.204 | 0.094 | 1 | ||
| GCall | 0.824 ** | 0.734 ** | 0.481 ** | 1 | |
| ENC | 0.182 | −0.216 | 0.545 ** | 0.177 | 1 |
| DNA Region | No. of Taxa | Aligned Length (bp) | No. Variable Characters (%) | No. Informative Characters (%) | Consistency Index | Retention Index |
|---|---|---|---|---|---|---|
| nrITS | 42 | 803 | 496 (61.77) | 320 (39.85) | 0.635 | 0.784 |
| plastid | 42 | 3679 | 1293 (35.15) | 828 (22.51) | 0.824 | 0.916 |
| combine | 42 | 4482 | 1789 (39.92) | 1148 (25.61) | 0.674 | 0.814 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Lu, X.; Li, S.; Sun, Y.; Shan, Y.; Wang, S.; Jiang, N.; Xiao, Y.; Wang, Q.; Yu, J.; et al. Unveiling a Meaningful Form of Cypripedium × ventricosum Sw. (Cypripedioideae, Orchidaceae) from Changbai Mountain, China: Insights from Morphological, Molecular, and Plastome Analyses. Plants 2025, 14, 772. https://doi.org/10.3390/plants14050772
Li Y, Lu X, Li S, Sun Y, Shan Y, Wang S, Jiang N, Xiao Y, Wang Q, Yu J, et al. Unveiling a Meaningful Form of Cypripedium × ventricosum Sw. (Cypripedioideae, Orchidaceae) from Changbai Mountain, China: Insights from Morphological, Molecular, and Plastome Analyses. Plants. 2025; 14(5):772. https://doi.org/10.3390/plants14050772
Chicago/Turabian StyleLi, Ying, Xi Lu, Shuang Li, Yue Sun, Yuze Shan, Shizhuo Wang, Nan Jiang, Yiting Xiao, Qi Wang, Jiahui Yu, and et al. 2025. "Unveiling a Meaningful Form of Cypripedium × ventricosum Sw. (Cypripedioideae, Orchidaceae) from Changbai Mountain, China: Insights from Morphological, Molecular, and Plastome Analyses" Plants 14, no. 5: 772. https://doi.org/10.3390/plants14050772
APA StyleLi, Y., Lu, X., Li, S., Sun, Y., Shan, Y., Wang, S., Jiang, N., Xiao, Y., Wang, Q., Yu, J., Cao, Q., Wu, S., Chen, L., & Dai, X. (2025). Unveiling a Meaningful Form of Cypripedium × ventricosum Sw. (Cypripedioideae, Orchidaceae) from Changbai Mountain, China: Insights from Morphological, Molecular, and Plastome Analyses. Plants, 14(5), 772. https://doi.org/10.3390/plants14050772






