Transcription Factor VvbHLH137 Positively Regulates Anthocyanin Accumulation in Grape (Vitis vinifera)
Abstract
:1. Introduction
2. Results
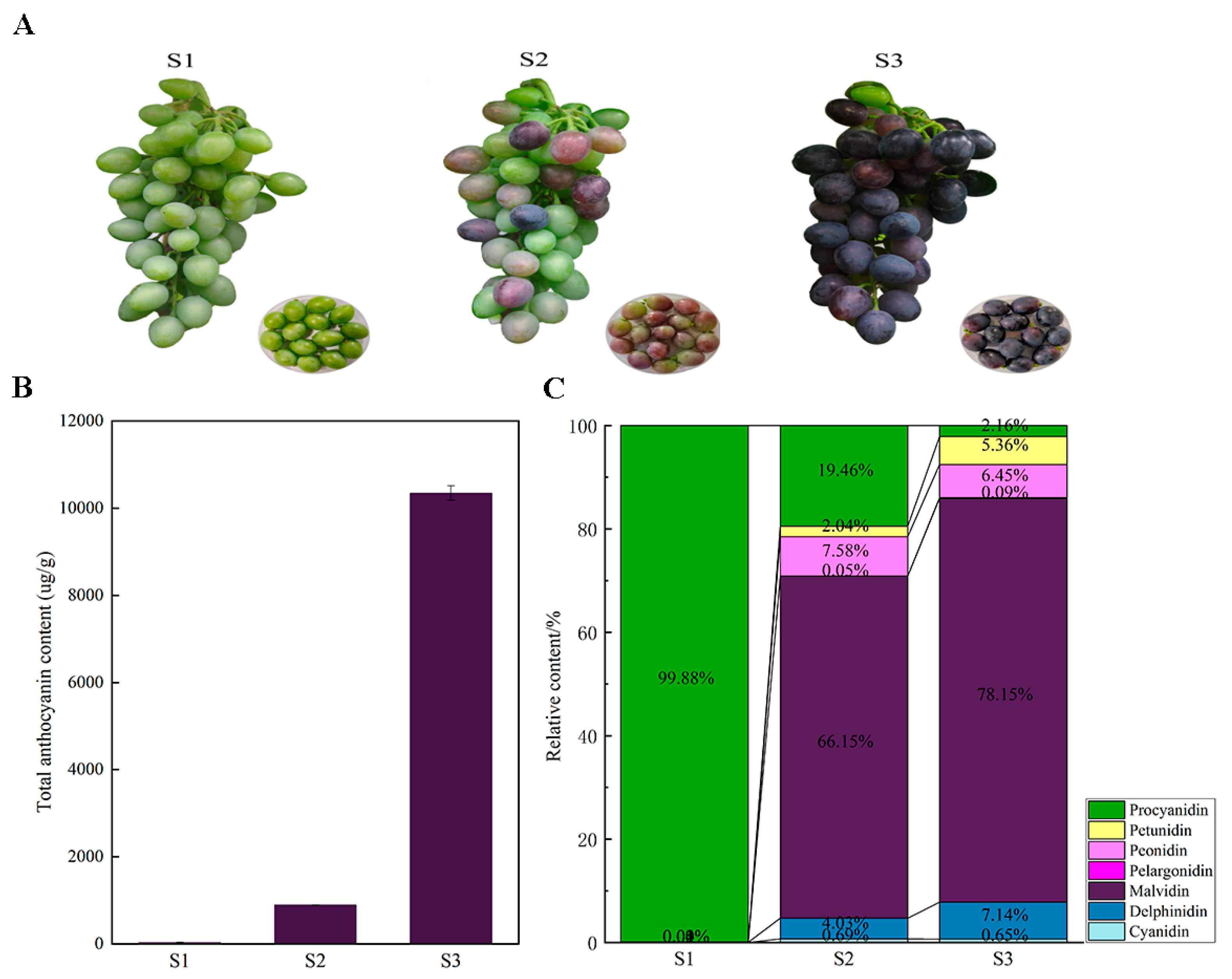
2.1. Changes in Anthocyanin Composition Drive Grape Coloration During Development
2.2. Transcriptome Profiling of Grape Berries During Development
2.3. Exploration of Differentially Expressed Genes (DEGs)
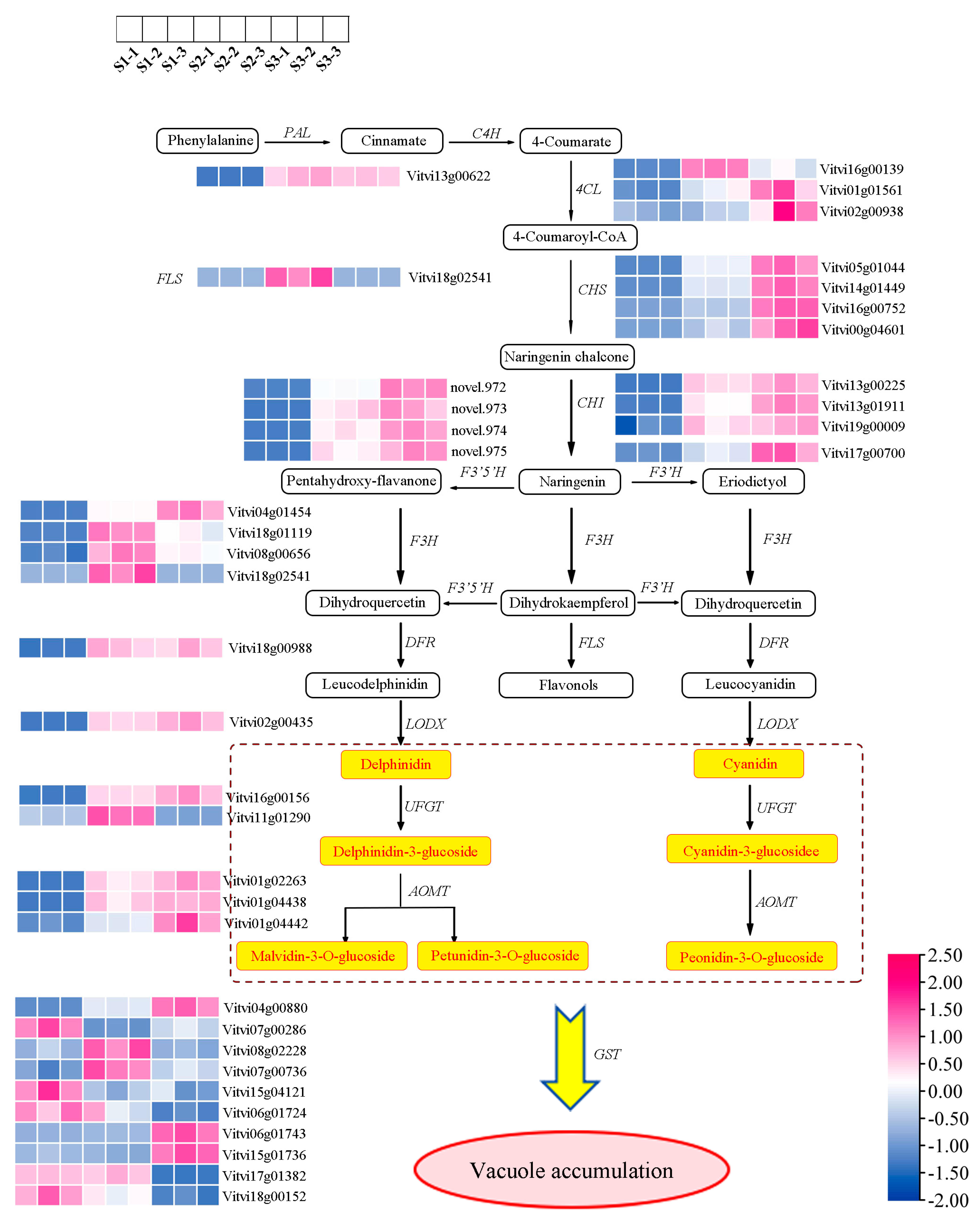
2.4. Identification of DEGs Responsible for Anthocyanin Synthesis
2.5. Identification of TFs Linked to Anthocyanin Synthesis
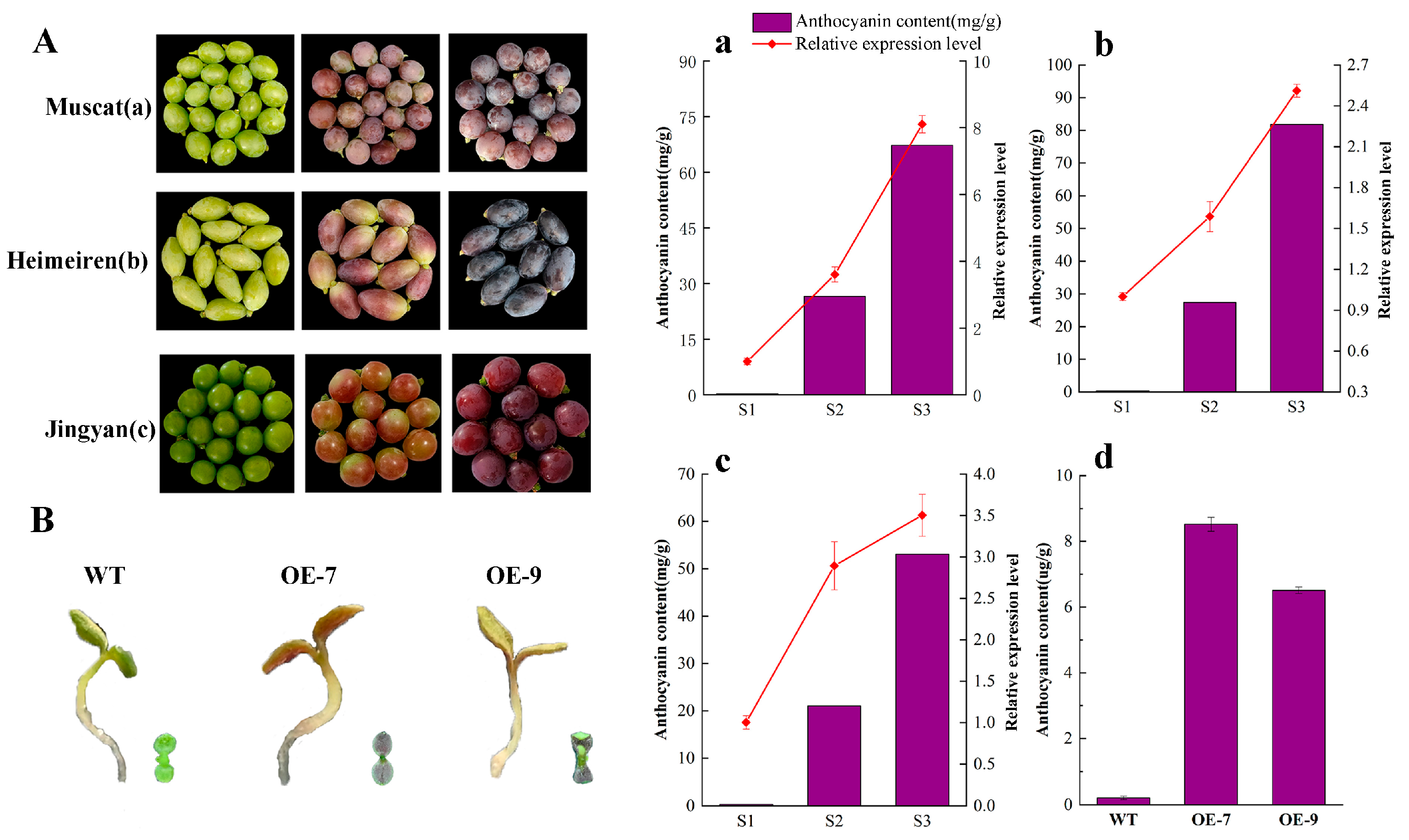
2.6. Expression of VvbHLH137 Correlates with Anthocyanin Accumulation in Grape Peels
2.7. Over-Expression of VvbHLH137 Increases Anthocyanin Accumulation in Arabidopsis thaliana
2.8. VvbHLH137 Directly Interacts with VvMYBs
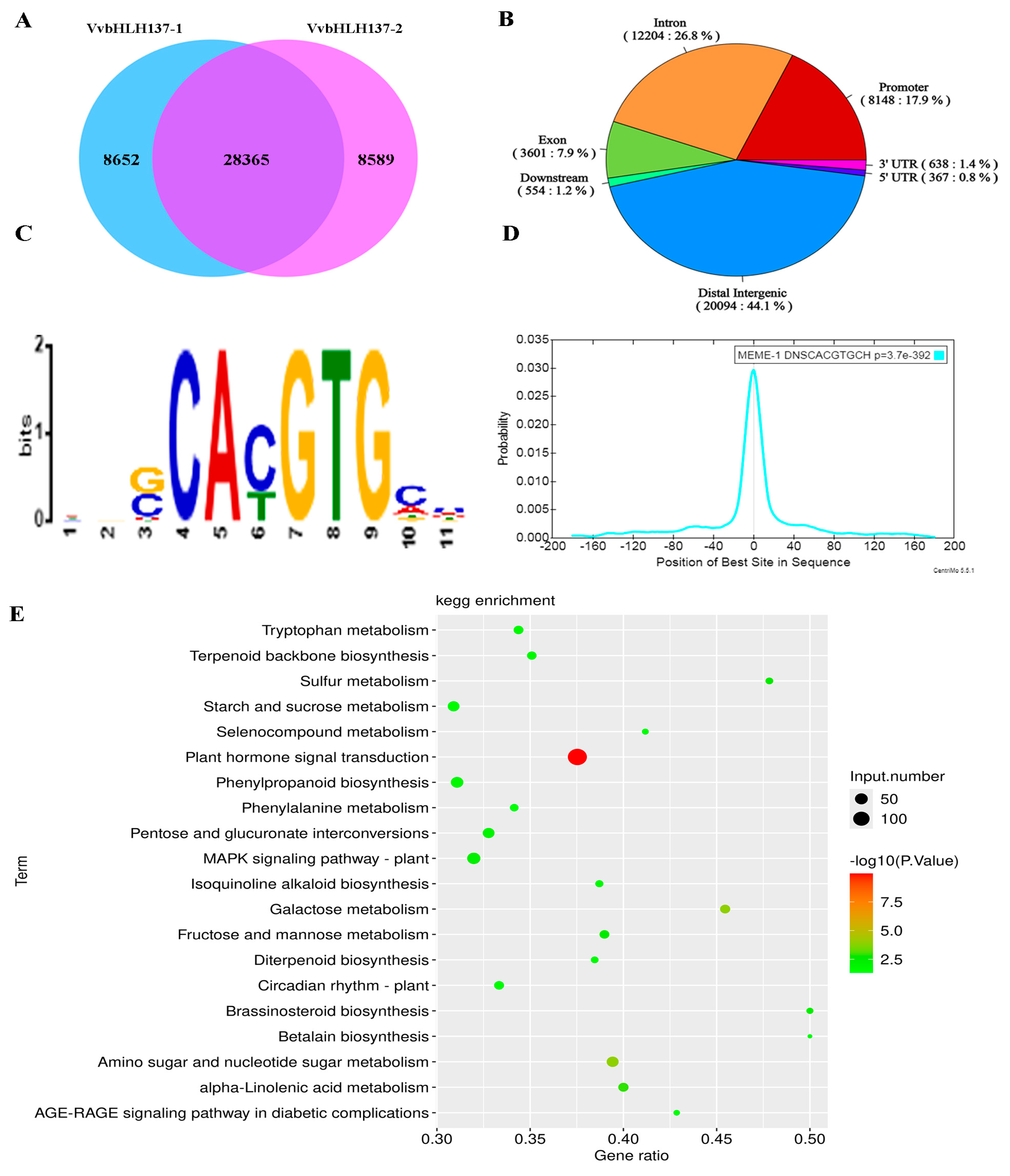
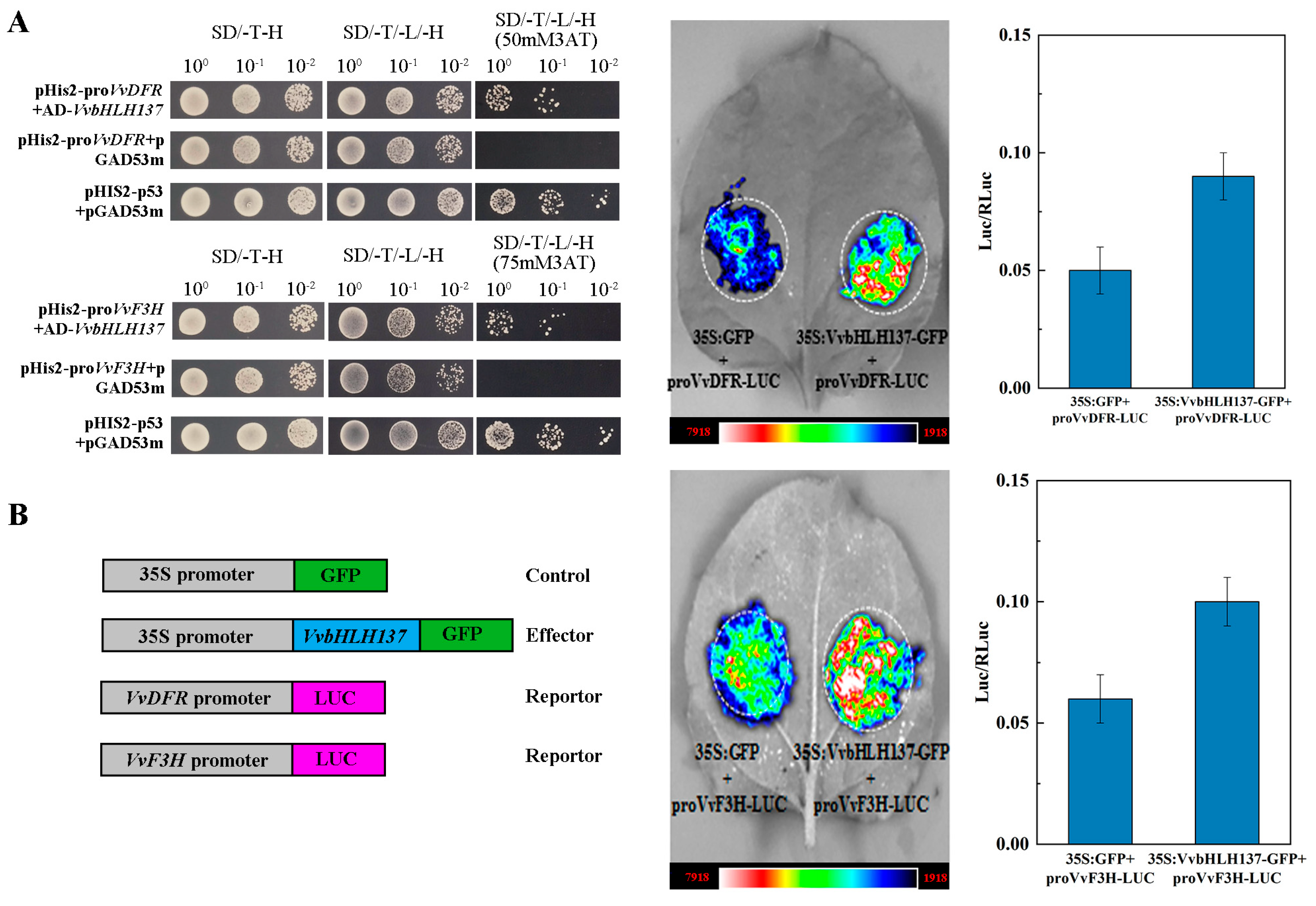
2.9. Detection and Verification of the Direct Downstream Genes of VvbHLH137
3. Discussion
4. Materials and Methods
4.1. Plants
4.2. Anthocyanin Content Determination
4.3. RNA Isolation and Sequencing
4.4. Quantitative Real-Time PCR (qRT-PCR)
4.5. Stable Transformation of Arabidopsis
4.6. Yeast Two-Hybrid (Y2H) Assays
4.7. Luciferase Complementation Assays (LCAs)
4.8. DNA Affinity Purification Sequencing (DAP-Seq)
4.9. Yeast One-Hybrid (Y1H) Assays
4.10. Transient Luciferase Expression Assays in Tobacco
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, Y.-M.; Sun, X.-R.; You, J.-L.; Li, X.-J.; Li, B.; Xie, Z.-S. Transcriptional Analysis of the Early Ripening of ‘Summer Black’ Grape in Response to Abscisic Acid Treatment. Sci. Hortic. 2022, 299, 111054. [Google Scholar] [CrossRef]
- Li, H.; Bai, Y.; Yang, Y.; Zheng, H.; Xu, X.; Li, H.; Wang, W.; Tao, J. Transcriptomic Analyses Reveal Light-Regulated Anthocyanin Accumulation in ‘ZhongShan-HongYu’ Grape Berries. Sci. Hortic. 2023, 309, 111669. [Google Scholar] [CrossRef]
- Jaakola, L. New Insights into the Regulation of Anthocyanin Biosynthesis in Fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef]
- Sun, T.; Xu, L.; Sun, H.; Yue, Q.; Zhai, H.; Yao, Y. VvVHP1; 2 Is Transcriptionally Activated by VvMYBA1 and Promotes Anthocyanin Accumulation of Grape Berry Skins via Glucose Signal. Front. Plant Sci. 2017, 8, 1811. [Google Scholar] [CrossRef] [PubMed]
- Mu, Z.; Yang, Y.; Yusuyin, A.; Xu, Y.; Yuan, H.; Liu, C. Transcriptome Analysis Provides Insights into Anthocyanin Synthesis in Blueberry. Horticulturae 2023, 9, 1036. [Google Scholar] [CrossRef]
- Fu, Z.; Shang, H.; Jiang, H.; Gao, J.; Dong, X.; Wang, H.; Li, Y.; Wang, L.; Zhang, J.; Shu, Q.; et al. Systematic Identification of the Light-Quality Responding Anthocyanin Synthesis-Related Transcripts in Petunia Petals. Hortic. Plant J. 2020, 6, 428–438. [Google Scholar] [CrossRef]
- Gao, Z.; Chen, Z.; Wang, J.; Liu, W. Integrated Transcriptome and Metabolome Analysis Revealed Mechanism Underlying Anthocyanin Biosynthesis During Flower Color Formation in Lagerstroemia Indica. Horticulturae 2024, 10, 1229. [Google Scholar] [CrossRef]
- Song, Z.; Pang, Q.; Lu, S.; Yu, L.; Pervaiz, T.; Fu, W.; Jia, H.; Fang, J. Transcriptomic and Metabolmic Approaches to Counter the Effect of Botrytis Cinerea in Grape Berry with the Application of Nitric Oxide. Sci. Hortic. 2022, 296, 110901. [Google Scholar] [CrossRef]
- Stommel, J.R.; Lightbourn, G.J.; Winkel, B.S.; Griesbach, R.J. Transcription Factor Families Regulate the Anthocyanin Biosynthetic Pathway in Capsicum Annuum. J. Am. Soc. Hortic. Sci. 2009, 134, 244–251. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the Anthocyanin Biosynthetic Pathway by the TTG1/bHLH/Myb Transcriptional Complex in Arabidopsis Seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thévenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and Robustness of the Flavonoid Transcriptional Regulatory Network Revealed by Comprehensive Analyses of MYB–bHLH–WDR Complexes and Their Targets in Arabidopsis Seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.R.; Lee, E.; Bogs, J.; McDavid, D.A.J.; Thomas, M.R.; Robinson, S.P. White Grapes Arose through the Mutation of Two Similar and Adjacent Regulatory Genes. Plant J. 2007, 49, 772–785. [Google Scholar] [CrossRef] [PubMed]
- Azuma, A.; Udo, Y.; Sato, A.; Mitani, N.; Kono, A.; Ban, Y.; Yakushiji, H.; Koshita, Y.; Kobayashi, S. Haplotype Composition at the Color Locus Is a Major Genetic Determinant of Skin Color Variation in Vitis × labruscana Grapes. Theor. Appl. Genet. 2011, 122, 1427–1438. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Su, L.; Gao, H.; Jiang, X.; Wu, X.; Li, Y.; Zhang, Q.; Wang, Y.; Ren, F. Genome-Wide Characterization of bHLH Genes in Grape and Analysis of Their Potential Relevance to Abiotic Stress Tolerance and Secondary Metabolite Biosynthesis. Front. Plant Sci. 2018, 9, 64. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, Y.; Kim, S.-U.; Chen, Z.; Nie, G.; Cheng, S.; Ye, J.; Xu, F. Genome-Wide Identification and Characterization of bHLH Family Genes from Ginkgo Biloba. Sci. Rep. 2020, 10, 13723. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, J.; Chen, Y.; Tang, H.; Wang, Y.; He, Y.; Ou, Y.; Sun, X.; Wang, S.; Yao, Y. Tomato SlAN11 Regulates Flavonoid Biosynthesis and Seed Dormancy by Interaction with bHLH Proteins but Not with MYB Proteins. Hortic. Res. 2018, 5, 27. [Google Scholar] [CrossRef]
- An, W.; Sun, Y.; Gao, Z.; Liu, X.; Guo, Q.; Sun, S.; Zhang, M.; Han, Y.; Irfan, M.; Chen, L.; et al. LvbHLH13 Regulates Anthocyanin Biosynthesis by Activating the LvMYB5 Promoter in Lily (Lilium ‘Viviana’). Horticulturae 2024, 10, 926. [Google Scholar] [CrossRef]
- Jin, W.; Wang, H.; Li, M.; Wang, J.; Yang, Y.; Zhang, X.; Yan, G.; Zhang, H.; Liu, J.; Zhang, K. The R2R3 MYB Transcription Factor PavMYB10.1 Involves in Anthocyanin Biosynthesis and Determines Fruit Skin Colour in Sweet Cherry (Prunus avium L.). Plant Biotechnol. J. 2016, 14, 2120–2133. [Google Scholar] [CrossRef]
- Ludwig, S.R.; Habera, L.F.; Dellaportat, S.L.; Wessler, S.R. Lc, a Member of the Maize R Gene Family Responsible for Tissue-Specific Anthocyanin Production, Encodes a Protein Similar to Transcriptional Activators and Contains the Myc-Homology Region. Proc. Natl. Acad. Sci. USA 1989, 86, 7092–7096. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red Colouration in Apple Fruit Is Due to the Activity of the MYB Transcription Factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef]
- Xie, X.; Li, S.; Zhang, R.; Zhao, J.; Chen, Y.; Zhao, Q.; Yao, Y.; You, C.; Zhang, X.; Hao, Y. The bHLH Transcription Factor MdbHLH3 Promotes Anthocyanin Accumulation and Fruit Colouration in Response to Low Temperature in Apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tang, W.; Hu, Y.; Zhang, Y.; Sun, J.; Guo, X.; Lu, H.; Yang, Y.; Fang, C.; Niu, X.; et al. A MYB/bHLH Complex Regulates Tissue-specific Anthocyanin Biosynthesis in the Inner Pericarp of Red-centered Kiwifruit Actinidia Chinensis Cv. Hongyang. Plant J. 2019, 99, 359–378. [Google Scholar] [CrossRef]
- Qi, Y.; Zhou, L.; Han, L.; Zou, H.; Miao, K.; Wang, Y. PsbHLH1, a Novel Transcription Factor Involved in Regulating Anthocyanin Biosynthesis in Tree Peony (Paeonia suffruticosa). Plant Physiol. Biochem. 2020, 154, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Lin, C.; Chen, B.; Lin, Y.; Su, H.; Du, Y.; Zhang, H.; Zhou, H.; Ji, R.; Zhang, L. A Light Responsive Transcription Factor CsbHLH89 Positively Regulates Anthocyanidin Synthesis in Tea (Camellia sinensis). Sci. Hortic. 2024, 327, 112784. [Google Scholar] [CrossRef]
- Ravaglia, D.; Espley, R.V.; Henry-Kirk, R.A.; Andreotti, C.; Ziosi, V.; Hellens, R.P.; Costa, G.; Allan, A.C. Transcriptional Regulation of Flavonoid Biosynthesis in Nectarine (Prunus persica) by a Set of R2R3 MYB Transcription Factors. BMC Plant Biol. 2013, 13, 68. [Google Scholar] [CrossRef] [PubMed]
- Lin-Wang, K.; McGhie, T.K.; Wang, M.; Liu, Y.; Warren, B.; Storey, R.; Espley, R.V.; Allan, A.C. Engineering the Anthocyanin Regulatory Complex of Strawberry (Fragaria vesca). Front. Plant Sci. 2014, 5, 651. [Google Scholar] [CrossRef]
- Liu, X.-F.; Yin, X.-R.; Allan, A.C.; Lin-Wang, K.; Shi, Y.-N.; Huang, Y.-J.; Ferguson, I.B.; Xu, C.-J.; Chen, K.-S. The Role of MrbHLH1 and MrMYB1 in Regulating Anthocyanin Biosynthetic Genes in Tobacco and Chinese Bayberry (Myrica rubra) during Anthocyanin Biosynthesis. Plant Cell Tissue Organ Cult. PCTOC 2013, 115, 285–298. [Google Scholar] [CrossRef]
- Li, X.; Cao, L.; Jiao, B.; Yang, H.; Ma, C.; Liang, Y. The bHLH Transcription Factor AcB2 Regulates Anthocyanin Biosynthesis in Onion (Allium cepa L.). Hortic. Res. 2022, 9, uhac128. [Google Scholar] [CrossRef]
- Zhao, R.; Song, X.; Yang, N.; Chen, L.; Xiang, L.; Liu, X.-Q.; Zhao, K. Expression of the Subgroup IIIf bHLH Transcription Factor CpbHLH1 from Chimonanthus praecox (L.) in Transgenic Model Plants Inhibits Anthocyanin Accumulation. Plant Cell Rep. 2020, 39, 891–907. [Google Scholar] [CrossRef]
- Hu, R.; Zhu, M.; Chen, S.; Li, C.; Zhang, Q.; Gao, L.; Liu, X.; Shen, S.; Fu, F.; Xu, X.; et al. BnbHLH92a Negatively Regulates Anthocyanin and Proanthocyanidin Biosynthesis in Brassica napus. Crop J. 2023, 11, 374–385. [Google Scholar] [CrossRef]
- Zhao, P.; Li, X.; Jia, J.; Yuan, G.; Chen, S.; Qi, D.; Cheng, L.; Liu, G. bHLH92 from Sheepgrass Acts as a Negative Regulator of Anthocyanin/Proanthocyandin Accumulation and Influences Seed Dormancy. J. Exp. Bot. 2019, 70, 269–284. [Google Scholar] [CrossRef]
- Hichri, I.; Heppel, S.C.; Pillet, J.; Léon, C.; Czemmel, S.; Delrot, S.; Lauvergeat, V.; Bogs, J. The Basic Helix-Loop-Helix Transcription Factor MYC1 Is Involved in the Regulation of the Flavonoid Biosynthesis Pathway in Grapevine. Mol. Plant 2010, 3, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhu, H.; Chen, D.; Li, Z.; Peng, R.; Yao, Q. A Grape bHLH Transcription Factor Gene, VvbHLH1, Increases the Accumulation of Flavonoids and Enhances Salt and Drought Tolerance in Transgenic Arabidopsis Thaliana. Plant Cell Tissue Organ Cult. PCTOC 2016, 125, 387–398. [Google Scholar] [CrossRef]
- Ge, M. Integrated Transcriptomic and Metabolic Analyses Unveil Anthocyanins Biosynthesis Metabolism in Three Different Color Cultivars of Grape (Vitis vinifera L.). Sci. Hortic. 2022, 305, 111418. [Google Scholar] [CrossRef]
- Wang, Q.; Zheng, H.; Gao, S.; Li, H.; Tao, J. Transcriptomic Analysis of Berry Development and a Corresponding Analysis of Anthocyanin Biosynthesis in Teinturier Grape. J. Plant Interact. 2019, 14, 617–629. [Google Scholar] [CrossRef]
- Poudel, P.R.; Azuma, A.; Kobayashi, S.; Koyama, K.; Goto-Yamamoto, N. VvMYBAs Induce Expression of a Series of Anthocyanin Biosynthetic Pathway Genes in Red Grapes (Vitis vinifera L.). Sci. Hortic. 2021, 283, 110121. [Google Scholar] [CrossRef]
- Dimitrovska, M.; Bocevska, M.; Dimitrovski, D.; Murkovic, M. Anthocyanin Composition of Vranec, Cabernet Sauvignon, Merlot and Pinot Noir Grapes as Indicator of Their Varietal Differentiation. Eur. Food Res. Technol. 2011, 232, 591–600. [Google Scholar] [CrossRef]
- Jiang, Q.; Ye, J.; Zhu, K.; Wu, F.; Chai, L.; Xu, Q.; Deng, X. Transcriptome and Co-Expression Network Analyses Provide Insights into Fruit Shading That Enhances Carotenoid Accumulation in Pomelo (Citrus grandis). Hortic. Plant J. 2022, 8, 423–434. [Google Scholar] [CrossRef]
- Bai, C.; Wu, C.; Ma, L.; Fu, A.; Zheng, Y.; Han, J.; Li, C.; Yuan, S.; Zheng, S.; Gao, L.; et al. Transcriptomics and Metabolomics Analyses Provide Insights into Postharvest Ripening and Senescence of Tomato Fruit under Low Temperature. Hortic. Plant J. 2023, 9, 109–121. [Google Scholar] [CrossRef]
- Zhang, Z.; Xing, Y.; Ramakrishnan, M.; Chen, C.; Xie, F.; Hua, Q.; Chen, J.; Zhang, R.; Zhao, J.; Hu, G.; et al. Transcriptomics-based Identification and Characterization of Genes Related to Sugar Metabolism in ‘Hongshuijing’ Pitaya. Hortic. Plant J. 2022, 8, 450–460. [Google Scholar] [CrossRef]
- Lian, H.; Wang, S. VvMYB15 and VvWRKY40 Positively Co-Regulated Anthocyanin Biosynthesis in Grape Berries in Response to Root Restriction. Front. Plant Sci. 2021, 12, 789002. [Google Scholar]
- Hu, X.; Liang, Z.; Sun, T.; Huang, L.; Wang, Y.; Chan, Z.; Xiang, L. The R2R3-MYB Transcriptional Repressor TgMYB4 Negatively Regulates Anthocyanin Biosynthesis in Tulips (Tulipa gesneriana L.). Int. J. Mol. Sci. 2024, 25, 563. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; De Vos, C.H.R.; Wein, M.; Sun, Z.; Greco, R.; Kroon, A.; Mol, J.N.M.; O’Connell, A.P. The Strawberry FaMYB1 Transcription Factor Suppresses Anthocyanin and Flavonol Accumulation in Transgenic Tobacco. Plant J. 2001, 28, 319–332. [Google Scholar] [CrossRef]
- Dong, Q.; Zhao, H.; Huang, Y.; Chen, Y.; Wan, M.; Zeng, Z.; Yao, P.; Li, C.; Wang, X.; Chen, H.; et al. FtMYB18 Acts as a Negative Regulator of Anthocyanin/Proanthocyanidin Biosynthesis in Tartary Buckwheat. Plant Mol. Biol. 2020, 104, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Lin-Wang, K.; Wang, F.; Espley, R.V.; Ren, F.; Zhao, J.; Ogutu, C.; He, H.; Jiang, Q.; Allan, A.C.; et al. Activator-type R2R3-MYB Genes Induce a Repressor-type R2R3-MYB Gene to Balance Anthocyanin and Proanthocyanidin Accumulation. New Phytol. 2019, 221, 1919–1934. [Google Scholar] [CrossRef]
- Xu, H.; Wang, N.; Liu, J.; Qu, C.; Wang, Y.; Jiang, S.; Lu, N.; Wang, D.; Zhang, Z.; Chen, X. The Molecular Mechanism Underlying Anthocyanin Metabolism in Apple Using the MdMYB16 and MdbHLH33 Genes. Plant Mol. Biol. 2017, 94, 149–165. [Google Scholar] [CrossRef]
- Hu, Y.; Cheng, H.; Zhang, Y.; Zhang, J.; Niu, S.; Wang, X.; Li, W.; Zhang, J.; Yao, Y. The MdMYB16/MdMYB1-miR7125-MdCCR Module Regulates the Homeostasis between Anthocyanin and Lignin Biosynthesis during Light Induction in Apple. New Phytol. 2021, 231, 1105–1122. [Google Scholar] [CrossRef]
- Ding, T.; Zhang, R.; Zhang, H.; Zhou, Z.; Liu, C.; Wu, M.; Wang, H.; Dong, H.; Liu, J.; Yao, J.-L.; et al. Identification of Gene Co-Expression Networks and Key Genes Regulating Flavonoid Accumulation in Apple (Malus × domestica) Fruit Skin. Plant Sci. 2021, 304, 110747. [Google Scholar] [CrossRef]
- Ni, J.; Bai, S.; Zhao, Y.; Qian, M.; Tao, R.; Yin, L.; Gao, L.; Teng, Y. Ethylene Response Factors Pp4ERF24 and Pp12ERF96 Regulate Blue Light-Induced Anthocyanin Biosynthesis in ‘Red Zaosu’ Pear Fruits by Interacting with MYB114. Plant Mol. Biol. 2019, 99, 67–78. [Google Scholar] [CrossRef]
- An, J.-P.; Zhang, X.-W.; Bi, S.-Q.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. The ERF Transcription Factor MdERF38 Promotes Drought Stress-induced Anthocyanin Biosynthesis in Apple. Plant J. 2020, 101, 573–589. [Google Scholar] [CrossRef]
- Mo, R.; Han, G.; Zhu, Z.; Essemine, J.; Dong, Z.; Li, Y.; Deng, W.; Qu, M.; Zhang, C.; Yu, C. The Ethylene Response Factor ERF5 Regulates Anthocyanin Biosynthesis in ‘Zijin’ Mulberry Fruits by Interacting with MYBA and F3H Genes. Int. J. Mol. Sci. 2022, 23, 7615. [Google Scholar] [CrossRef]
- Alabd, A.; Ahmad, M.; Zhang, X.; Gao, Y.; Peng, L.; Zhang, L.; Ni, J.; Bai, S.; Teng, Y. Light-Responsive Transcription Factor PpWRKY44 Induces Anthocyanin Accumulation by Regulating PpMYB10 Expression in Pear. Hortic. Res. 2022, 9, uhac199. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY Transcription Factors in Plant Responses to Stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Mao, Z.; Jiang, H.; Wang, S.; Wang, Y.; Yu, L.; Zou, Q.; Liu, W.; Jiang, S.; Wang, N.; Zhang, Z.; et al. The MdHY5-MdWRKY41-MdMYB Transcription Factor Cascade Regulates the Anthocyanin and Proanthocyanidin Biosynthesis in Red-Fleshed Apple. Plant Sci. 2021, 306, 110848. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wu, J.; Hu, K.-D.; Wei, S.-W.; Sun, H.-Y.; Hu, L.-Y.; Han, Z.; Yao, G.-F.; Zhang, H. PyWRKY26 and PybHLH3 Cotargeted the PyMYB114 Promoter to Regulate Anthocyanin Biosynthesis and Transport in Red-Skinned Pears. Hortic. Res. 2020, 7, 37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, C.; Jiang, C.; Lin, H.; Zhao, Y.; Guo, Y. VvWRKY5 Positively Regulates Wounding-Induced Anthocyanin Accumulation in Grape by Interplaying with VvMYBA1 and Promoting Jasmonic Acid Biosynthesis. Hortic. Res. 2024, 11, uhae083. [Google Scholar] [CrossRef] [PubMed]
- An, J.-P.; Zhang, X.-W.; You, C.-X.; Bi, S.-Q.; Wang, X.-F.; Hao, Y.-J. MdWRKY40 Promotes Wounding-induced Anthocyanin Biosynthesis in Association with MdMYB1 and Undergoes MdBT2-mediated Degradation. New Phytol. 2019, 224, 380–395. [Google Scholar] [CrossRef]
- Yu, D.; Wei, W.; Fan, Z.; Chen, J.; You, Y.; Huang, W.; Zhan, J. VabHLH137 Promotes Proanthocyanidin and Anthocyanin Biosynthesis and Enhances Resistance to Colletotrichum gloeosporioides in Grapevine. Hortic. Res. 2023, 10, uhac261. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Fang, Y.; Wang, D.; Xiao, L.; Quan, M.; Qi, W.; Song, F.; Zhou, J.; Liu, X.; Qin, S.; Du, Q.; et al. Allelic Variation in Transcription Factor PtoWRKY68 Contributes to Drought Tolerance in Populus. Plant Physiol. 2023, 193, 736–755. [Google Scholar] [CrossRef] [PubMed]
- Vasimuddin, M.; Misra, S.; Li, H.; Aluru, S. Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems. In Proceedings of the 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS), Rio de Janeiro, Brazil, 20–24 May 2019; pp. 314–324. [Google Scholar]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-Based Analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Machanick, P.; Bailey, T.L. MEME-ChIP: Motif Analysis of Large DNA Datasets. Bioinformatics 2011, 27, 1696–1697. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, Z.; Zhang, Z.; Zhao, Y.; Xuan, L.; Chen, Z.; Yang, L. Transcription Factor VvbHLH137 Positively Regulates Anthocyanin Accumulation in Grape (Vitis vinifera). Plants 2025, 14, 871. https://doi.org/10.3390/plants14060871
Niu Z, Zhang Z, Zhao Y, Xuan L, Chen Z, Yang L. Transcription Factor VvbHLH137 Positively Regulates Anthocyanin Accumulation in Grape (Vitis vinifera). Plants. 2025; 14(6):871. https://doi.org/10.3390/plants14060871
Chicago/Turabian StyleNiu, Zaozhu, Zhichao Zhang, Yanzhuo Zhao, Lifeng Xuan, Zhan Chen, and Lili Yang. 2025. "Transcription Factor VvbHLH137 Positively Regulates Anthocyanin Accumulation in Grape (Vitis vinifera)" Plants 14, no. 6: 871. https://doi.org/10.3390/plants14060871
APA StyleNiu, Z., Zhang, Z., Zhao, Y., Xuan, L., Chen, Z., & Yang, L. (2025). Transcription Factor VvbHLH137 Positively Regulates Anthocyanin Accumulation in Grape (Vitis vinifera). Plants, 14(6), 871. https://doi.org/10.3390/plants14060871



