Exploring Alternative Splicing in Response to Salinity: A Tissue-Level Comparative Analysis Using Arabidopsis thaliana Public Transcriptomic Data
Abstract
1. Introduction
2. Results
2.1. Summary of the Selected Datasets
2.2. Alternative Splicing as a Regulatory Mechanism in Salinity Responses
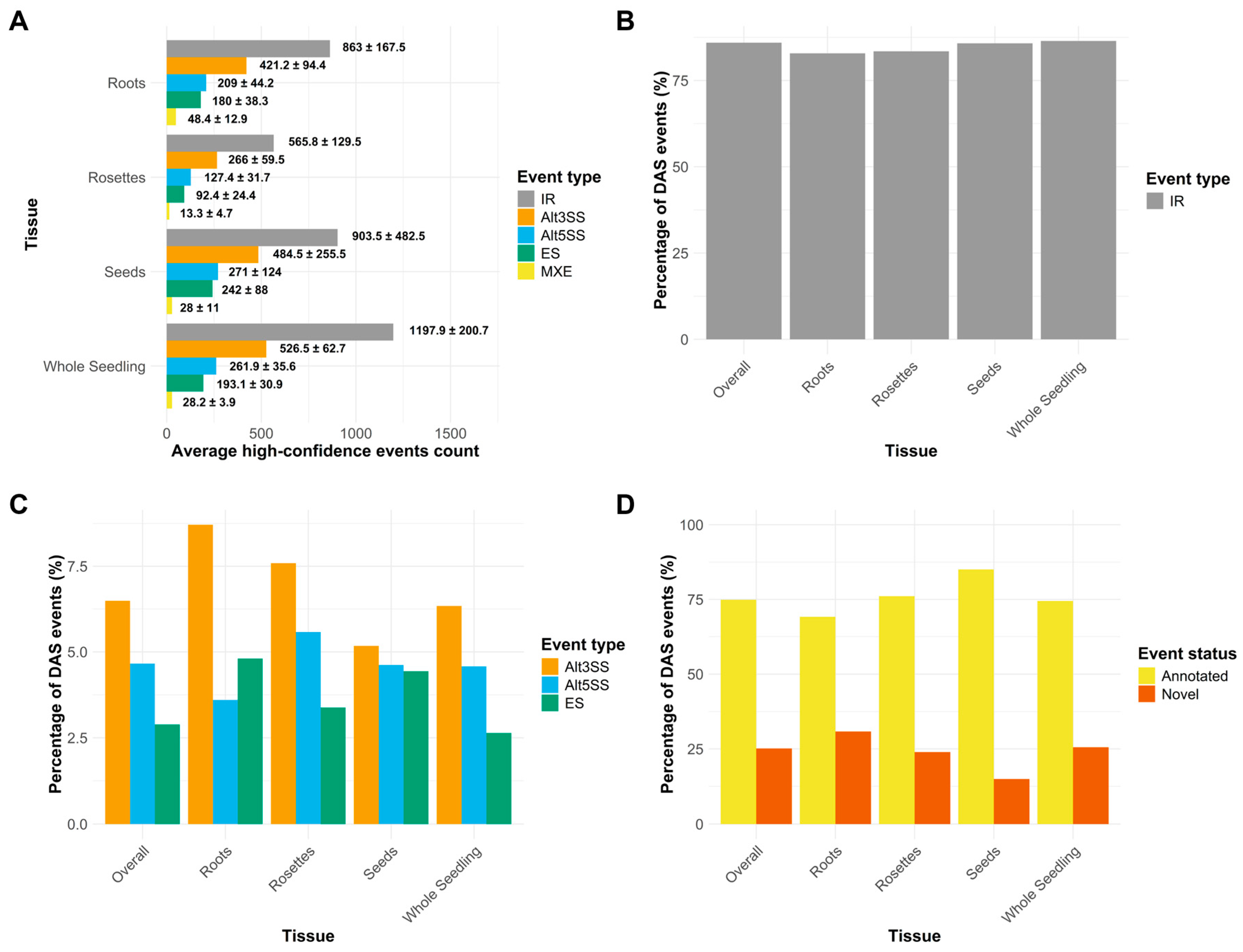
2.3. Characteristics of Alternative Splicing in Salinity
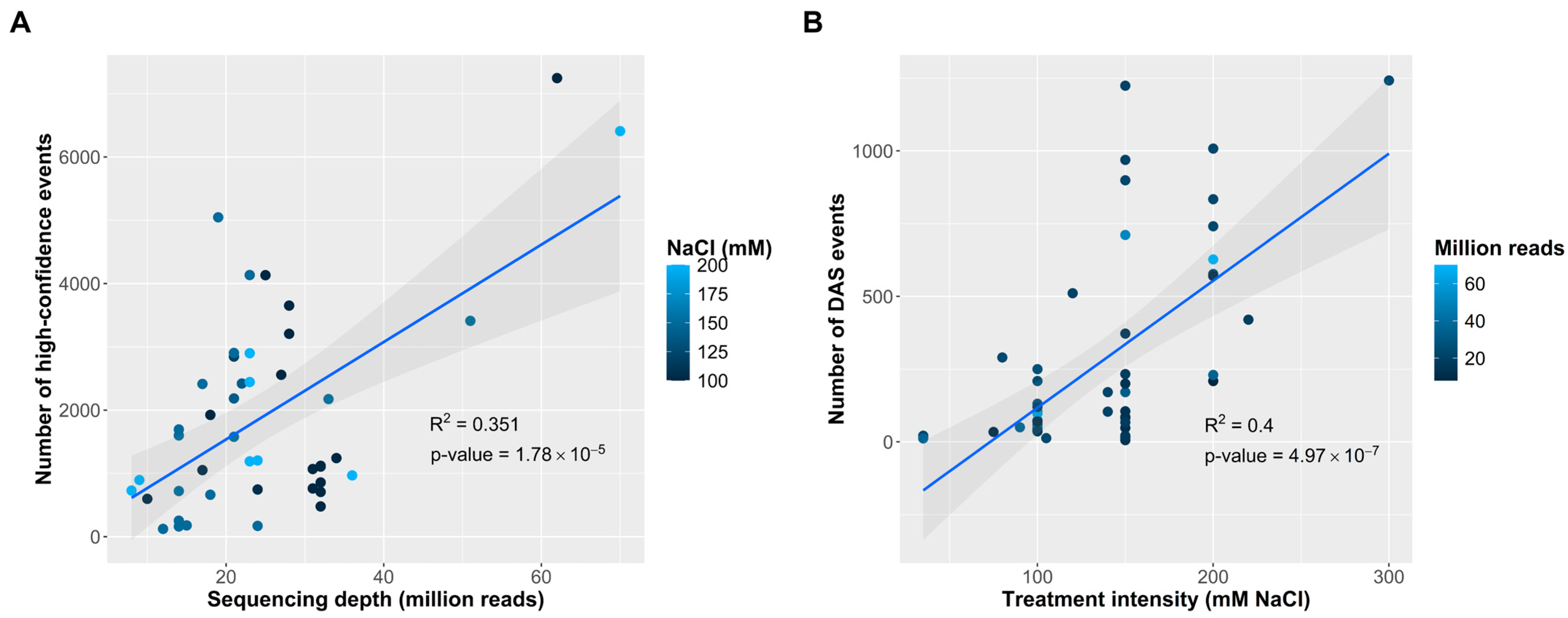
2.4. Determinants of AS Events Detection and Generation
2.5. Tissue-Specific AS Regulation
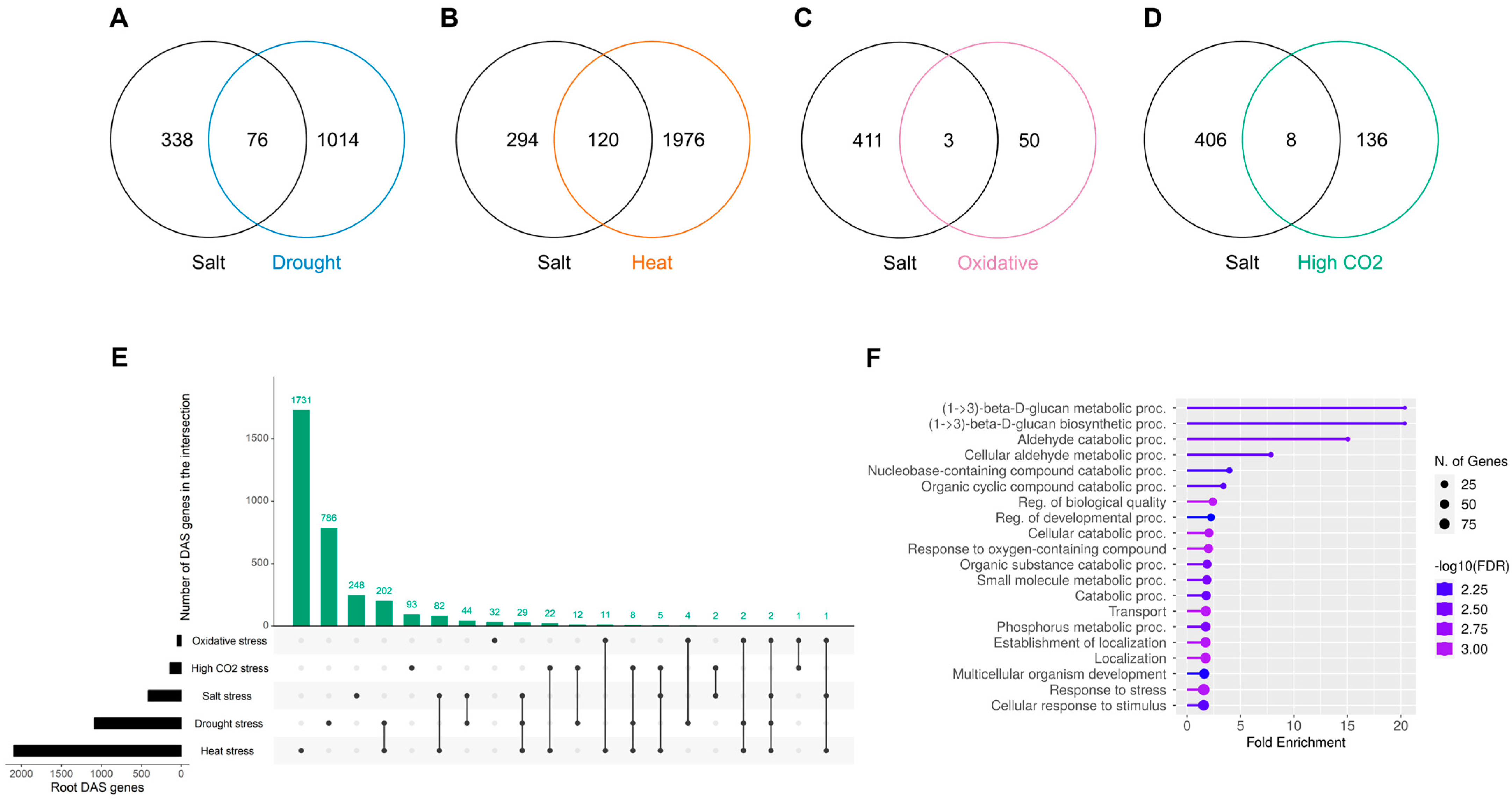
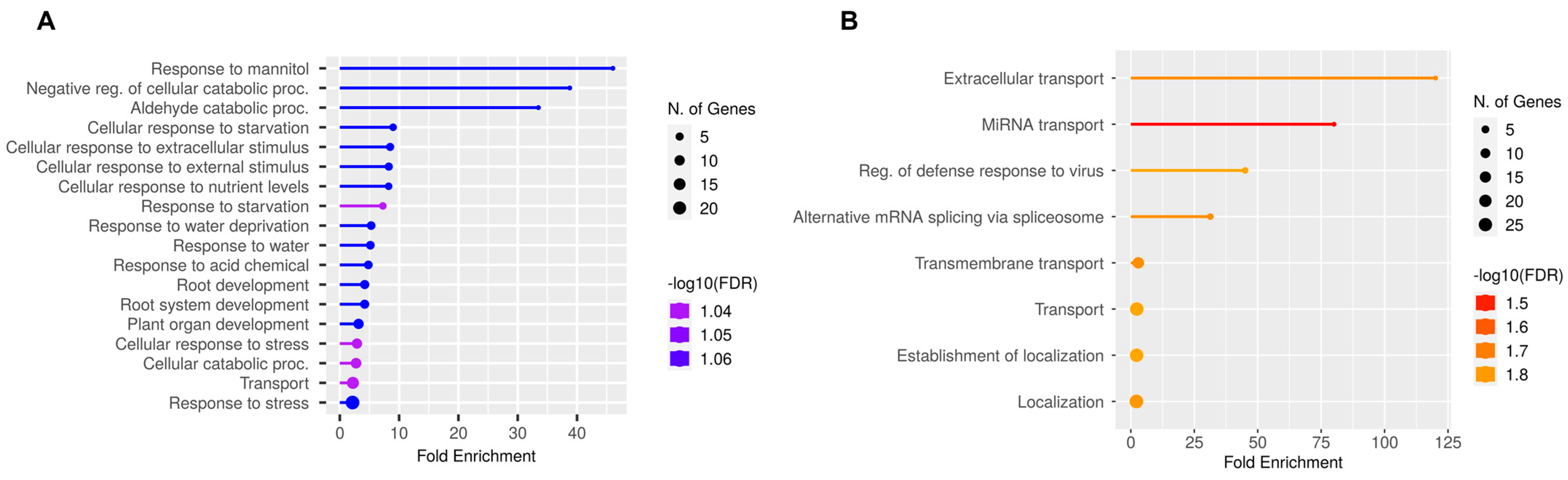
2.6. Cross-Stress Alternative Splicing Regulation in the Roots
3. Discussion
3.1. Alternative Splicing as a Regulatory Layer in Salinity
3.2. Mechanistic Insights into Alternative Splicing Regulation in Salinity
3.3. Experimental and Biological Considerations in Alternative Splicing Studies
3.4. Tissue-Dependent Nature of Alternative Splicing Regulation in Salinity
3.5. Shared and Unique Alternative Splicing Regulated Genes Across Stress Conditions
4. Materials and Methods
4.1. Data Retrieval
4.2. Pre-Processing and Differential Expression Analysis
4.3. Detection of Alternative Splicing and Differential Alternative Splicing
4.4. Data Exploration, Parameters Evaluation, and Data Integration
4.5. Gene Ontology (GO) Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Machado, R.M.A.; Serralheiro, R.P. Soil Salinity: Effect on Vegetable Crop Growth. Management Practices to Prevent and Mitigate Soil Salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Cabot, C.; Sibole, J.V.; Barceló, J.; Poschenrieder, C. Lessons from crop plants struggling with salinity. Plant Sci. 2014, 226, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Carillo, P.; Annunziata, M.G.; Pontecorvo, G.; Fuggi, A.; Woodrow, P. Salinity Stress and Salt Tolerance. In Abiotic Stress in Plants—Mechanisms and Adaptations; IntechOpen: London, UK, 2011. [Google Scholar]
- Park, H.J.; Kim, W.Y.; Yun, D.J. A New Insight of Salt Stress Signaling in Plant. Mol. Cells 2016, 39, 447–459. [Google Scholar]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.B.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Al Mahmud, J.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef] [PubMed]
- Fahad, S.; Hussain, S.; Matloob, A.; Khan, F.A.; Khaliq, A.; Saud, S.; Hassan, S.; Shan, D.; Khan, F.; Ullah, N.; et al. Phytohormones and plant responses to salinity stress: A review. Plant Growth Regul. 2015, 75, 391–404. [Google Scholar] [CrossRef]
- Shabala, S.; Wu, H.; Bose, J. Salt stress sensing and early signalling events in plant roots: Current knowledge and hypothesis. Plant Sci. 2015, 241, 109–119. [Google Scholar] [CrossRef]
- Palovaara, J.; de Zeeuw, T.; Weijers, D. Tissue and Organ Initiation in the Plant Embryo: A First Time for Everything. Annu. Rev. Cell Dev. Biol. 2016, 32, 47–75. [Google Scholar] [CrossRef] [PubMed]
- Kreps, J.A.; Wu, Y.; Chang, H.-S.; Zhu, T.; Wang, X.; Harper, J.F. Transcriptome Changes for Arabidopsis in Response to Salt, Osmotic, and Cold Stress. Plant Physiol. 2002, 130, 2129–2141. [Google Scholar] [CrossRef]
- Denby, K.; Gehring, C. Engineering drought and salinity tolerance in plants: Lessons from genome-wide expression profiling in Arabidopsis. Trends Biotechnol. 2005, 23, 547–552. [Google Scholar] [CrossRef]
- Laloum, T.; Martín, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Tang, Z.; Liu, F.; Mao, F.; Yujuan, G.; Wang, Z.; Zhao, X. Normal, novel or none: Versatile regulation from alternative splicing. Plant Signal. Behav. 2021, 16, 1917170. [Google Scholar] [CrossRef] [PubMed]
- Wahl, M.C.; Will, C.L.; Lührmann, R. The Spliceosome: Design Principles of a Dynamic RNP Machine. Cell 2009, 136, 701–718. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Moore, M.J. Spliceosomes. Curr. Biol. 2015, 25, R181–R183. [Google Scholar] [CrossRef]
- Guo, Y.; Shang, X.; Ma, L.; Cao, Y. RNA-Binding Protein-Mediated Alternative Splicing Regulates Abiotic Stress Responses in Plants. Int. J. Mol. Sci. 2024, 25, 10548. [Google Scholar] [CrossRef]
- Kelemen, O.; Convertini, P.; Zhang, Z.; Wen, Y.; Shen, M.; Falaleeva, M.; Stamm, S. Function of alternative splicing. Gene 2013, 514, 1–30. [Google Scholar]
- Ding, F.; Cui, P.; Wang, Z.; Zhang, S.; Ali, S.; Xiong, L. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genom. 2014, 15, 431. [Google Scholar] [CrossRef]
- Feng, J.; Li, J.; Gao, Z.; Lu, Y.; Yu, J.; Zheng, Q.; Yan, S.; Zhang, W.; He, H.; Ma, L.; et al. SKIP Confers Osmotic Tolerance during Salt Stress by Controlling Alternative Gene Splicing in Arabidopsis. Mol. Plant 2015, 8, 1038–1052. [Google Scholar]
- Xu, Z.; Zhang, N.; Fu, H.; Wang, F.; Wen, M.; Chang, H.; Wu, J.; Abdelaala, W.B.; Luo, Q.; Li, Y.; et al. Salt Stress Modulates the Landscape of Transcriptome and Alternative Splicing in Date Palm (Phoenix dactylifera L.). Front. Plant Sci. 2022, 12, 807739. [Google Scholar] [CrossRef]
- Jin, Z.; Lv, X.; Sun, Y.; Fan, Z.; Xiang, G.; Yao, Y. Comprehensive discovery of salt-responsive alternative splicing events based on Iso-Seq and RNA-seq in grapevine roots. Environ. Exp. Bot. 2021, 192, 104645. [Google Scholar] [CrossRef]
- Zhu, G.; Li, W.; Zhang, F.; Guo, W. RNA-seq analysis reveals alternative splicing under salt stress in cotton, Gossypium davidsonii. BMC Genom. 2018, 19, 73. [Google Scholar] [CrossRef] [PubMed]
- Ganie, S.A.; Reddy, A.S.N. Stress-Induced Changes in Alternative Splicing Landscape in Rice: Functional Significance of Splice Isoforms in Stress Tolerance. Biology 2021, 10, 309. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Shen, Q.; Kuang, L.; Wu, D.; Zhang, G. Transcriptomic and alternative splicing analyses reveal mechanisms of the difference in salt tolerance between barley and rice. Environ. Exp. Bot. 2019, 166, 103810. [Google Scholar] [CrossRef]
- Guo, W.; Yu, K.; Han, L.; Li, X.; Wang, H.; Liu, Y.; Zhang, Y. Global profiling of alternative splicing landscape responsive to salt stress in wheat (Triticum aestivum L.). Plant Growth Regul. 2020, 92, 107–116. [Google Scholar] [CrossRef]
- Seo, P.J.; Park, M.-J.; Park, C.-M. Alternative splicing of transcription factors in plant responses to low temperature stress: Mechanisms and functions. Planta 2013, 237, 1415–1424. [Google Scholar] [CrossRef]
- Lin, J.; Zhu, Z. Plant responses to high temperature: A view from pre-mRNA alternative splicing. Plant Mol. Biol. 2021, 105, 575–583. [Google Scholar] [CrossRef]
- Yang, H.; Li, P.; Jin, G.; Gui, D.; Liu, L.; Zhang, C. Temporal regulation of alternative splicing events in rice memory under drought stress. Plant Divers. 2022, 44, 116–125. [Google Scholar] [CrossRef]
- Duque, P. A role for SR proteins in plant stress responses. Plant Signal. Behav. 2011, 6, 49–54. [Google Scholar] [CrossRef]
- Kim, M.; Tagkopoulos, I. Data integration and predictive modeling methods for multi-omics datasets. Mol. Omics 2018, 14, 8–25. [Google Scholar] [CrossRef]
- Szakonyi, D.; Duque, P. Alternative Splicing as a Regulator of Early Plant Development. Front. Plant Sci. 2018, 9, 1174. [Google Scholar] [CrossRef]
- Li, H.; Li, A.; Shen, W.; Ye, N.; Wang, G.; Zhang, J. Global Survey of Alternative Splicing in Rice by Direct RNA Sequencing During Reproductive Development: Landscape and Genetic Regulation. Rice 2021, 14, 75. [Google Scholar] [CrossRef] [PubMed]
- Punzo, P.; Grillo, S.; Batelli, G. Alternative splicing in plant abiotic stress responses. Biochem. Soc. Trans. 2020, 48, 2117–2126. [Google Scholar] [CrossRef]
- Kufel, J.; Diachenko, N.; Golisz, A. Alternative splicing as a key player in the fine-tuning of the immunity response in Arabidopsis. Mol. Plant Pathol. 2022, 23, 1226–1238. [Google Scholar] [CrossRef]
- Seok, H.-Y.; Lee, S.-Y.; Sarker, S.; Bayzid, M.; Moon, Y.-H. Genome-Wide Analysis of Stress-Responsive Genes and Alternative Splice Variants in Arabidopsis Roots under Osmotic Stresses. Int. J. Mol. Sci. 2023, 24, 14580. [Google Scholar] [CrossRef] [PubMed]
- Rosenkranz, R.R.E.; Ullrich, S.; Löchli, K.; Simm, S.; Fragkostefanakis, S. Relevance and Regulation of Alternative Splicing in Plant Heat Stress Response: Current Understanding and Future Directions. Front. Plant Sci. 2022, 13, 911277. [Google Scholar] [CrossRef]
- Prall, W.; Sharma, B.; Gregory, B.D. Transcription Is Just the Beginning of Gene Expression Regulation: The Functional Significance of RNA-Binding Proteins to Post-transcriptional Processes in Plants. Plant Cell Physiol. 2019, 60, 1939–1952. [Google Scholar] [CrossRef]
- Floris, M.; Mahgoub, H.; Lanet, E.; Robaglia, C.; Menand, B. Post-transcriptional Regulation of Gene Expression in Plants during Abiotic Stress. Int. J. Mol. Sci. 2009, 10, 3168–3185. [Google Scholar] [CrossRef]
- Drechsel, G.; Kahles, A.; Kesarwani, A.K.; Stauffer, E.; Behr, J.; Drewe, P.; Rätsch, G.; Wachter, A. Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 2013, 25, 3726–3742. [Google Scholar] [CrossRef]
- Remy, E.; Cabrito, T.R.; Batista, R.A.; Hussein, M.A.M.; Teixeira, M.C.; Athanasiadis, A.; Sá-Correia, I.; Duque, P. Intron Retention in the 5′UTR of the Novel ZIF2 Transporter Enhances Translation to Promote Zinc Tolerance in Arabidopsis. PLoS Genet. 2014, 10, e1004375. [Google Scholar] [CrossRef]
- Kashkan, I.; Hrtyan, M.; Retzer, K.; Humpolíčková, J.; Jayasree, A.; Filepová, R.; Vondráková, Z.; Simon, S.; Rombaut, D.; Jacobs, T.B.; et al. Mutually opposing activity of PIN7 splicing isoforms is required for auxin-mediated tropic responses in Arabidopsis thaliana. New Phytol. 2022, 233, 329–343. [Google Scholar] [CrossRef]
- Remy, E.; Cabrito, T.R.; Baster, P.; Batista, R.A.; Teixeira, M.C.; Friml, J.; Sá-Correia, I.; Duque, P. A Major Facilitator Superfamily Transporter Plays a Dual Role in Polar Auxin Transport and Drought Stress Tolerance in Arabidopsis The Plant Cell. Plant Cell 2013, 25, 901–926. [Google Scholar] [CrossRef] [PubMed]
- Albaqami, M.; Laluk, K.; Reddy, A.S.N. The Arabidopsis splicing regulator SR45 confers salt tolerance in a splice isoform-dependent manner. Plant Mol. Biol. 2019, 100, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Li, X.-H.; Guo, Q.-H.; Liu, P.; Li, Y.; Wu, C.-A.; Yang, G.-D.; Huang, J.-G.; Zhang, S.-Z.; Zheng, C.-C.; et al. Salt responsive alternative splicing of a RING finger E3 ligase modulates the salt stress tolerance by fine-tuning the balance of COP9 signalosome subunit 5A. PLoS Genet. 2021, 17, e1009898. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; Ji, X.-L.; Du, Y.-P.; He, X.; Zhao, X.-J.; Zhai, H. Identification of a Novel Alternative Splicing Variant of VvPMA1 in Grape Root under Salinity. Front. Plant Sci. 2017, 8, 605. [Google Scholar] [CrossRef]
- Jian, G.; Mo, Y.; Hu, Y.; Huang, Y.; Ren, L.; Zhang, Y.; Hu, H.; Zhou, S.; Liu, G.; Guo, J.; et al. Variety-Specific Transcriptional and Alternative Splicing Regulations Modulate Salt Tolerance in Rice from Early Stage of Stress. Rice 2022, 15, 56. [Google Scholar] [CrossRef]
- Mei, W.; Boatwright, L.; Feng, G.; Schnable, J.C.; Barbazuk, W.B. Evolutionarily Conserved Alternative Splicing Across Monocots. Genetics 2017, 207, 465–480. [Google Scholar] [CrossRef]
- Marquez, Y.; Brown, J.W.; Simpson, C.; Barta, A.; Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef]
- John, S.; Olas, J.J.; Mueller-Roeber, B. Regulation of alternative splicing in response to temperature variation in plants. J. Exp. Bot. 2021, 72, 6150–6163. [Google Scholar] [CrossRef]
- Marasco, L.E.; Kornblihtt, A.R. The physiology of alternative splicing. Nat. Rev. Mol. Cell Biol. 2023, 24, 242–254. [Google Scholar] [CrossRef]
- Martín, G.; Márquez, Y.; Mantica, F.; Duque, P.; Irimia, M. Alternative splicing landscapes in Arabidopsis thaliana across tissues and stress conditions highlight major functional differences with animals. Genome Biol. 2021, 22, 35. [Google Scholar] [CrossRef]
- Shaul, O. Unique Aspects of Plant Nonsense-Mediated mRNA Decay. Trends Plant Sci. 2015, 20, 767–779. [Google Scholar] [CrossRef] [PubMed]
- Alyahya, N.; Taybi, T. Transcriptome-wide characterization of alternative splicing regulation in Najran wheat (Triticum aestivum) under salt stress. Curr. Plant Biol. 2024, 38, 100334. [Google Scholar] [CrossRef]
- Gan, J.; Qiu, Y.; Tao, Y.; Zhang, L.; Okita, T.W.; Yan, Y.; Tian, L. RNA-seq analysis reveals transcriptome reprogramming and alternative splicing during early response to salt stress in tomato root. Front. Plant Sci. 2024, 15, 1394223. [Google Scholar] [CrossRef]
- Chaudhary, S.; Khokhar, W.; Jabre, I.; Reddy, A.S.N.; Byrne, L.J.; Wilson, C.M.; Syed, N.H. Alternative Splicing and Protein Diversity: Plants Versus Animals. Front. Plant Sci. 2019, 10, 708. [Google Scholar] [CrossRef] [PubMed]
- Tsoy, O.; Ameling, S.; Franzenburg, S.; Hoffmann, M.; Liv-Willuth, L.; Lee, H.K.; Knabl, L.; Furth, P.A.; Voelker, U.; Hennighausen, L.; et al. RNA sequencing depth guidelines for the study of alternative splicing. bioRxiv 2024. [Google Scholar] [CrossRef]
- Williams, A.G.; Thomas, S.; Wyman, S.K.; Holloway, A.K. RNA-seq Data: Challenges in and Recommendations for Experimental Design and Analysis. Curr. Protoc. Hum. Genet. 2014, 83, 11.13.1–11.13.20. [Google Scholar] [CrossRef]
- Consortium, E. ENCODE Guidelines and Best Practices for RNA-Seq: Revised December 2016. 2016. Available online: https://www.encodeproject.org/documents/cede0cbe-d324-4ce7-ace4-f0c3eddf5972/@@download/attachment/ENCODE%20Best%20Practices%20for%20RNA_v2.pdf (accessed on 19 February 2025).
- Illumina, Inc. RNA Sequencing: Methods and Workflows; Illumina, Inc.: San Diego, CA, USA, 2023. [Google Scholar]
- Jamil, A.; Riaz, S.; Ashraf, M.; Foolad, M.R. Gene Expression Profiling of Plants under Salt Stress. Crit. Rev. Plant Sci. 2011, 30, 435–458. [Google Scholar] [CrossRef]
- Li, S.; Yamada, M.; Han, X.; Ohler, U.; Benfey, P.N. High-Resolution Expression Map of the Arabidopsis Root Reveals Alternative Splicing and lincRNA Regulation. Dev. Cell 2016, 39, 508–522. [Google Scholar] [CrossRef]
- Misra, C.S.; Sousa, A.G.G.; Barros, P.M.; Kermanov, A.; Becker, J.D. Cell-type-specific alternative splicing in the Arabidopsis germline. Plant Physiol. 2022, 192, 85–101. [Google Scholar] [CrossRef]
- Verhage, L.; Severing, E.I.; Bucher, J.; Lammers, M.; Busscher-Lange, J.; Bonnema, G.; Rodenburg, N.; Proveniers, M.C.G.; Angenent, G.C.; Immink, R.G.H. Splicing-related genes are alternatively spliced upon changes in ambient temperatures in plants. PLoS ONE 2017, 12, e0172950. [Google Scholar] [CrossRef]
- Guo, Q.; Liu, L.; Barkla, B.J. Membrane Lipid Remodeling in Response to Salinity. Int. J. Mol. Sci. 2019, 20, 4264. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Liu, L.; Rupasinghe, T.W.T.; Roessner, U.; Barkla, B.J. Salt stress alters membrane lipid content and lipid biosynthesis pathways in the plasma membrane and tonoplast. Plant Physiol. 2022, 189, 805–826. [Google Scholar] [CrossRef]
- Xiao, F.; Zhou, H. Plant salt response: Perception, signaling, and tolerance. Front. Plant Sci. 2023, 13, 1053699. [Google Scholar] [CrossRef]
- Ma, L.; Liu, X.; Lv, W.; Yang, Y. Molecular Mechanisms of Plant Responses to Salt Stress. Front. Plant Sci. 2022, 13, 934877. [Google Scholar] [CrossRef]
- Kawa, D.; Julkowska, M.M.; Sommerfeld, H.M.; Ter Horst, A.; Haring, M.A.; Testerink, C. Phosphate-Dependent Root System Architecture Responses to Salt Stress. Plant Physiol. 2016, 172, 690–706. [Google Scholar] [CrossRef] [PubMed]
- Galvan-Ampudia, C.S.; Testerink, C. Salt stress signals shape the plant root. Curr. Opin. Plant Biol. 2011, 14, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Ding, R.; Kang, S.; Du, T.; Tong, L.; Zhang, Y.; Chen, J.; Shukla, M.K. Chapter Three—Drought, salt, and combined stresses in plants: Effects, tolerance mechanisms, and strategies. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: Cambridge, MA, USA, 2023; pp. 107–163. [Google Scholar]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Lam, P.Y.; Wang, L.; Lo, C.; Zhu, F.-Y. Alternative Splicing and Its Roles in Plant Metabolism. Int. J. Mol. Sci. 2022, 23, 7355. [Google Scholar] [CrossRef]
- He, B.; Meng, L.; Tang, L.; Qi, W.; Hu, F.; Lv, Y.; Song, W. The Landscape of Alternative Splicing Regulating Potassium Use Efficiency in Nicotiana tabacum. Front. Plant Sci. 2021, 12, 774829. [Google Scholar] [CrossRef]
- Taji, T.; Ohsumi, C.; Iuchi, S.; Seki, M.; Kasuga, M.; Kobayashi, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 2002, 29, 417–426. [Google Scholar] [CrossRef]
- Contreras-López, O.; Moyano, T.C.; Soto, D.C.; Gutiérrez, R.A. Step-by-Step Construction of Gene Co-expression Networks from High-Throughput Arabidopsis RNA Sequencing Data. In Root Development: Methods and Protocols; Ristova, D., Barbez, E., Eds.; Springer: New York, NY, USA, 2018; pp. 275–301. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 25 June 2023).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef]
- Zhang, R.; Calixto, C.P.G.; Marquez, Y.; Venhuizen, P.; Tzioutziou, N.A.; Guo, W.; Spensley, M.; Entizne, J.C.; Lewandowska, D.; Have, S.T.; et al. A high quality Arabidopsis transcriptome for accurate transcript-level analysis of alternative splicing. Nucleic Acids Res. 2017, 45, 5061–5073. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wang, Y.; Xie, Z.; Kutschera, E.; Adams, J.I.; Kadash-Edmondson, K.E.; Xing, Y. rMATS-turbo: An efficient and flexible computational tool for alternative splicing analysis of large-scale RNA-seq data. Nat. Protoc. 2024, 19, 1083–1104. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef]
- Mancini, E.; Rabinovich, A.; Iserte, J.; Yanovsky, M.; Chernomoretz, A. ASpli: Integrative analysis of splicing landscapes through RNA-Seq assays. Bioinformatics 2021, 37, 2609–2616. [Google Scholar] [CrossRef]
- Wickham, H.; François, R.; Henry, L.; Müller, K. dplyr: A Grammar of Data Manipulation. 2022. Available online: https://CRAN.R-project.org/package=dplyr (accessed on 1 June 2023).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Gao, C.-H.; Yu, G.; Cai, P. ggVennDiagram: An Intuitive, Easy-to-Use, and Highly Customizable R Package to Generate Venn Diagram. Front. Genet. 2021, 12, 706907. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Khan, A.; Mathelier, A. Intervene: A tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinform. 2017, 18, 287. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2019, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]


| Condition | Number of Categories | Categories |
|---|---|---|
| Tissue | 4 | Roots, whole seedlings, rosettes, seeds |
| Treatment intensity | 12 | From 35 to 300 mM NaCl |
| Treatment duration | 19 | From 15 min to 16 days |
| Treatment method | 5 | Solidified media, hydroponic, imbibition, irrigation |
| Plant age at treatment | 13 | Pre-germination, from 4 dpg 1 to 4 weeks dpg |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Urrieta, J.; Álvarez, J.M.; O’Brien, J.A. Exploring Alternative Splicing in Response to Salinity: A Tissue-Level Comparative Analysis Using Arabidopsis thaliana Public Transcriptomic Data. Plants 2025, 14, 1064. https://doi.org/10.3390/plants14071064
Hernández-Urrieta J, Álvarez JM, O’Brien JA. Exploring Alternative Splicing in Response to Salinity: A Tissue-Level Comparative Analysis Using Arabidopsis thaliana Public Transcriptomic Data. Plants. 2025; 14(7):1064. https://doi.org/10.3390/plants14071064
Chicago/Turabian StyleHernández-Urrieta, Jesús, José Miguel Álvarez, and José Antonio O’Brien. 2025. "Exploring Alternative Splicing in Response to Salinity: A Tissue-Level Comparative Analysis Using Arabidopsis thaliana Public Transcriptomic Data" Plants 14, no. 7: 1064. https://doi.org/10.3390/plants14071064
APA StyleHernández-Urrieta, J., Álvarez, J. M., & O’Brien, J. A. (2025). Exploring Alternative Splicing in Response to Salinity: A Tissue-Level Comparative Analysis Using Arabidopsis thaliana Public Transcriptomic Data. Plants, 14(7), 1064. https://doi.org/10.3390/plants14071064






