Genome-Wide Identification and Expression Profiling of ABA-Stress-Ripening (ASR) Gene Family in Barley (Hordeum vulgare L.)
Abstract
:1. Introduction
2. Results
2.1. Barley HvASRs Identification
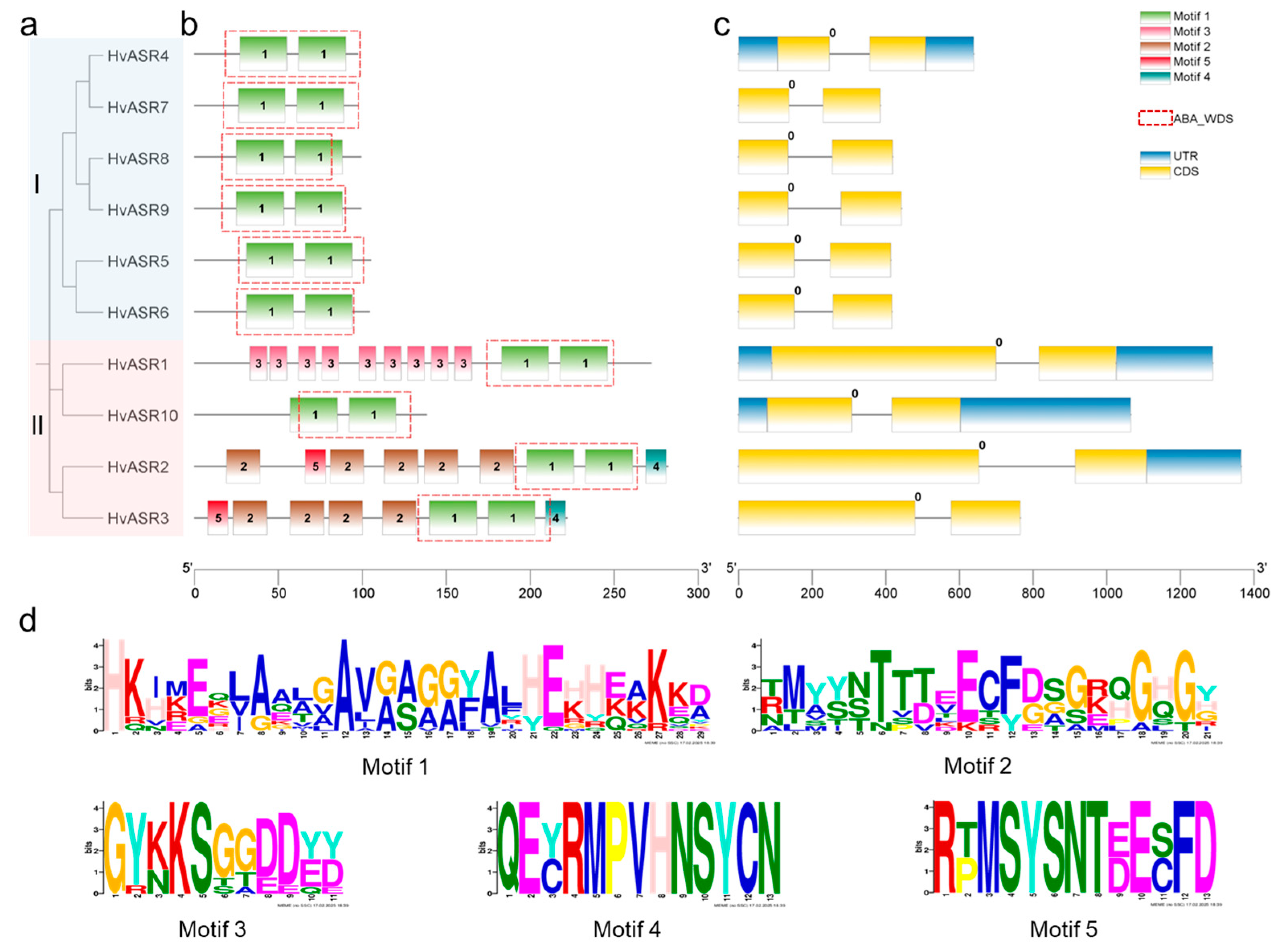
2.2. Phylogenetic Relationships and Gene Structures of HvASRs
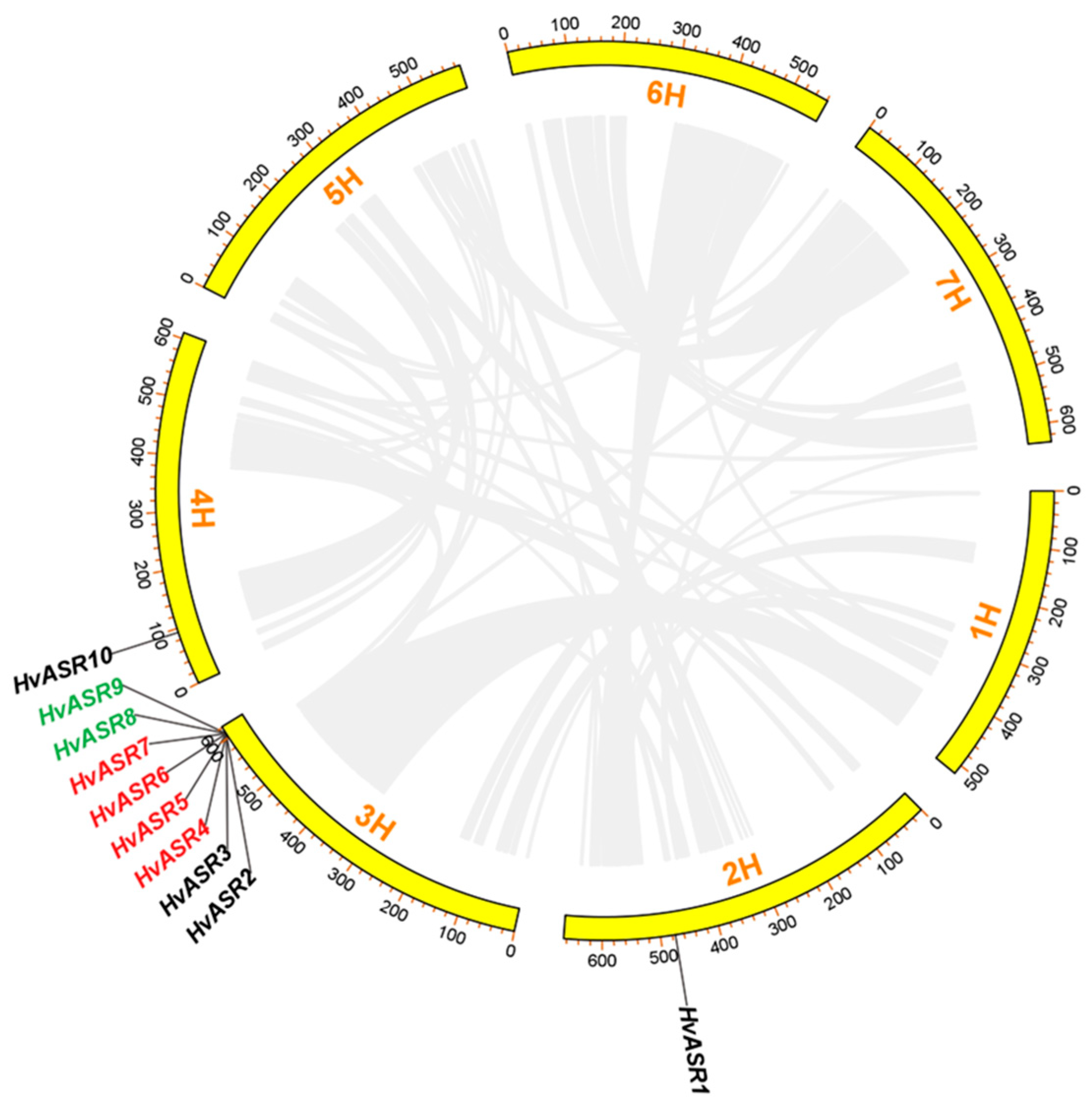
2.3. HvASRs Distribution and Duplication
2.4. Cis-Acting Elements
2.5. Syntenic Relationships of ASRs Between Barley and Other Species
2.6. Tissue-Specific Expression of HvASRs
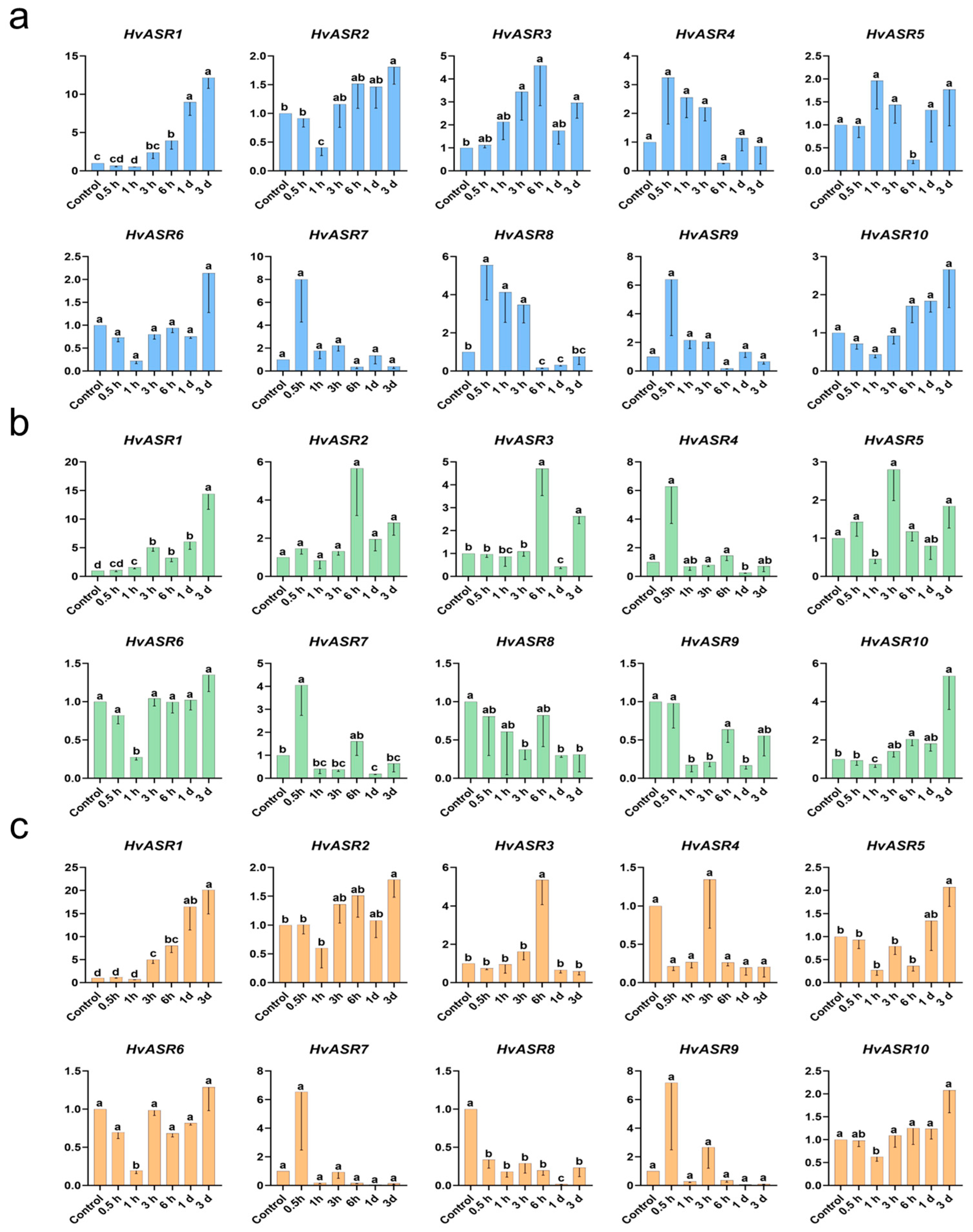
2.7. Responses of HvASRs to Salt and Osmotic Stress and Exogenous ABA Treatment
2.8. Transcriptional Activation Capacity of HvASRs
3. Discussion
4. Materials and Methods
4.1. Identification of HvASRs
4.2. Properties and Localizations of HvASRs
4.3. Phylogeny, Synteny, and Duplication Analyses
4.4. Sequence, Promoter, and Expression Analyses
4.5. Plant Growth, Treatments, and qRT-PCR
4.6. Transcriptional Activation Capacity Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yacoubi, I.; Hamdi, K.; Fourquet, P.; Bignon, C.; Longhi, S. Structural and Functional Characterization of the ABA-Water Deficit Stress Domain from Wheat and Barley: An Intrinsically Disordered Domain Behind the Versatile Functions of the Plant Abscissic Acid, Stress and Ripening Protein Family. Int. J. Mol. Sci. 2021, 22, 2314. [Google Scholar] [CrossRef] [PubMed]
- Iusem, N.D.; Bartholomew, D.M.; Hitz, W.D.; Scolnik, P.A. Tomato (Lycopersicon esculentum) Transcript Induced by Water Deficit and Ripening. Plant Physiol. 1993, 102, 1353–1354. [Google Scholar] [CrossRef] [PubMed]
- González, R.M.; Iusem, N.D. Twenty Years of Research on Asr (ABA-Stress-Ripening) Genes and Proteins. Planta 2014, 239, 941–949. [Google Scholar] [CrossRef]
- Konrad, Z.; Bar-Zvi, D. Synergism Between the Chaperone-like Activity of the Stress Regulated ASR1 Protein and the Osmolyte Glycine-Betaine. Planta 2008, 227, 1213–1219. [Google Scholar] [CrossRef]
- Dai, J.-R.; Liu, B.; Feng, D.-R.; Liu, H.; He, Y.; Qi, K.; Wang, H.-B.; Wang, J.-F. MpAsr Encodes an Intrinsically Unstructured Protein and Enhances Osmotic Tolerance in Transgenic Arabidopsis. Plant Cell Rep. 2011, 30, 1219–1230. [Google Scholar] [CrossRef]
- Hsu, Y.-F.; Yu, S.-C.; Yang, C.-Y.; Wang, C.-S. Lily ASR Protein-Conferred Cold and Freezing Resistance in Arabidopsis. Plant Physiol. Biochem. PPB 2011, 49, 937–945. [Google Scholar] [CrossRef]
- Goldgur, Y.; Rom, S.; Ghirlando, R.; Shkolnik, D.; Shadrin, N.; Konrad, Z.; Bar-Zvi, D. Desiccation and Zinc Binding Induce Transition of Tomato Abscisic Acid Stress Ripening 1, a Water Stress- and Salt Stress-Regulated Plant-Specific Protein, from Unfolded to Folded State. Plant Physiol. 2006, 143, 617–628. [Google Scholar] [CrossRef]
- Wang, L.; Hu, W.; Feng, J.; Yang, X.; Huang, Q.; Xiao, J.; Liu, Y.; Yang, G.; He, G. Identification of the ASR Gene Family from Brachypodium distachyon and Functional Characterization of BdASR1 in Response to Drought Stress. Plant Cell Rep. 2016, 35, 1221–1234. [Google Scholar] [CrossRef]
- Kim, S.-J.; Lee, S.-C.; Hong, S.K.; An, K.; An, G.; Kim, S.-R. Ectopic Expression of a Cold-Responsive OsAsr1 cDNA Gives Enhanced Cold Tolerance in Transgenic Rice Plants. Mol. Cells 2009, 27, 449–458. [Google Scholar] [CrossRef]
- Park, S.I.; Kim, J.J.; Shin, S.Y.; Kim, Y.S.; Yoon, H.S. ASR Enhances Environmental Stress Tolerance and Improves Grain Yield by Modulating Stomatal Closure in Rice. Front. Plant Sci. 2020, 10, 1752. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Yin, Z.; Jiang, J.; Zhang, M.; Guo, X.; Ye, Z.; Zhao, Y.; Xiong, H.; Zhang, Z.; et al. OsASR5 Enhances Drought Tolerance through a Stomatal Closure Pathway Associated with ABA and H2O2 Signalling in Rice. Plant Biotechnol. J. 2017, 15, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, Y.; Jiang, Y.; Li, A.; Cheng, B.; Wu, J. OsASR6 Enhances Salt Stress Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 9340. [Google Scholar] [CrossRef]
- Feng, Z.J.; Xu, Z.S.; Sun, J.; Li, L.C.; Chen, M.; Yang, G.X.; He, G.Y.; Ma, Y.Z. Investigation of the ASR Family in Foxtail Millet and the Role of ASR1 in Drought/Oxidative Stress Tolerance. Plant Cell Rep. 2016, 35, 115–128. [Google Scholar] [CrossRef]
- Li, J.; Dong, Y.; Li, C.; Pan, Y.; Yu, J. SiASR4, the Target Gene of SiARDP from Setaria italica, Improves Abiotic Stress Adaption in Plants. Front. Plant Sci. 2017, 7, 2053. [Google Scholar] [CrossRef]
- Qiu, D.; Hu, W.; Zhou, Y.; Xiao, J.; Hu, R.; Wei, Q.; Zhang, Y.; Feng, J.; Sun, F.; Sun, J.; et al. TaASR1-D Confers Abiotic Stress Resistance by Affecting ROS Accumulation and ABA Signalling in Transgenic Wheat. Plant Biotechnol. J. 2021, 19, 1588–1601. [Google Scholar] [CrossRef]
- Yoon, J.S.; Kim, J.Y.; Kim, D.Y.; Seo, Y.W. A Novel Wheat ASR Gene, TaASR2D, Enhances Drought Tolerance in Brachypodium distachyon. Plant Physiol. Biochem. 2021, 159, 400–414. [Google Scholar] [CrossRef]
- Liang, Y.; Jiang, Y.; Du, M.; Li, B.; Chen, L.; Chen, M.; Jin, D.; Wu, J. ZmASR3 from the Maize ASR Gene Family Positively Regulates Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2019, 20, 2278. [Google Scholar] [CrossRef]
- Yoon, J.S.; Kim, J.Y.; Lee, M.B.; Seo, Y.W. Over-Expression of the Brachypodium ASR Gene, BdASR4, Enhances Drought Tolerance in Brachypodium distachyon. Plant Cell Rep. 2019, 38, 1109–1125. [Google Scholar] [CrossRef]
- Kalifa, Y.; Perlson, E.; Gilad, A.; Konrad, Z.; Scolnik, P.A.; Bar-Zvi, D. Over-Expression of the Water and Salt Stress-Regulated Asr1 Gene Confers an Increased Salt Tolerance. Plant Cell Environ. 2004, 27, 1459–1468. [Google Scholar] [CrossRef]
- Tiwari, V.; Chaturvedi, A.K.; Mishra, A.; Jha, B. Introgression of the SbASR-1 Gene Cloned from a Halophyte Salicornia brachiata Enhances Salinity and Drought Endurance in Transgenic Groundnut (Arachis hypogaea) and Acts as a Transcription Factor. PLoS ONE 2015, 10, e0131567. [Google Scholar] [CrossRef]
- Jha, B.; Lal, S.; Tiwari, V.; Yadav, S.K.; Agarwal, P.K. The SbASR-1 Gene Cloned from an Extreme Halophyte Salicornia brachiata Enhances Salt Tolerance in Transgenic Tobacco. Mar. Biotechnol. 2012, 14, 782–792. [Google Scholar] [CrossRef]
- Wu, M.; Liu, R.; Gao, Y.; Xiong, R.; Shi, Y.; Xiang, Y. PheASR2, a Novel Stress-Responsive Transcription Factor from Moso Bamboo (Phyllostachys edulis), Enhances Drought Tolerance in Transgenic Rice via Increased Sensitivity to Abscisic Acid. Plant Physiol. Biochem. 2020, 154, 184–194. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, H.; Zhou, T.; Zhu, Z.; Zhang, Y.; Zhao, X.; Wang, C. ThASR3 Confers Salt and Osmotic Stress Tolerances in Transgenic Tamarix and Arabidopsis. BMC Plant Biol. 2022, 22, 586. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.H.; Ren, W.; Gao, H.J.; Lü, X.P.; Zhao, Q.; Zhang, H.; Rensing, C.; Zhang, J.L. HaASR2 from Haloxylon ammodendron Confers Drought and Salt Tolerance in Plants. Plant Sci. 2023, 328, 111572. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, Q.; Yu, H.; Li, L.; Zhang, G.; Chen, X.; Jiang, M.; Tan, M. Comprehensive Analysis of the Cadmium Tolerance of Abscisic Acid-, Stress-and Ripening-Induced Proteins (ASRs) in Maize. Int. J. Mol. Sci. 2019, 20, 133. [Google Scholar] [CrossRef]
- Arenhart, R.A.; Schunemann, M.; Bucker Neto, L.; Margis, R.; Wang, Z.Y.; Margis-Pinheiro, M. Rice ASR1 and ASR5 Are Complementary Transcription Factors Regulating Aluminium Responsive Genes. Plant Cell Environ. 2016, 39, 645–651. [Google Scholar] [CrossRef]
- Pérez-Díaz, J.; Wu, T.M.; Pérez-Díaz, R.; Ruíz-Lara, S.; Hong, C.Y.; Casaretto, J.A. Organ- and Stress-Specific Expression of the ASR Genes in Rice. Plant Cell Rep. 2014, 33, 61–73. [Google Scholar] [CrossRef]
- Zhang, L.; Yi, X.; Guo, J.; Zhao, H.; Xia, Q.; He, P.; Huo, S.; Qu, J.; Cao, Y.; Tan, Y.; et al. Expression Patterns of the ZmASR Family During Simulated Drought Stress in Maize. Agron. J. 2021, 113, 894–905. [Google Scholar] [CrossRef]
- Zan, T.; Li, L.; Xie, T.; Zhang, L.; Li, X. Genome-Wide Identification and Abiotic Stress Response Patterns of Abscisic Acid Stress Ripening Protein Family Members in Triticum aestivum L. Genomics 2020, 112, 3794–3802. [Google Scholar] [CrossRef]
- Li, H.; Guan, H.; Zhuo, Q.; Wang, Z.; Li, S.; Si, J.; Zhang, B.; Feng, B.; Kong, L.A.; Wang, F.; et al. Genome-Wide Characterization of the Abscisic Acid-, Stress- and Ripening-Induced (ASR) Gene Family in Wheat (Triticum aestivum L.). Biol. Res. 2020, 53, 23. [Google Scholar] [CrossRef]
- Cao, X.N.; Shen, L.H.; Song, J.; Wang, J.J.; Wang, H.G.; Chen, L.; Pei, Y.X.; Liu, S.C.; Qiao, Z.J. Analysis of Cloned Sequences and Expression of Asr Gene Family in Millet. J. Anim. Plant Sci. 2021, 31, 1309–1318. [Google Scholar] [CrossRef]
- Jia, H.; Jiu, S.; Zhang, C.; Wang, C.; Tariq, P.; Liu, Z.; Wang, B.; Cui, L.; Fang, J. Abscisic Acid and Sucrose Regulate Tomato and Strawberry Fruit Ripening through the Abscisic Acid-Stress-Ripening Transcription Factor. Plant Biotechnol. J. 2016, 14, 2045–2065. [Google Scholar] [CrossRef] [PubMed]
- Golan, I.; Guadalupe Dominguez, P.; Konrad, Z.; Shkolnik-Inbar, D.; Carrari, F.; Bar-Zvi, D. Tomato ABSCISIC ACID STRESS RIPENING (ASR) Gene Family Revisited. PLoS ONE 2014, 9, e107117. [Google Scholar] [CrossRef] [PubMed]
- Guruprasad, K.; Reddy, B.V.B.; Pandit, M.W. Correlation Between Stability of a Protein and Its Dipeptide Composition: A Novel Approach for Predicting in Vivo Stability of a Protein from Its Primary Sequence. Protein Eng. Des. Sel. 1990, 4, 155–161. [Google Scholar] [CrossRef]
- Hamdi, K.; Salladini, E.; O’Brien, D.P.; Brier, S.; Chenal, A.; Yacoubi, I.; Longhi, S. Structural Disorder and Induced Folding within Two Cereal, ABA Stress and Ripening (ASR) Proteins. Sci. Rep. 2017, 7, 15544. [Google Scholar] [CrossRef]
- Pérez-Díaz, J.; Pérez-Díaz, J.R.; Medeiros, D.B.; Zuther, E.; Hong, C.Y.; Nunes-Nesi, A.; Hincha, D.K.; Ruiz-Lara, S.; Casaretto, J.A. Transcriptome Analysis Reveals Potential Roles of a Barley ASR Gene That Confers Stress Tolerance in Transgenic Rice. J. Plant Physiol. 2019, 238, 29–39. [Google Scholar] [CrossRef]
- Rogozin, I.B.; Carmel, L.; Csuros, M.; Koonin, E.V. Origin and Evolution of Spliceosomal Introns. Biol. Direct 2012, 7, 11. [Google Scholar] [CrossRef]
- Kuo, Y.T.; Chao, Y.T.; Chen, W.C.; Shih, M.C.; Chang, S.B. Segmental and Tandem Chromosome Duplications Led to Divergent Evolution of the Chalcone synthase Gene Family in Phalaenopsis orchids. Ann. Bot. 2019, 123, 69–77. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) Expansin Gene Superfamily Origins: Segmental and Tandem Duplication Events Followed by Divergent Selection among Subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef]
- Sadowski, I.; Ma, J.; Triezenberg, S.; Ptashne, M. GAL4-VP16 Is an Unusually Potent Transcriptional Activator. Nature 1988, 335, 563–564. [Google Scholar] [CrossRef]
- Yu, Y.; Li, M.; Song, T.; Zhang, S.; Wang, T. Genome-Wide Identification of Miscanthus ASR Gene Family Reveals That MsASR4 Is Linked to NaCl Tolerance. Ind. Crops Prod. 2024, 219, 119113. [Google Scholar] [CrossRef]
- Liu, H.; Ding, Q.; Cao, L.; Huang, Z.; Wang, Z.; Zhang, M.; Jian, S. Identification of the Abscisic Acid-, Stress-, and Ripening-Induced (ASR) Family Involved in the Adaptation of Tetragonia tetragonoides (Pall.) Kuntze to Saline–Alkaline and Drought Habitats. Int. J. Mol. Sci. 2023, 24, 15815. [Google Scholar] [CrossRef] [PubMed]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly Regulated Genes Are Intron Poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef]
- Xiao, F.; Zhou, H. Plant Salt Response: Perception, Signaling, and Tolerance. Front. Plant Sci. 2023, 13, 1053699. [Google Scholar] [CrossRef]
- Meena, R.P.; Vishwakarma, H.; Ghosh, G.; Gaikwad, K.; Chellapilla, T.S.; Singh, M.P.; Padaria, J.C. Novel ASR Isolated from Drought Stress Responsive SSH Library in Pearl Millet Confers Multiple Abiotic Stress Tolerance in PgASR3 Transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 156, 7–19. [Google Scholar] [CrossRef]
- Hu, Y.-X.; Yang, X.; Li, X.-L.; Yu, X.-D.; Li, Q.-L. The SlASR Gene Cloned from the Extreme Halophyte Suaeda liaotungensis K. Enhances Abiotic Stress Tolerance in Transgenic Arabidopsis thaliana. Gene 2014, 549, 243–251. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005. [Google Scholar]
- Ishida, T.; Kinoshita, K. PrDOS: Prediction of Disordered Protein Regions from Amino Acid Sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An Integrative Web Server to Predict Subcellular Localization of Proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and Sequence Analysis Tools Services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Zeng, F.; Wang, J.; Zhang, G. Identification and Characterization of HAK/KUP/KT Potassium Transporter Gene Family in Barley and Their Expression under Abiotic Stress. BMC Genom. 2021, 22, 317. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Jiang, W.; Tong, T.; Li, W.; Huang, Z.; Chen, G.; Zeng, F.; Riaz, A.; Amoanimaa-Dede, H.; Pan, R.; Zhang, W.; et al. Molecular Evolution of Plant 14-3-3 Proteins and Function of Hv14-3-3A in Stomatal Regulation and Drought Tolerance. Plant Cell Physiol. 2022, 63, 1857–1872. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Colmsee, C.; Beier, S.; Himmelbach, A.; Schmutzer, T.; Stein, N.; Scholz, U.; Mascher, M. BARLEX—The Barley Draft Genome Explorer. Mol. Plant 2015, 8, 964–966. [Google Scholar] [CrossRef]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A Chromosome Conformation Capture Ordered Sequence of the Barley Genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef]
- Cai, K.; Kuang, L.; Yue, W.; Xie, S.; Xia, X.; Zhang, G.; Wang, J. Calmodulin and Calmodulin-like Gene Family in Barley: Identification, Characterization and Expression Analyses. Front. Plant Sci. 2022, 13, 964888. [Google Scholar] [CrossRef]
- Cai, K.; Song, X.; Yue, W.; Liu, L.; Ge, F.; Wang, J. Identification and Functional Characterization of Abiotic Stress Tolerance-Related PLATZ Transcription Factor Family in Barley (Hordeum vulgare L.). Int. J. Mol. Sci. 2024, 25, 10191. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]






| Gene Name | Gene ID | Length (aas) | pI | MW (kDa) | Instability Index | Aliphatic Index | GRAVY | Disordered aas (%) | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| HvASR1 | HORVU.MOREX.r3.2HG0168120 | 272 | 5.21 | 29.59 | 41.95 | 18.05 | −1.68 | 89.34 | nucleus |
| HvASR2 | HORVU.MOREX.r3.3HG0325150 | 282 | 6.59 | 29.95 | 50.62 | 35.07 | −0.89 | 59.57 | chloroplast |
| HvASR3 | HORVU.MOREX.r3.3HG0325170 | 222 | 6.40 | 23.86 | 38.19 | 29.14 | −1.00 | 67.57 | nucleus |
| HvASR4 | HORVU.MOREX.r3.3HG0325200 | 97 | 9.82 | 10.76 | 42.80 | 48.45 | −1.22 | 73.20 | nucleus |
| HvASR5 | HORVU.MOREX.r3.3HG0325210 | 105 | 9.52 | 11.11 | 36.32 | 67.24 | −0.78 | 76.19 | nucleus |
| HvASR6 | HORVU.MOREX.r3.3HG0325230 | 104 | 9.40 | 11.06 | 35.54 | 65.10 | −0.84 | 75.00 | nucleus |
| HvASR7 | HORVU.MOREX.r3.3HG0325240 | 97 | 9.87 | 10.65 | 42.05 | 49.59 | −1.13 | 71.13 | nucleus |
| HvASR8 | HORVU.MOREX.r3.3HG0325300 | 99 | 9.99 | 11.04 | 34.67 | 49.60 | −1.34 | 73.74 | nucleus |
| HvASR9 | HORVU.MOREX.r3.3HG0325310 | 99 | 9.94 | 11.08 | 38.93 | 46.67 | −1.29 | 74.75 | nucleus |
| HvASR10 | HORVU.MOREX.r3.4HG0349450 | 138 | 6.14 | 15.43 | 28.89 | 43.33 | −1.20 | 84.78 | nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, J.; Cai, K.; Song, X.; Yue, W.; Liu, L.; Ge, F.; Wang, Q.; Wang, J. Genome-Wide Identification and Expression Profiling of ABA-Stress-Ripening (ASR) Gene Family in Barley (Hordeum vulgare L.). Plants 2025, 14, 970. https://doi.org/10.3390/plants14060970
Ren J, Cai K, Song X, Yue W, Liu L, Ge F, Wang Q, Wang J. Genome-Wide Identification and Expression Profiling of ABA-Stress-Ripening (ASR) Gene Family in Barley (Hordeum vulgare L.). Plants. 2025; 14(6):970. https://doi.org/10.3390/plants14060970
Chicago/Turabian StyleRen, Jie, Kangfeng Cai, Xiujuan Song, Wenhao Yue, Lei Liu, Fangying Ge, Qiuyu Wang, and Junmei Wang. 2025. "Genome-Wide Identification and Expression Profiling of ABA-Stress-Ripening (ASR) Gene Family in Barley (Hordeum vulgare L.)" Plants 14, no. 6: 970. https://doi.org/10.3390/plants14060970
APA StyleRen, J., Cai, K., Song, X., Yue, W., Liu, L., Ge, F., Wang, Q., & Wang, J. (2025). Genome-Wide Identification and Expression Profiling of ABA-Stress-Ripening (ASR) Gene Family in Barley (Hordeum vulgare L.). Plants, 14(6), 970. https://doi.org/10.3390/plants14060970






