Genomic Prediction of Wheat Grain Yield Using Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset Description
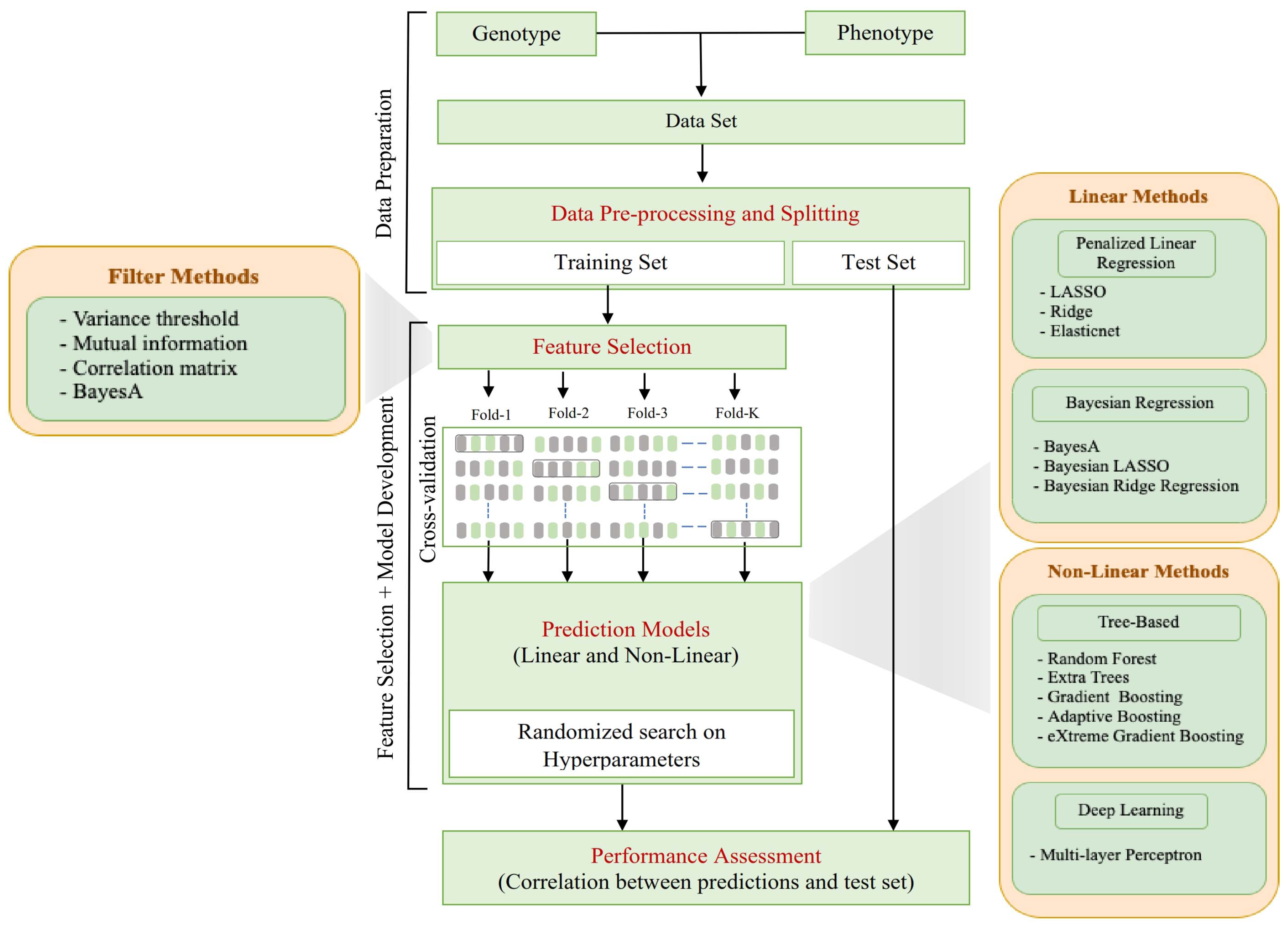
2.2. Computational Pipeline Overview
2.3. Data Pre-Preprocessing and Splitting
2.4. Feature Selection
2.5. Prediction Models
- 1.
- 2.
- Penalized linear regression: LASSO_py, ridge_py, and elasticnet_py with the scikit-learn linear model library [30];
- 3.
- Deep learning: Multilayer Perceptron (mlp_py) with Keras library [45];
- 4.
- Bayesian Methods: BayesA_r, Bayesian Ridge Regression (BRR_r), and Bayesian LASSO (BL_r) were implemented with the BGLR R library [36].
2.6. Performance Assessment
3. Results and Discussion
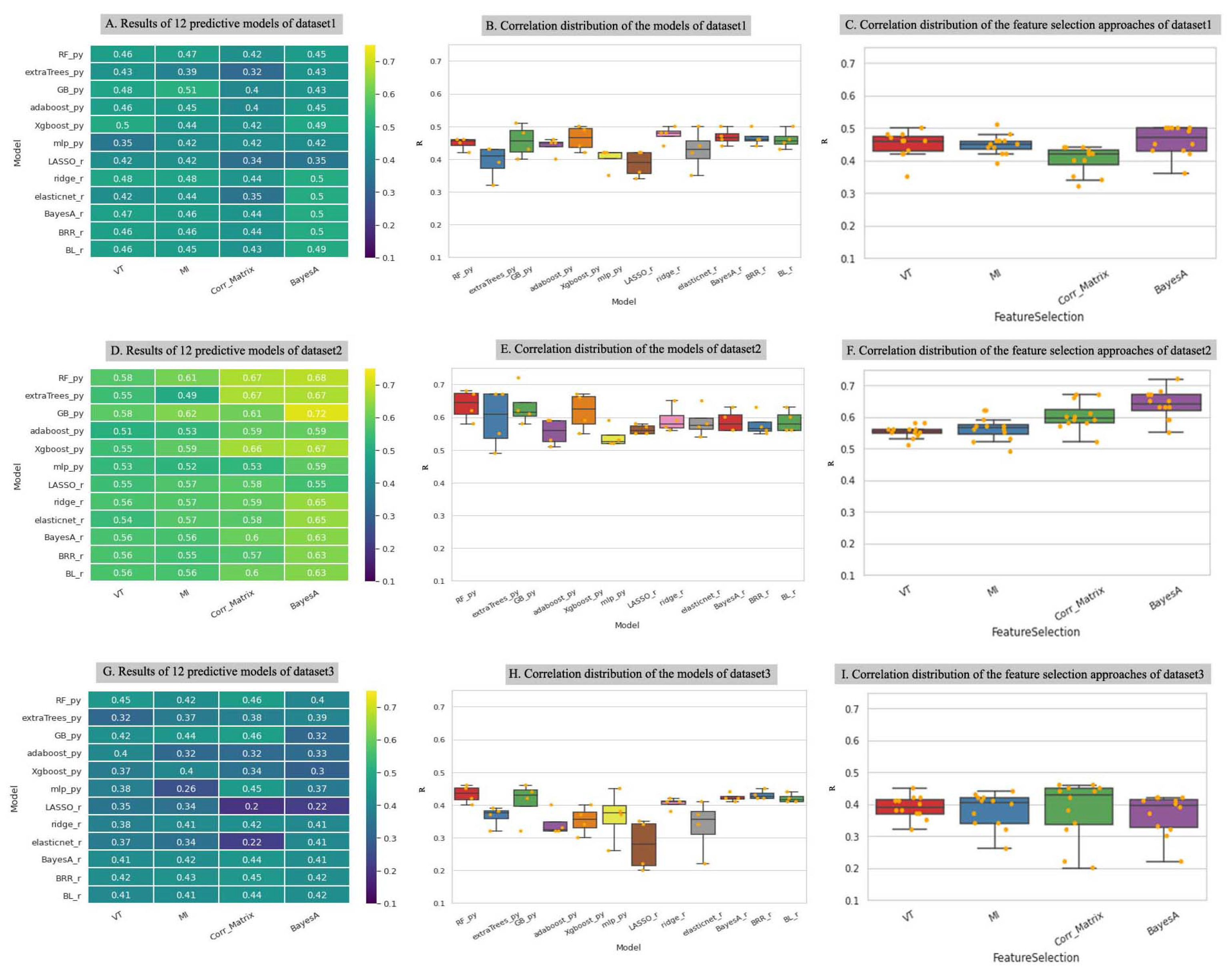
3.1. Feature Selection Methods Vary Widely in the Number and Identity of the Selected SNPs
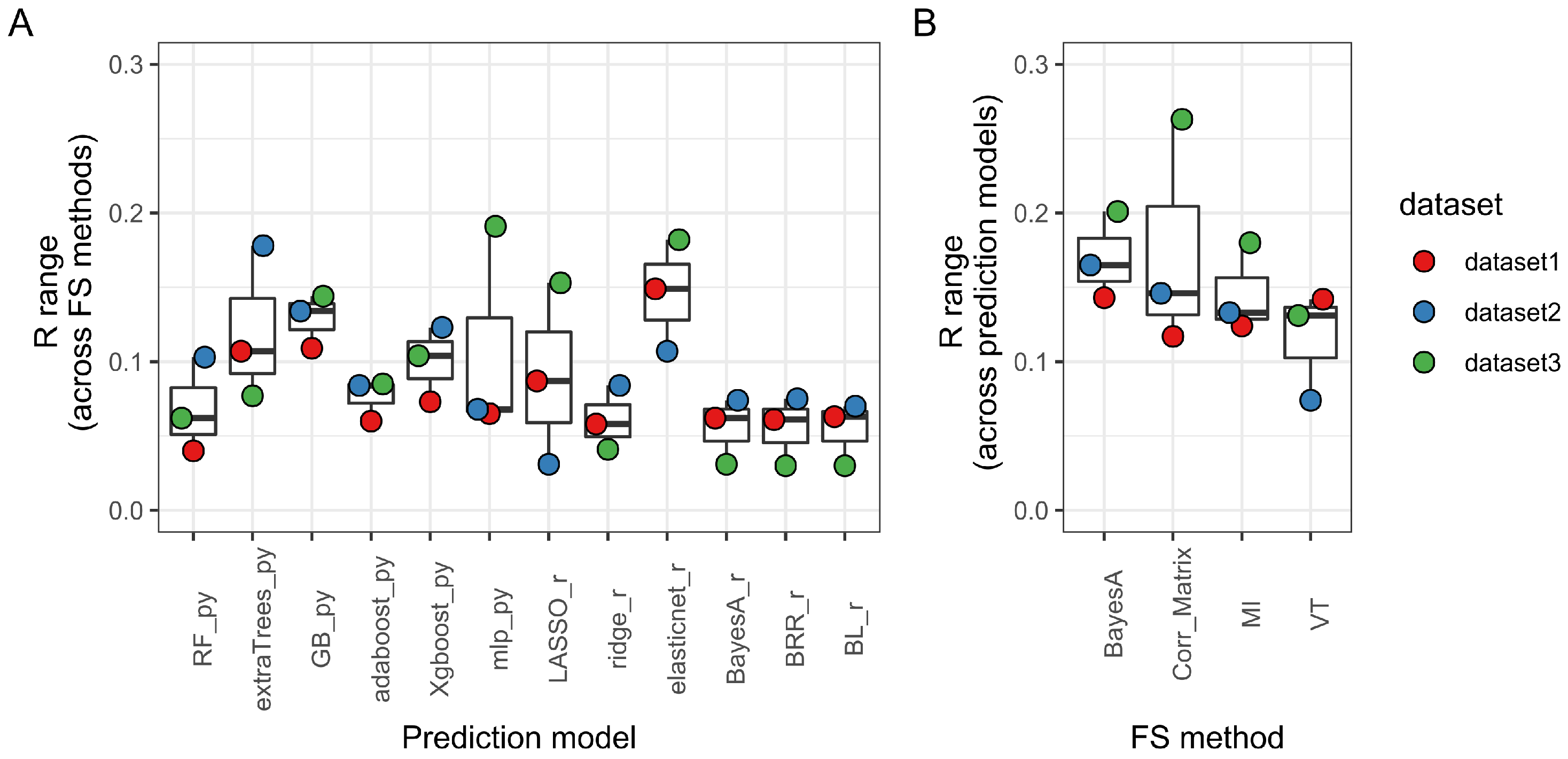
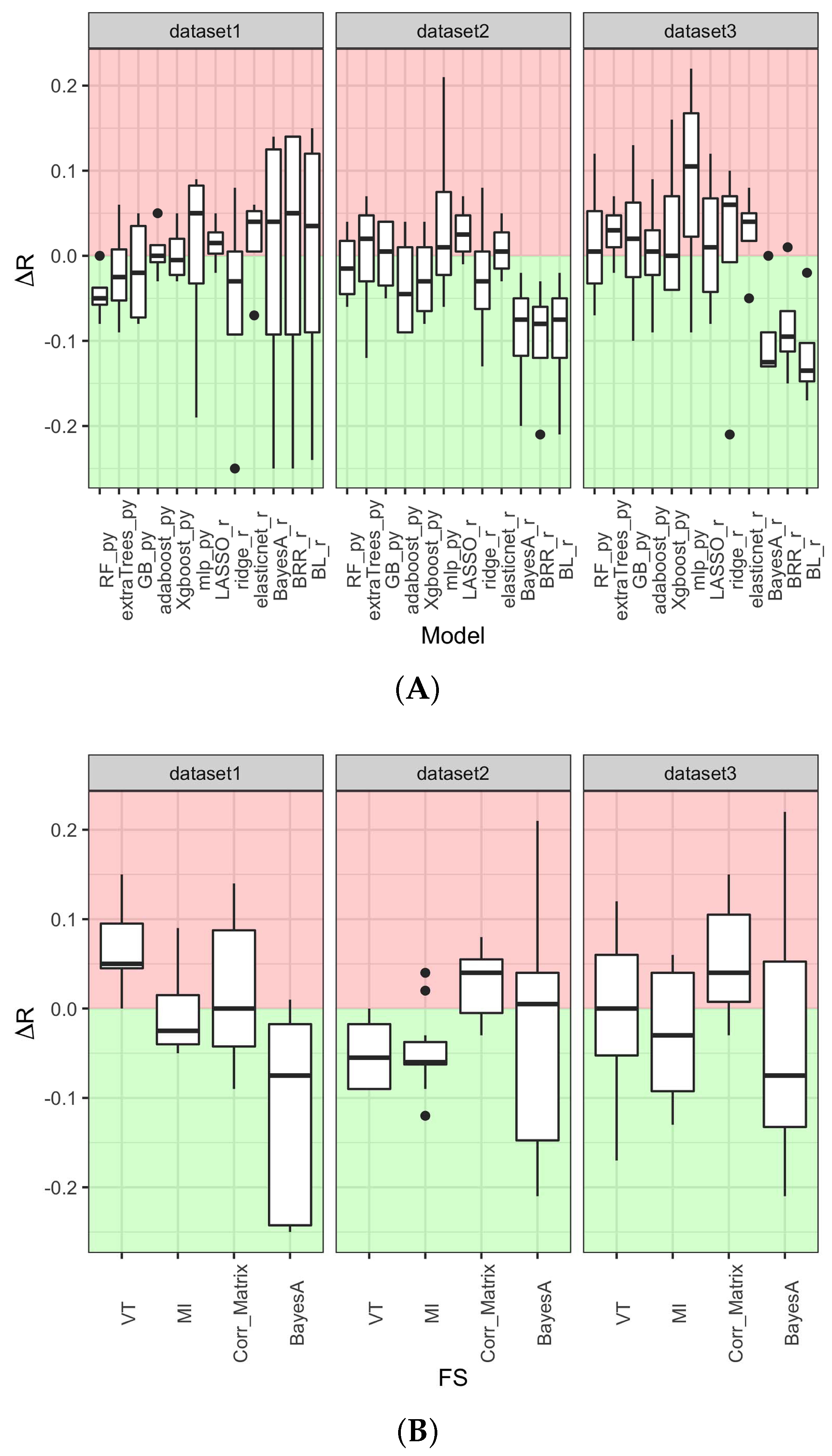
3.2. Prediction Method Is the Main Determinant of Model Performance
3.3. Machine Learning Tree-Based Prediction Methods Maximize Model Performance While Minimizing Over- and Under-Fitting Issues
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| adaboost | adaptive boosting |
| Corr_Matrix | correlation matrix |
| CV | cross-validation |
| DL | deep learning |
| DR | dimensionality reduction |
| ExtraTree | extremely randomized tree |
| GB | gradient boosting |
| gBLUP | genomic best linear unbiased predictor |
| GBS | genotyping-by-sequencing |
| FS | feature selection |
| GP | genomic prediction |
| LASSO | least absolute shrinkage and selection operator |
| MI | mutual information |
| ML | machine learning |
| MLP | multilayer perceptrons |
| R | Pearson’s correlation coefficient |
| RF | random forests |
| SNP | single nucleotide polymorphism |
| VT | variance threshold |
| Xgboost | eXtreme gradient boosting |
References
- Hayes, B.; Goddard, M.; Goddard, M. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Bernardo, R. Molecular markers and selection for complex traits in plants: Learning from the last 20 years. Crop Sci. 2008, 48, 1649–1664. [Google Scholar] [CrossRef]
- Scheben, A.; Yuan, Y.; Edwards, D. Advances in genomics for adapting crops to climate change. Curr. Plant Biol. 2016, 6, 2–10. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, X.; Fu, J.; Wang, H.; Wang, J.; Huang, C.; Prasanna, B.M.; Olsen, M.S.; Wang, G.; Zhang, A. Enhancing genetic gain through genomic selection: From livestock to plants. Plant Commun. 2020, 1, 100005. [Google Scholar] [CrossRef] [PubMed]
- González-Camacho, J.M.; Ornella, L.; Pérez-Rodríguez, P.; Gianola, D.; Dreisigacker, S.; Crossa, J. Applications of machine learning methods to genomic selection in breeding wheat for rust resistance. Plant Genome 2018, 11, 170104. [Google Scholar] [CrossRef]
- Sandhu, K.S.; Patil, S.S.; Aoun, M.; Carter, A.H. Multi-Trait Multi-Environment Genomic Prediction for End-Use Quality Traits in Winter Wheat. Front. Genet. 2022, 13, 831020. [Google Scholar] [CrossRef]
- Farooq, M.; van Dijk, A.D.; Nijveen, H.; Mansoor, S.; de Ridder, D. Genomic prediction in plants: Opportunities for machine learning-based approaches. F1000Research 2022. [Google Scholar] [CrossRef]
- Crossa, J.; Campos, G.D.L.; Pérez, P.; Gianola, D.; Burgueno, J.; Araus, J.L.; Makumbi, D.; Singh, R.P.; Dreisigacker, S.; Yan, J.; et al. Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics 2010, 186, 713–724. [Google Scholar] [CrossRef]
- Habier, D.; Fernando, R.L.; Kizilkaya, K.; Garrick, D.J. Extension of the bayesian alphabet for genomic selection. BMC Bioinform. 2011, 12, 186. [Google Scholar] [CrossRef]
- Saini, D.K.; Chopra, Y.; Singh, J.; Sandhu, K.S.; Kumar, A.; Bazzer, S.; Srivastava, P. Comprehensive evaluation of mapping complex traits in wheat using genome-wide association studies. Mol. Breed. 2022, 42, 1–52. [Google Scholar] [CrossRef]
- Meher, P.K.; Rustgi, S.; Kumar, A. Performance of Bayesian and BLUP alphabets for genomic prediction: Analysis, comparison and results. Heredity 2022, 128, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, K.; Patil, S.S.; Pumphrey, M.; Carter, A. Multitrait machine-and deep-learning models for genomic selection using spectral information in a wheat breeding program. Plant Genome 2021, 14, e20119. [Google Scholar] [CrossRef] [PubMed]
- Montesinos-López, O.A.; Gonzalez, H.N.; Montesinos-López, A.; Daza-Torres, M.; Lillemo, M.; Montesinos-López, J.C.; Crossa, J. Comparing gradient boosting machine and Bayesian threshold BLUP for genome-based prediction of categorical traits in wheat breeding. Plant Genome 2022, e20214. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, K.S.; Aoun, M.; Morris, C.F.; Carter, A.H. Genomic selection for end-use quality and processing traits in soft white winter wheat breeding program with machine and deep learning models. Biology 2021, 10, 689. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, K.S.; Lozada, D.N.; Zhang, Z.; Pumphrey, M.O.; Carter, A.H. Deep learning for predicting complex traits in spring wheat breeding program. Front. Plant Sci. 2021, 11, 613325. [Google Scholar] [CrossRef] [PubMed]
- Bellman, R.E. Adaptive Control Processes: A Guided Tour; Princeton University Press: Princeton, NJ, USA, 2015; Volume 2045. [Google Scholar]
- Chandrashekar, G.; Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Van Der Maaten, L.; Postma, E.; Van den Herik, J. Dimensionality reduction: A comparative. J. Mach. Learn Res. 2009, 10, 13. [Google Scholar]
- Jain, R.; Xu, W. HDSI: High dimensional selection with interactions algorithm on feature selection and testing. PLoS ONE 2021, 16, e0246159. [Google Scholar] [CrossRef]
- Zhou, W.; Bellis, E.S.; Stubblefield, J.; Causey, J.; Qualls, J.; Walker, K.; Huang, X. Minor QTLs mining through the combination of GWAS and machine learning feature selection. bioRxiv 2019. [Google Scholar] [CrossRef]
- Azodi, C.B.; Bolger, E.; McCarren, A.; Roantree, M.; de Los Campos, G.; Shiu, S.H. Benchmarking parametric and machine learning models for genomic prediction of complex traits. G3 Genes Genomes Genet. 2019, 9, 3691–3702. [Google Scholar] [CrossRef]
- Grinberg, N.F.; Orhobor, O.I.; King, R.D. An evaluation of machine-learning for predicting phenotype: Studies in yeast, rice, and wheat. Mach. Learn. 2020, 109, 251–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Mouël, C.; Lattre-Gasquet, D.; Mora, O. Land Use and Food Security in 2050: A Narrow Road; Éditions Quae: Paris, France, 2018. [Google Scholar]
- Lozada, D.N.; Ward, B.P.; Carter, A.H. Gains through selection for grain yield in a winter wheat breeding program. PLoS ONE 2020, 15, e0221603. [Google Scholar] [CrossRef] [PubMed]
- Pandas—Python Data Analysis Library. Available online: https://pandas.pydata.org/ (accessed on 2 April 2021).
- McKinney, W.; Team, P. Pandas-Powerful Python Data Analysis Toolkit. Pandas—Powerful Python Data Anal Toolkit 2015, 1625. Available online: https://pandas.pydata.org/ (accessed on 2 April 2021).
- Géron, A. Hands-On Machine Learning with Scikit-Learn, Keras, and TensorFlow: Concepts, Tools, and Techniques to Build Intelligent Systems; O’Reilly Media: Sebastopol, CA, USA, 2019. [Google Scholar]
- Duch, W. Filter methods. In Feature Extraction; Springer: Berlin/Heidelberg, Germany, 2006; pp. 89–117. [Google Scholar] [CrossRef]
- Bermingham, M.L.; Pong-Wong, R.; Spiliopoulou, A.; Hayward, C.; Rudan, I.; Campbell, H.; Wright, A.F.; Wilson, J.F.; Agakov, F.; Navarro, P.; et al. Application of high-dimensional feature selection: Evaluation for genomic prediction in man. Sci. Rep. 2015, 5, 10312. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Variance Threshold Feature Selection Using Sklearn. Available online: https://scikit-learn.org/stable/modules/generated/sklearn.feature_selection.VarianceThreshold.html (accessed on 22 June 2021).
- Plotting a Diagonal Correlation Matrix. Available online: https://seaborn.pydata.org/examples/many_pairwise_correlations.html (accessed on 29 June 2021).
- Kraskov, A.; Stögbauer, H.; Grassberger, P. Estimating mutual information. Phys. Rev. E 2004, 69, 066138. [Google Scholar] [CrossRef] [PubMed]
- Vergara, J.R.; Estévez, P.A. A review of feature selection methods based on mutual information. Neural Comput. Appl. 2014, 24, 175–186. [Google Scholar] [CrossRef]
- O’Hara, R.B.; Sillanpää, M.J. A review of Bayesian variable selection methods: What, how and which. Bayesian Anal. 2009, 4, 85–117. [Google Scholar] [CrossRef]
- Pérez, P.; de los Campos, G. BGLR: A statistical package for whole genome regression and prediction. Genetics 2014, 198, 483–495. [Google Scholar] [CrossRef]
- de los Campos, G.; Pataki, A.; Pérez, P. The BGLR (Bayesian Generalized Linear Regression) R-Package. 2015. Available online: http://bglr.r-forge.r-project.org/ (accessed on 8 April 2022).
- Bengio, Y.; Grandvalet, Y. No unbiased estimator of the variance of k-fold cross-validation. J. Mach. Learn. Res. 2004, 5, 1089–1105. [Google Scholar]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Probst, P.; Boulesteix, A.L.; Bischl, B. Tunability: Importance of hyperparameters of machine learning algorithms. J. Mach. Learn. Res. 2019, 20, 1934–1965. [Google Scholar]
- Sanner, M.F. Python: A programming language for software integration and development. J. Mol. Graph Model. 1999, 17, 57–61. [Google Scholar]
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- Scikit-Learn Machine Learning in Python. Available online: https://scikit-learn.org/stable/ (accessed on 15 July 2020).
- XGBoost Documentation. Available online: https://xgboost.readthedocs.io/en/latest/ (accessed on 15 July 2020).
- Gulli, A.; Pal, S. Deep Learning with Keras; Packt Publishing Ltd.: Birmingham, UK, 2017. [Google Scholar]
- SPSS Tutorials: Pearson Correlation. Available online: https://libguides.library.kent.edu/SPSS/PearsonCorr (accessed on 1 July 2020).
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; Heisterkamp, S.; Van Willigen, B.; Maintainer, R. Package ‘nlme’. Linear Nonlinear Mixed Eff. Model. Version 2017, 3. Available online: https://CRAN.R-project.org/package=nlme (accessed on 12 June 2022).
- González-Recio, O.; Forni, S. Genome-wide prediction of discrete traits using Bayesian regressions and machine learning. Genet. Sel. Evol. 2011, 43, 7. [Google Scholar] [CrossRef] [PubMed]
- Montesinos-López, O.A.; Montesinos-López, A.; Pérez-Rodríguez, P.; Barrón-López, J.A.; Martini, J.W.; Fajardo-Flores, S.B.; Gaytan-Lugo, L.S.; Santana-Mancilla, P.C.; Crossa, J. A review of deep learning applications for genomic selection. BMC Genom. 2021, 22, 19. [Google Scholar] [CrossRef]
- Belkin, M.; Hsu, D.; Ma, S.; Mandal, S. Reconciling modern machine-learning practice and the classical bias–variance trade-off. Proc. Natl. Acad. Sci. USA 2019, 116, 15849–15854. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Nikoloski, Z. Machine learning approaches for crop improvement: Leveraging phenotypic and genotypic big data. J. Plant Physiol. 2021, 257, 153354. [Google Scholar] [CrossRef]
- Mendes-Moreira, J.; Soares, C.; Jorge, A.M.; Sousa; Jorge, F. Ensemble approaches for regression: A survey. ACM Comput. Surv. (CSUR) 2012, 45, 1–40. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests; Springer: Berlin/Heidelberg, Germany, 2001; Volume 45, pp. 5–32. [Google Scholar]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Freund, Y.; Schapire, R.E. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med. 1997, 4, 385–395. [Google Scholar] [CrossRef]
- Park, T.; Casella, G. The bayesian lasso. J. Am. Stat. Assoc. 2008, 103, 681–686. [Google Scholar] [CrossRef]
- de Los Campos, G.; Naya, H.; Gianola, D.; Crossa, J.; Legarra, A.; Manfredi, E.; Weigel, K.; Cotes, J.M. Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 2009, 182, 375–385. [Google Scholar] [CrossRef] [Green Version]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef]
|
Feature Selection | Dataset1 | Dataset2 | Dataset3 |
|---|---|---|---|
| VT | 3235 | 2417 | 4700 |
| MI | 1109 | ||
| Corr_Matrix | 4714 | 3835 | 3169 |
| BayesA | 5500 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sirsat, M.S.; Oblessuc, P.R.; Ramiro, R.S. Genomic Prediction of Wheat Grain Yield Using Machine Learning. Agriculture 2022, 12, 1406. https://doi.org/10.3390/agriculture12091406
Sirsat MS, Oblessuc PR, Ramiro RS. Genomic Prediction of Wheat Grain Yield Using Machine Learning. Agriculture. 2022; 12(9):1406. https://doi.org/10.3390/agriculture12091406
Chicago/Turabian StyleSirsat, Manisha Sanjay, Paula Rodrigues Oblessuc, and Ricardo S. Ramiro. 2022. "Genomic Prediction of Wheat Grain Yield Using Machine Learning" Agriculture 12, no. 9: 1406. https://doi.org/10.3390/agriculture12091406
APA StyleSirsat, M. S., Oblessuc, P. R., & Ramiro, R. S. (2022). Genomic Prediction of Wheat Grain Yield Using Machine Learning. Agriculture, 12(9), 1406. https://doi.org/10.3390/agriculture12091406





