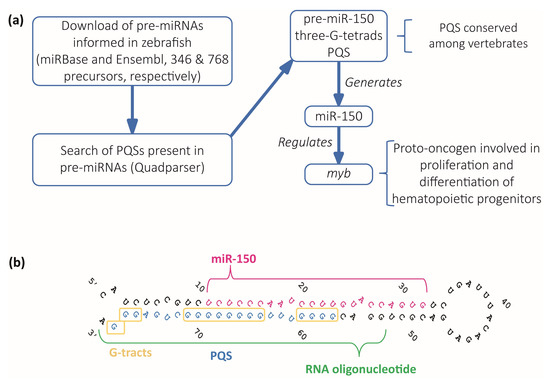
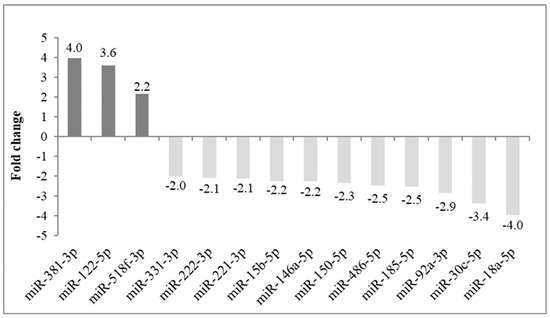
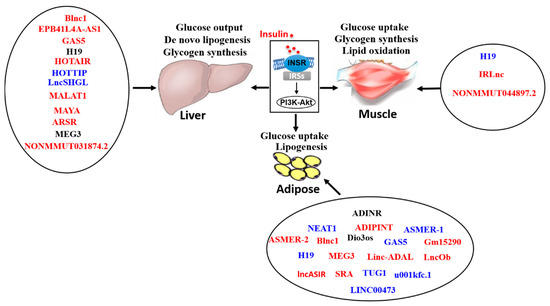
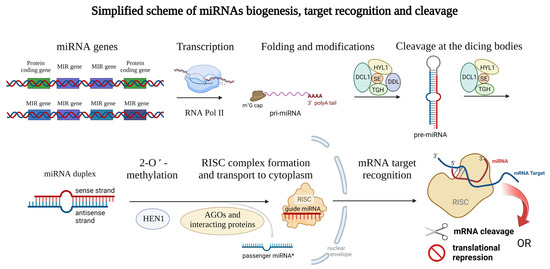
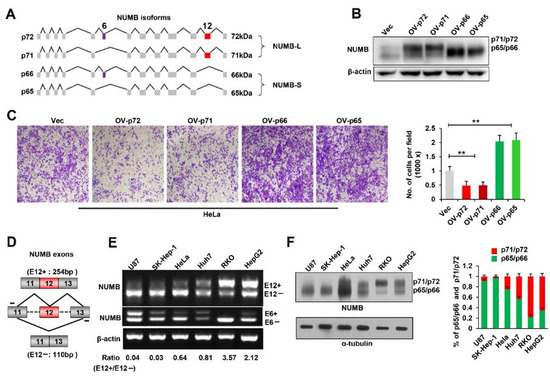
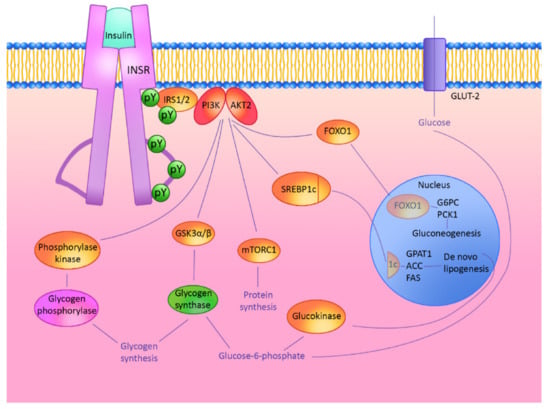
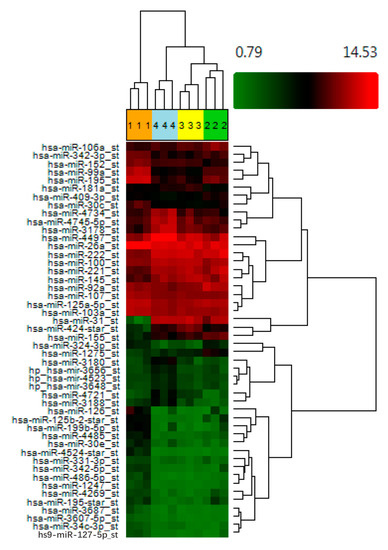
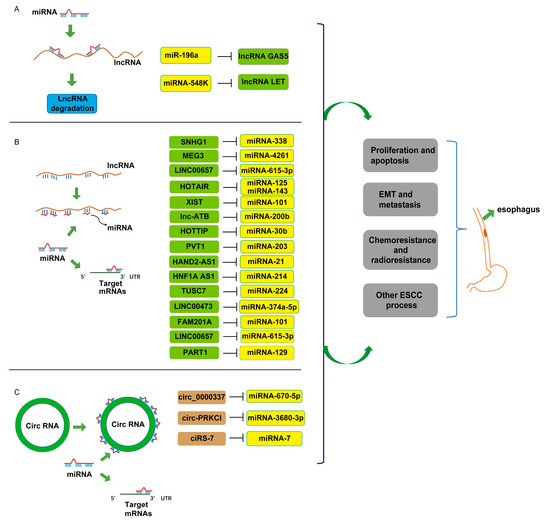
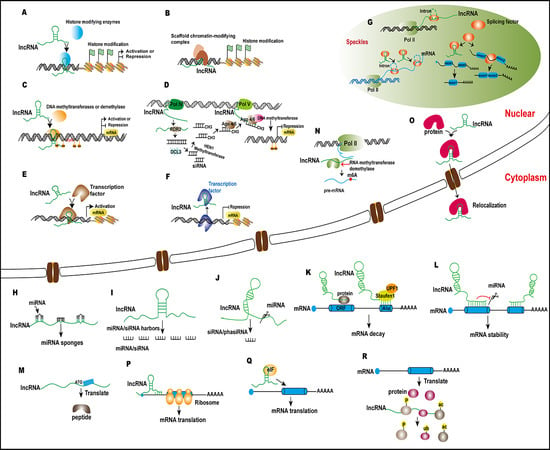
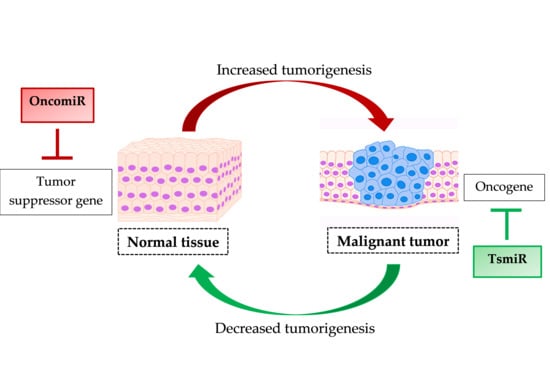
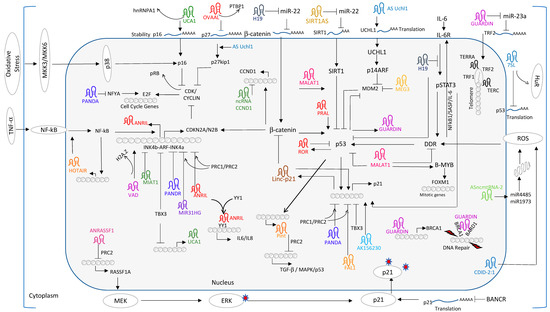
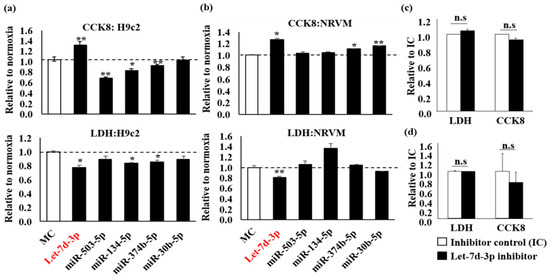
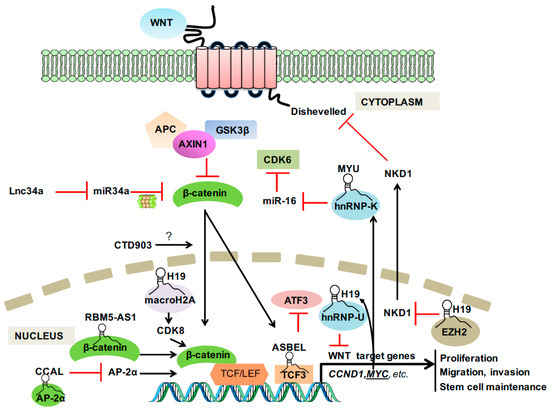
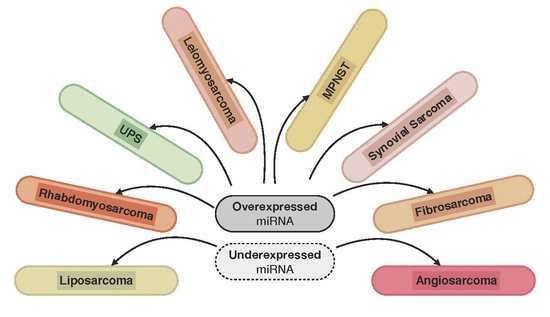
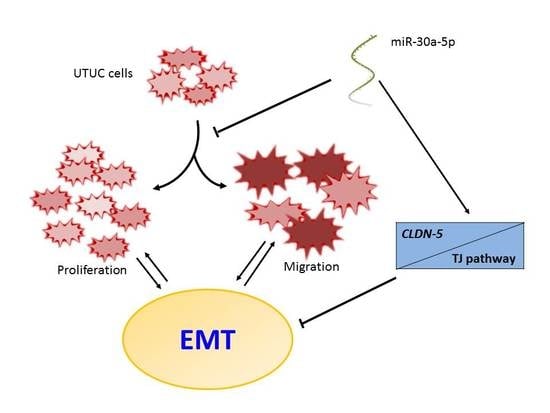
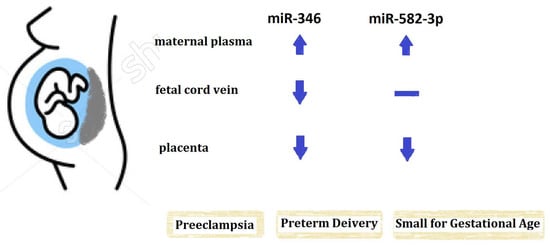
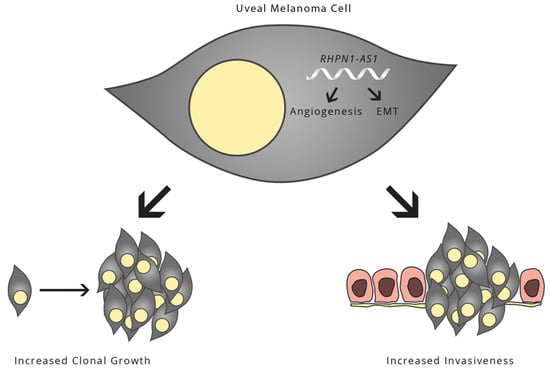
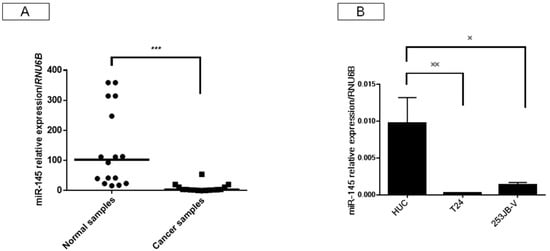
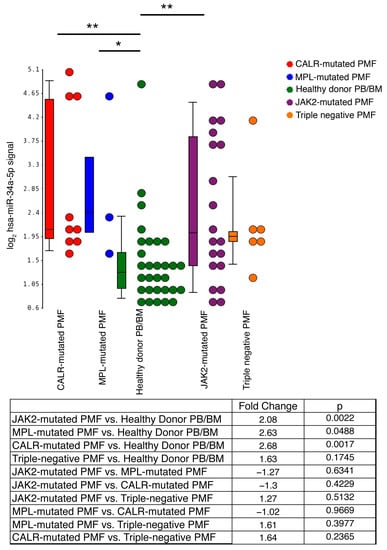
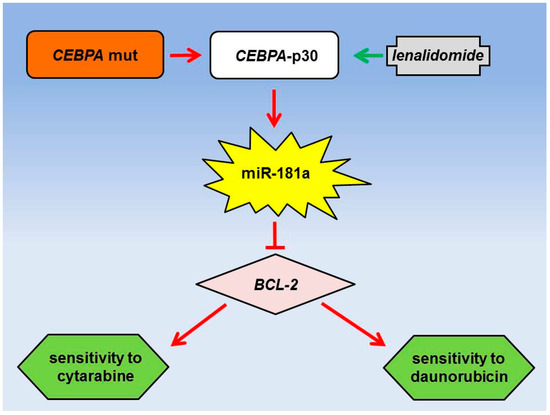
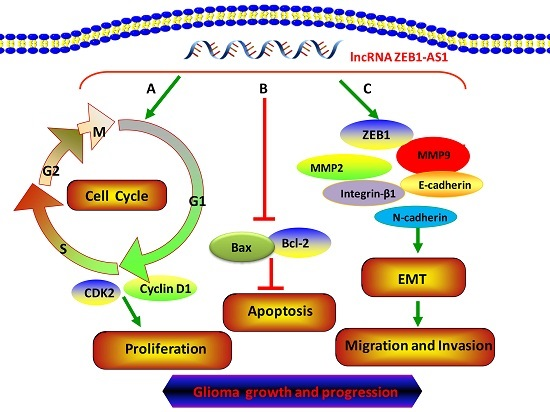
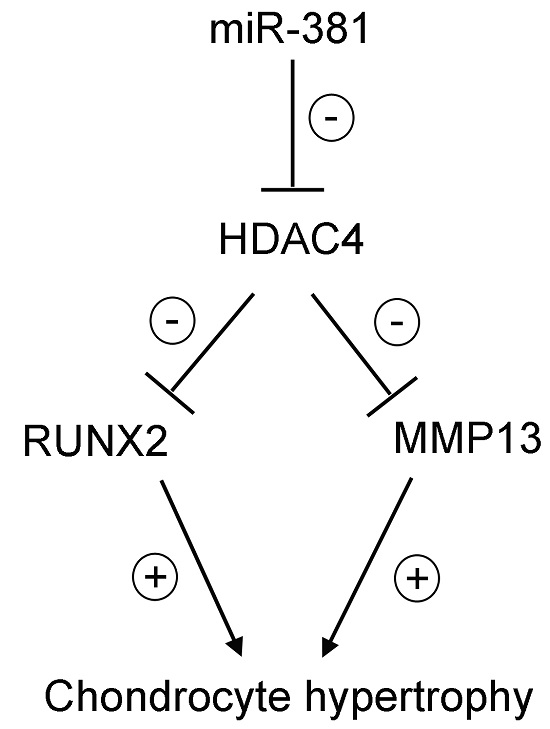
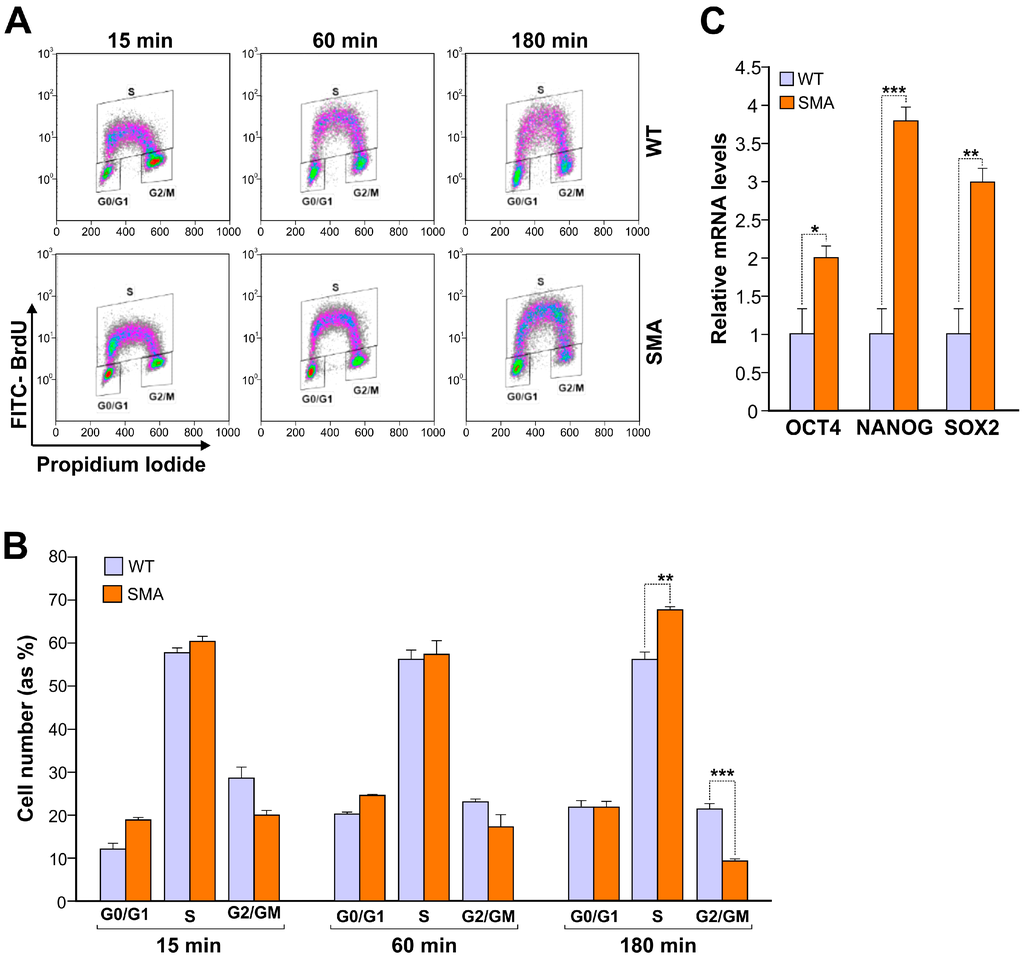
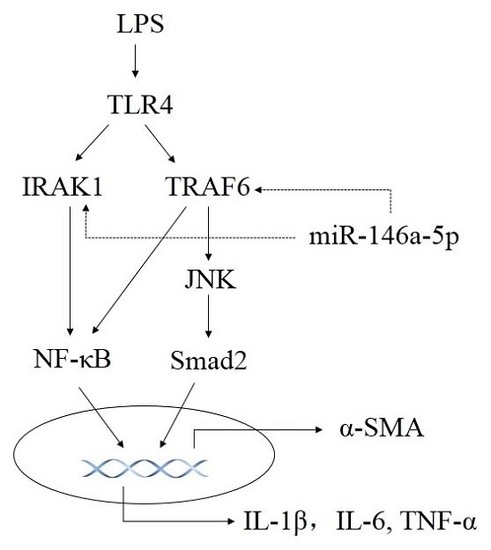
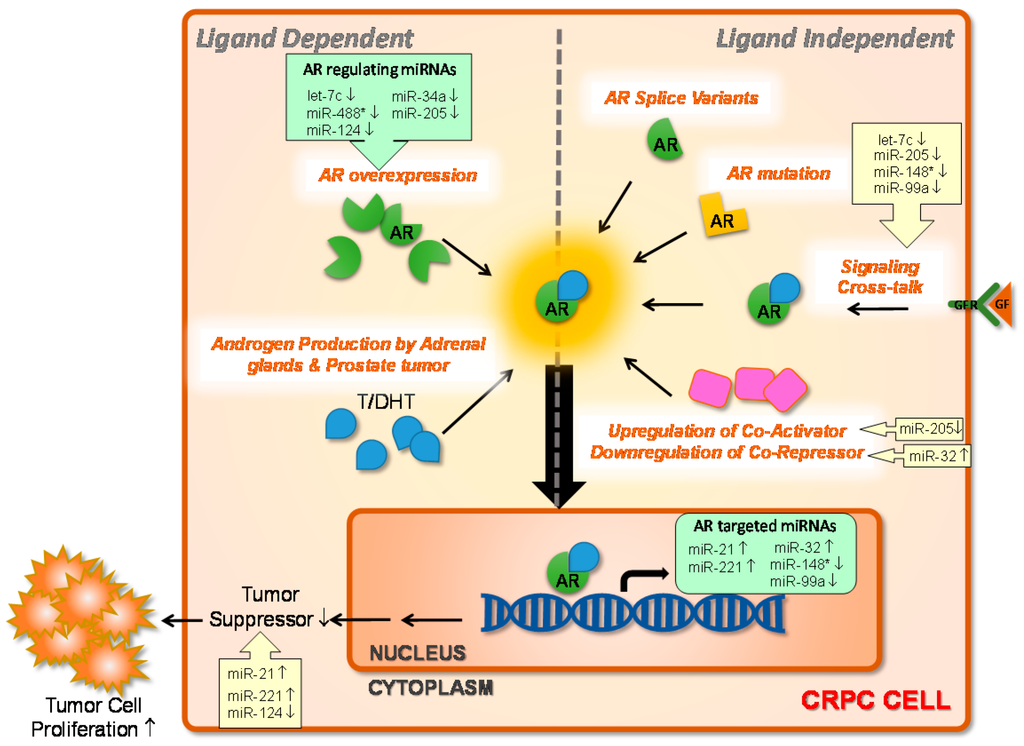
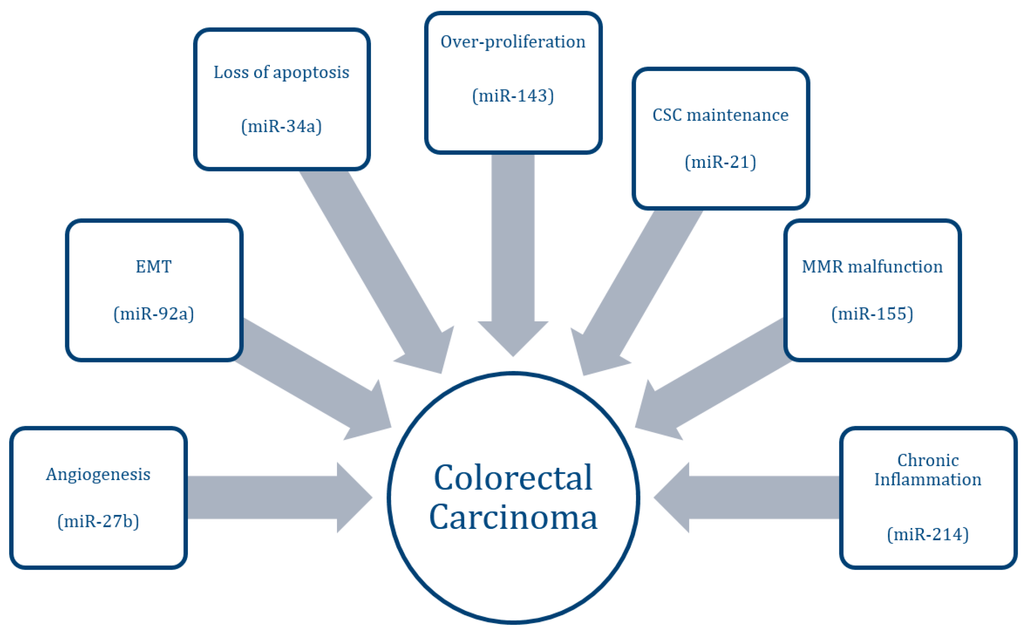
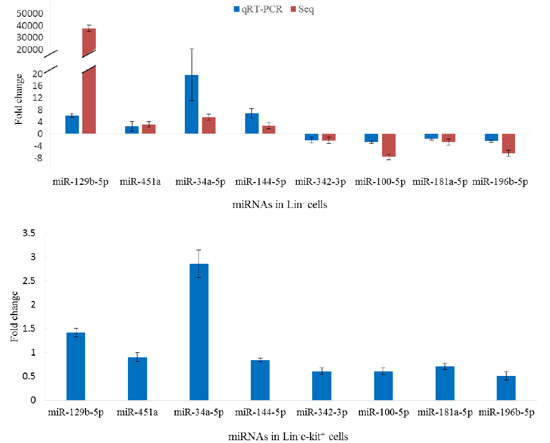
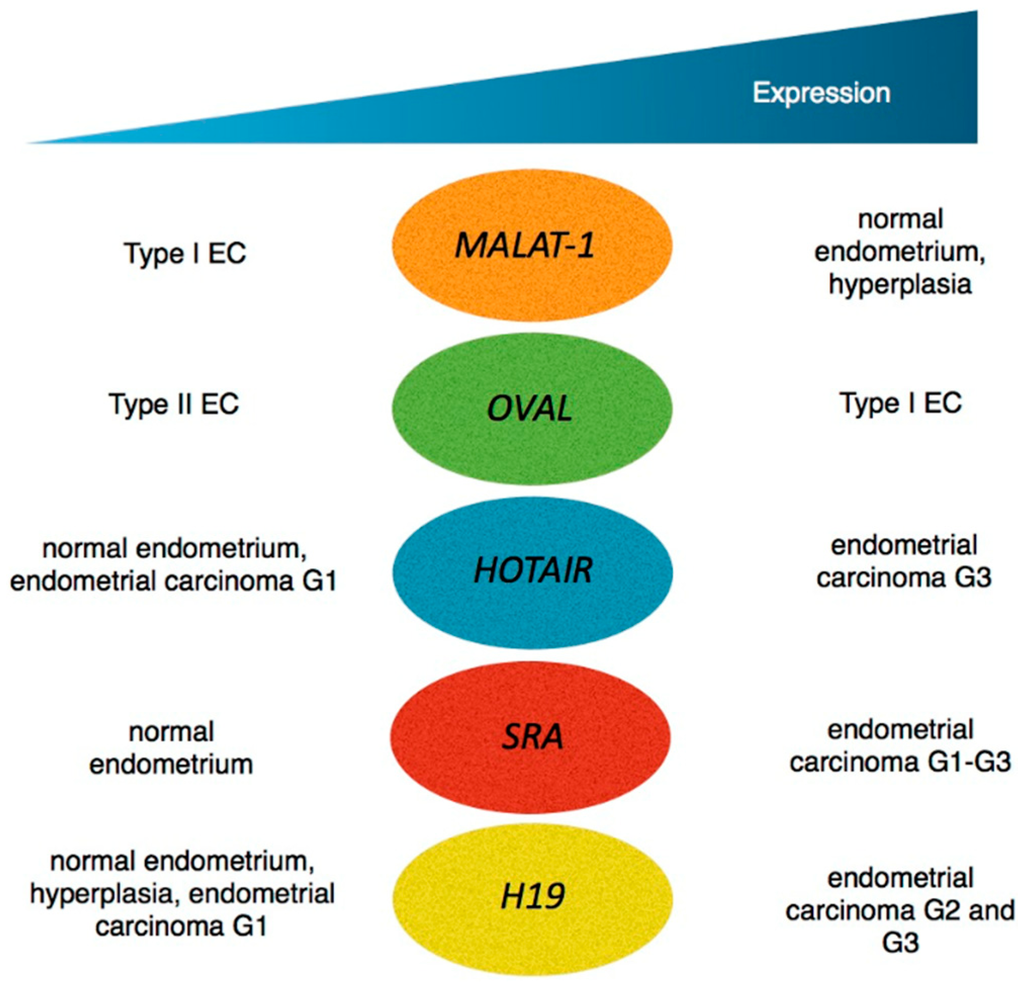
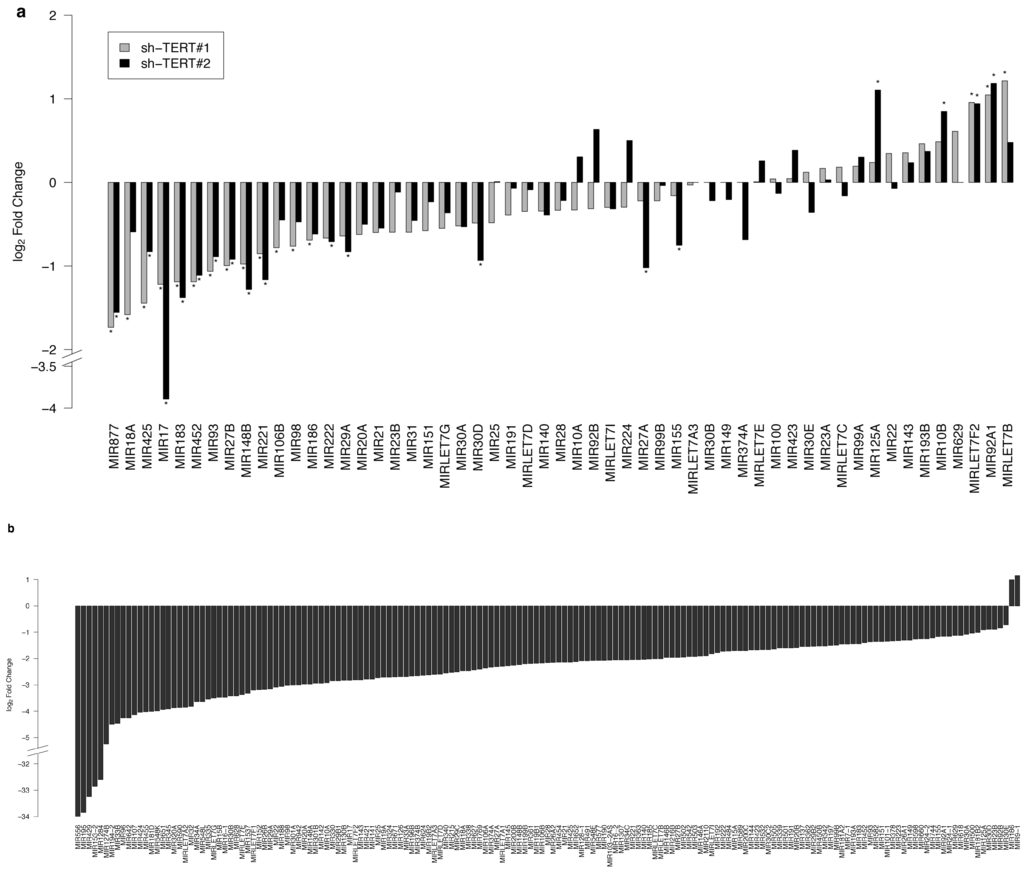
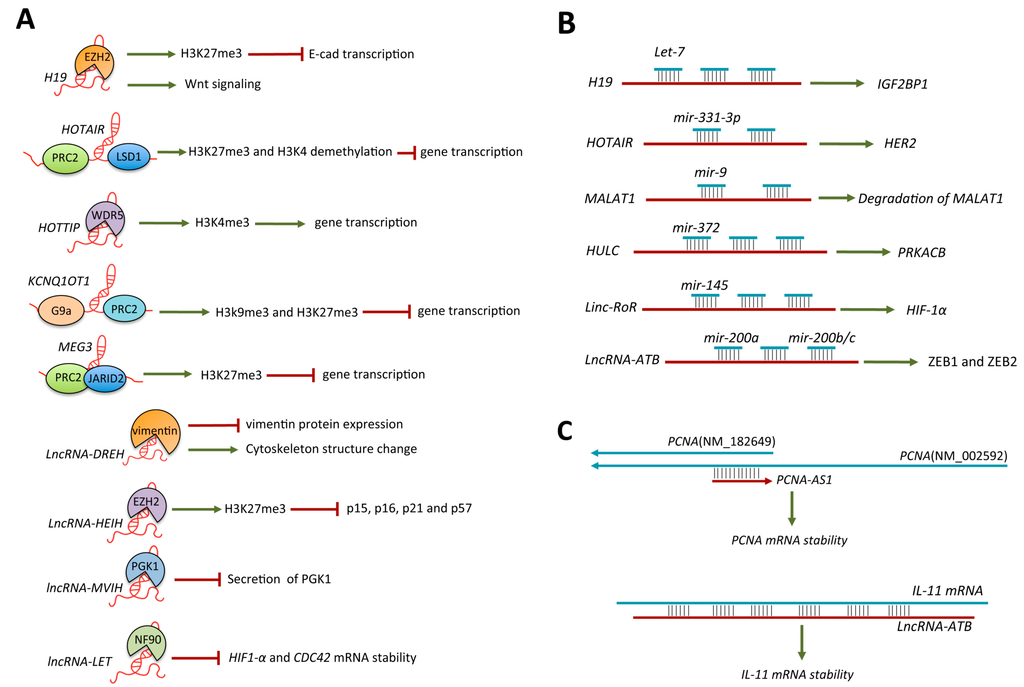
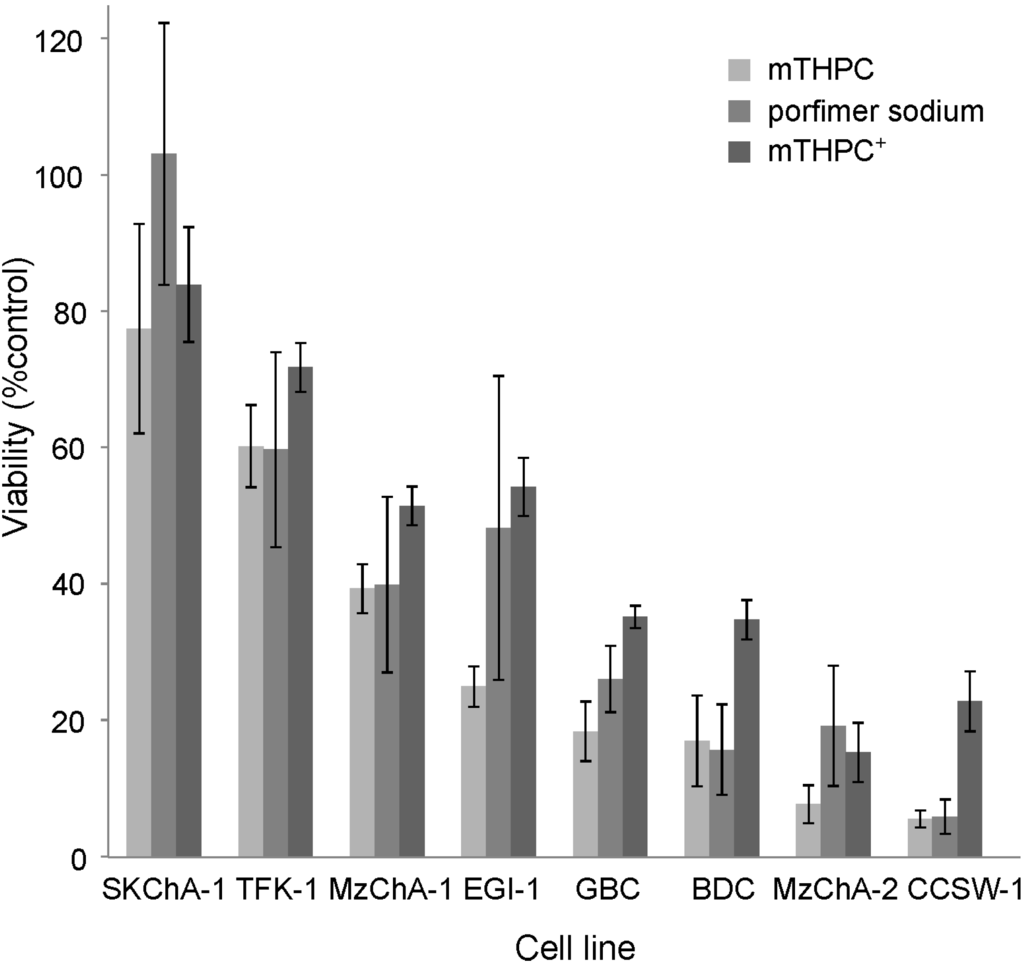
Regulation by Non-coding RNAs
A topical collection in International Journal of Molecular Sciences (ISSN 1422-0067). This collection belongs to the section "Molecular Biology".
Viewed by 1027089Editor
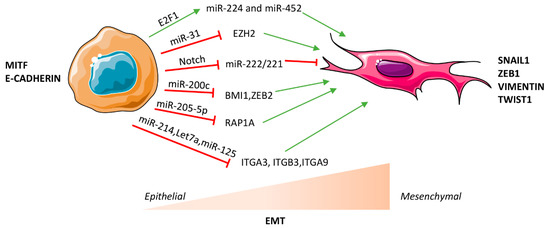
Interests: non-coding RNAs; microRNAs; cancer; inflammation; metabolism; gene expression; stem cells; epithelial-mesenchymal transition
* Non-Coding RNAs in Cancer: An Interview with Dr. Martin Pichler https://www.mdpi.com/1422-0067/17/4/605/htm
Topical Collection Information
Dear Colleagues,
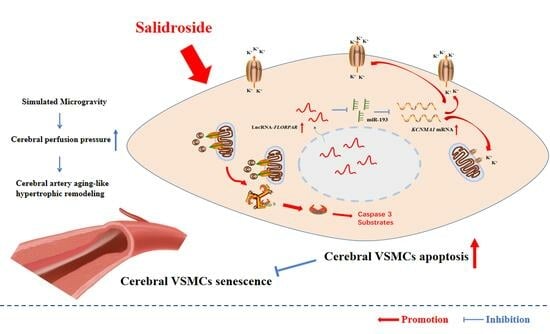
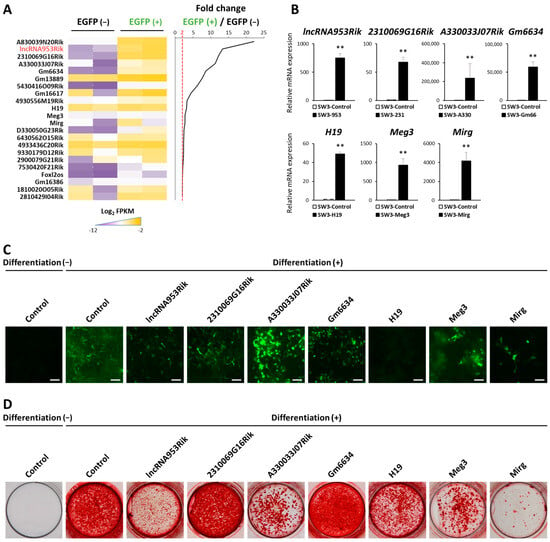
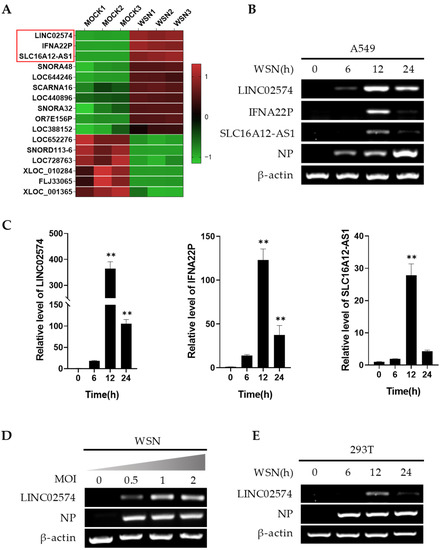
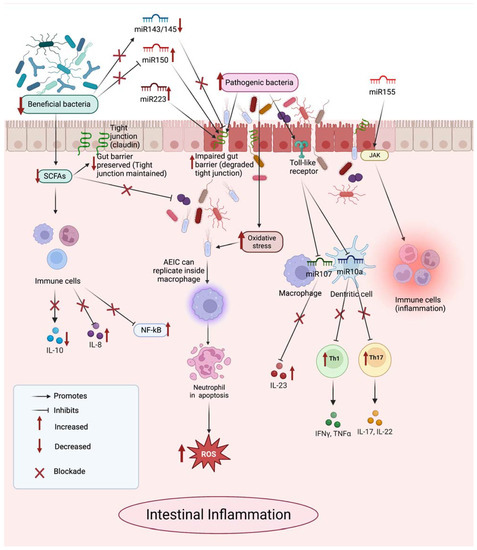
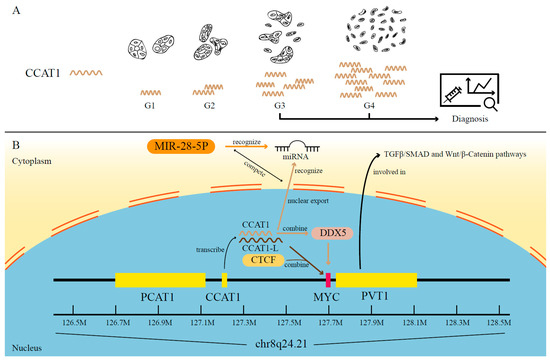
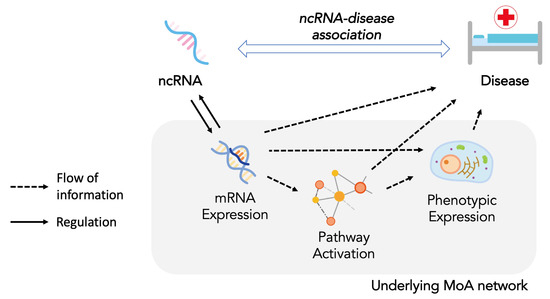
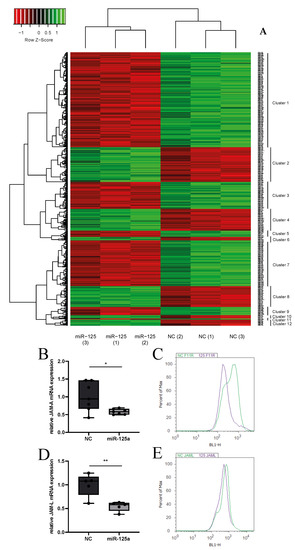
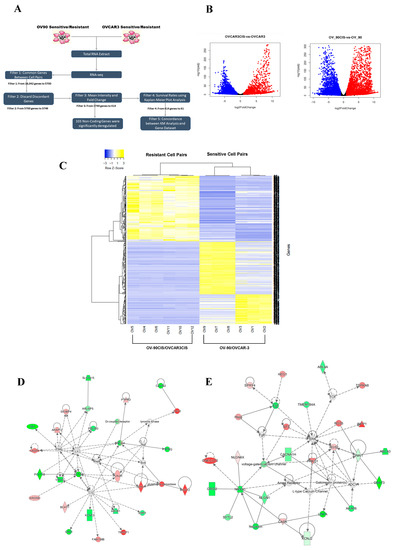
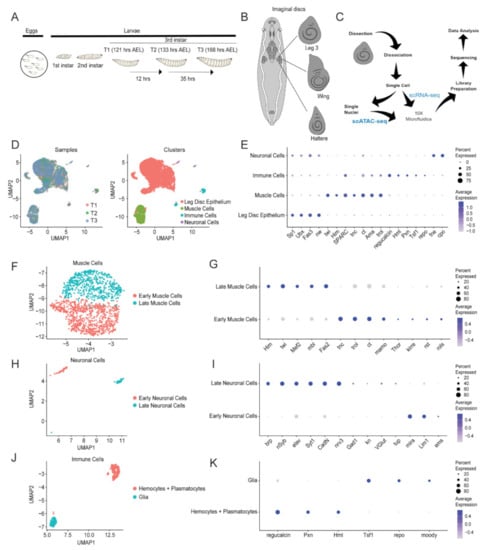
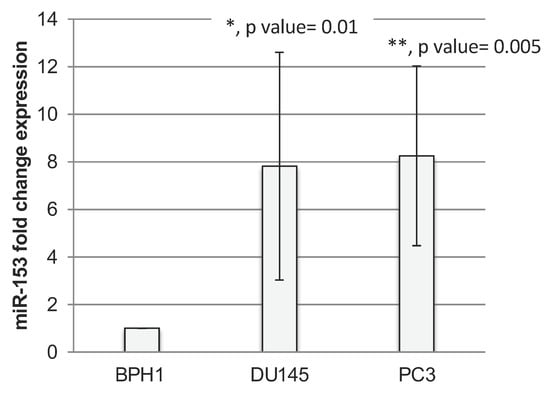
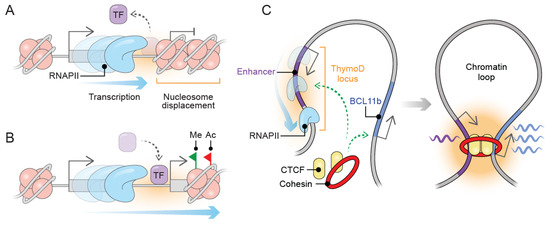
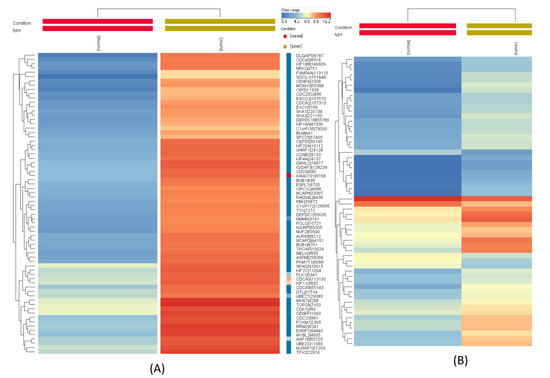
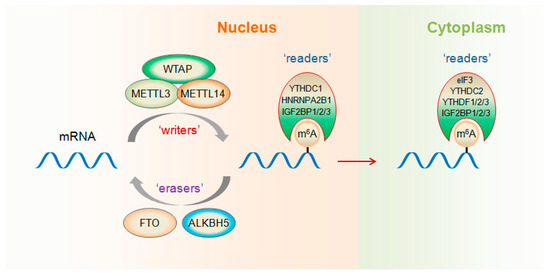
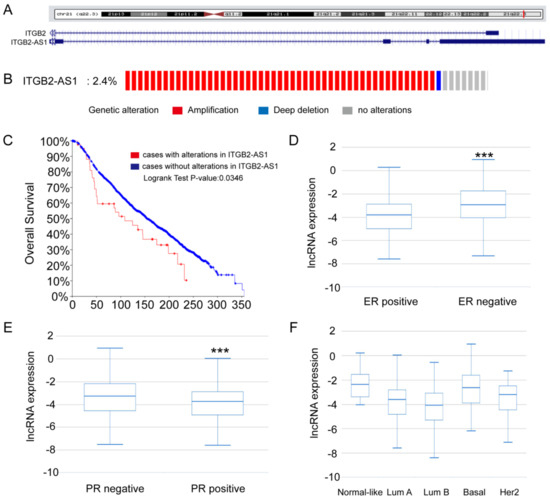
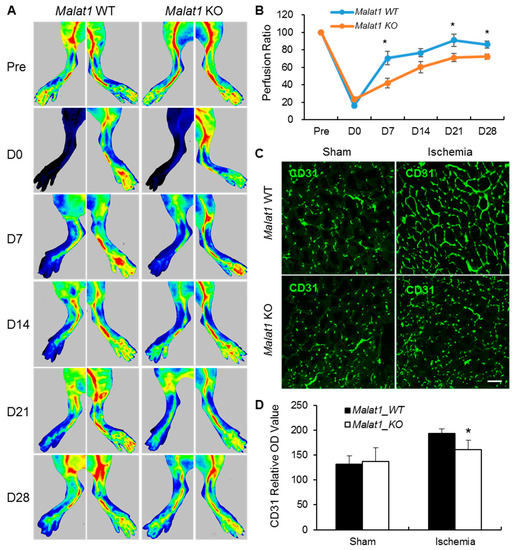
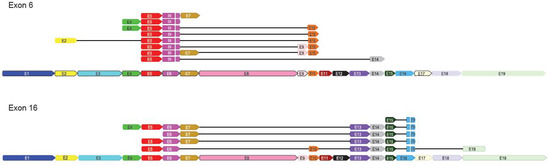
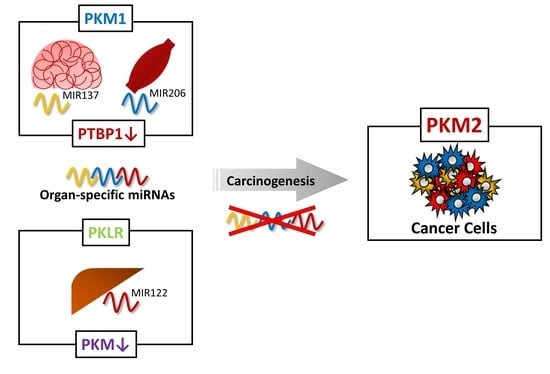
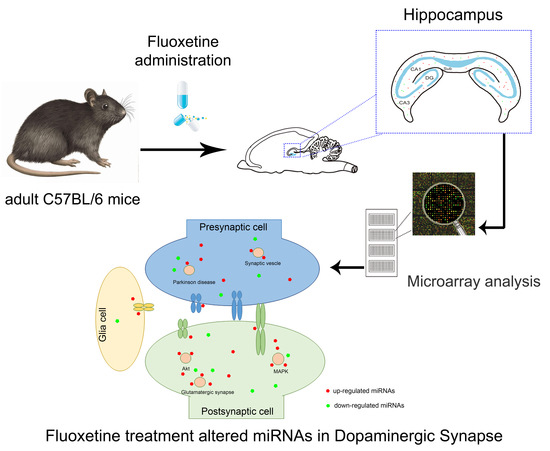
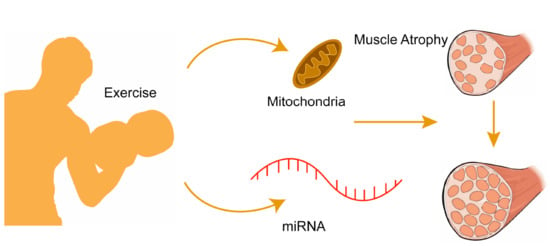
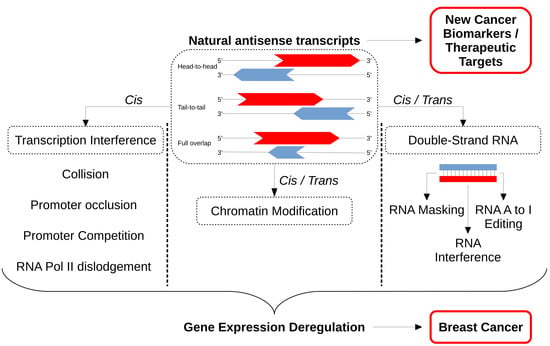
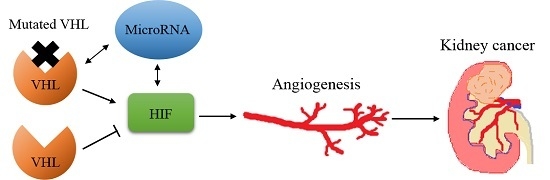
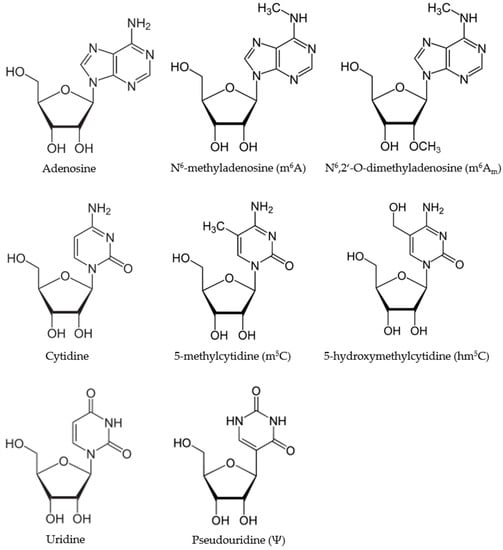
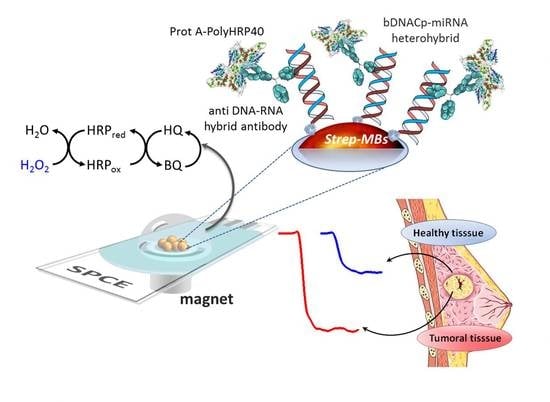
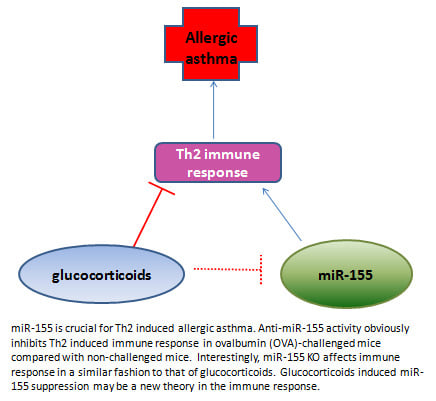
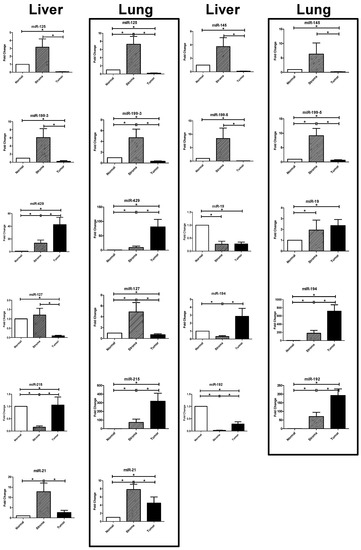
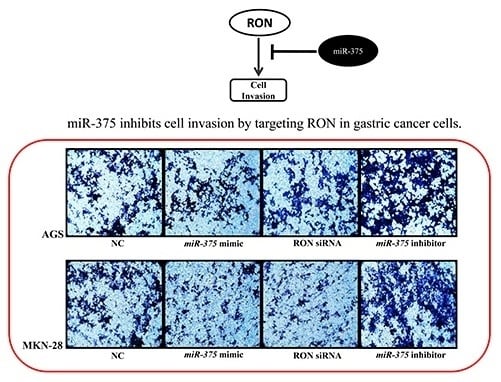
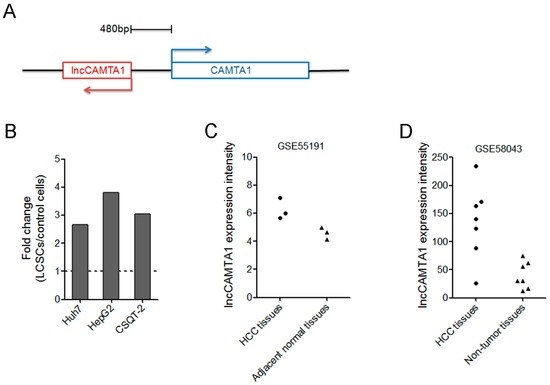
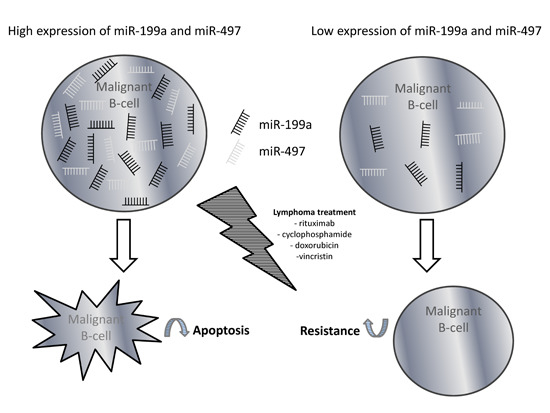
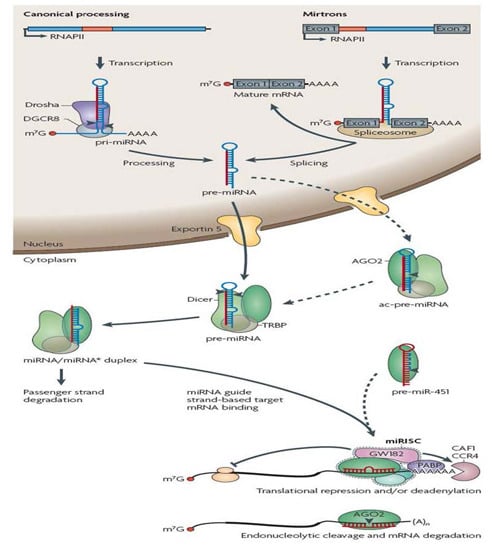
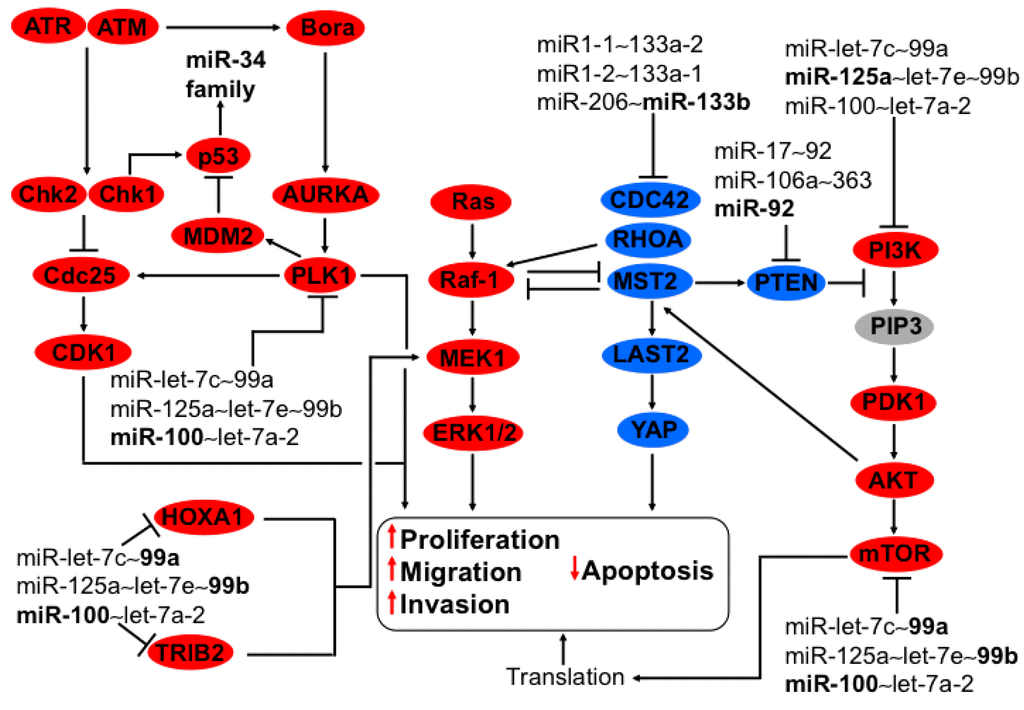
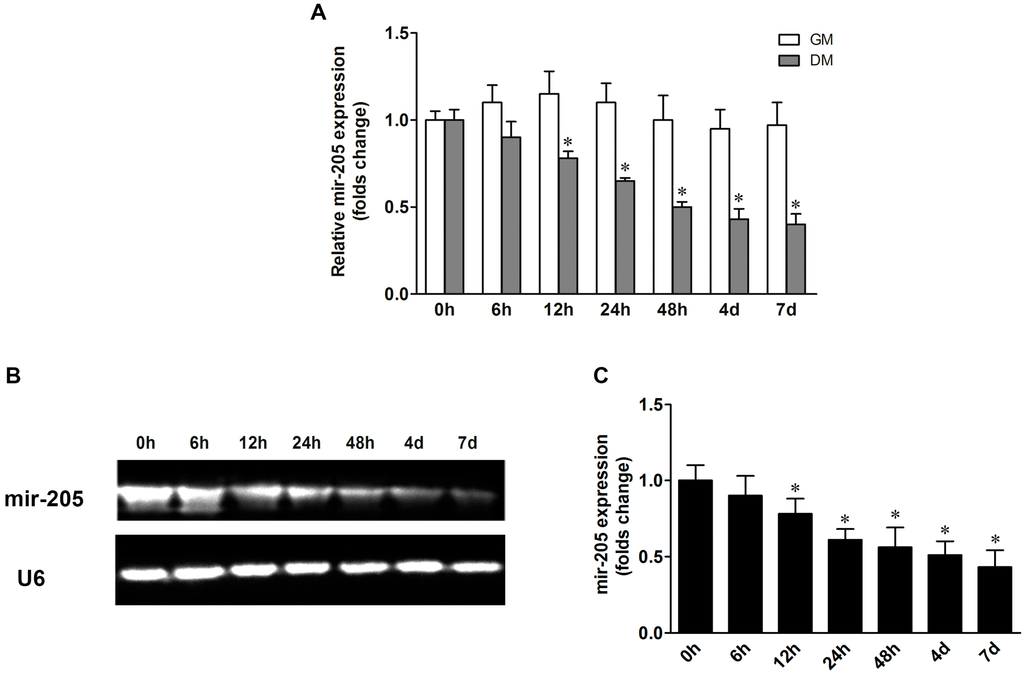
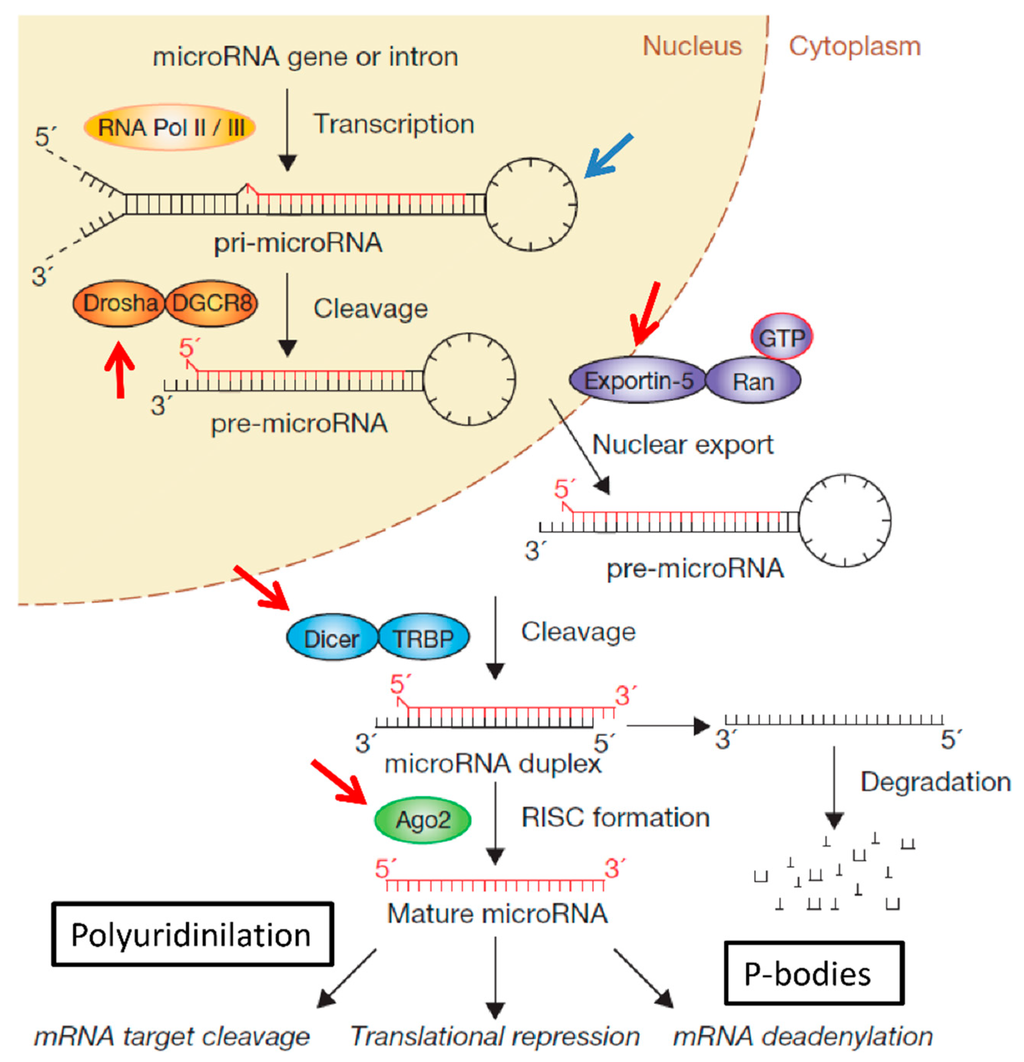
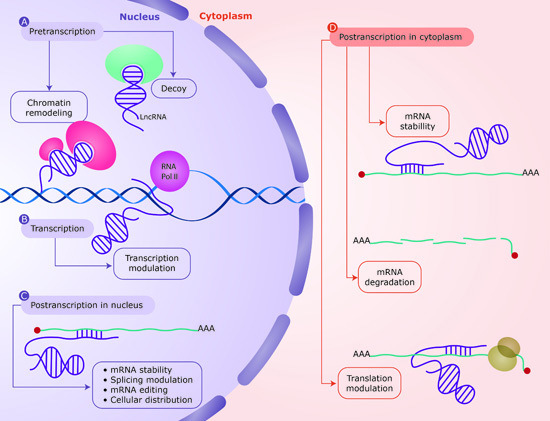
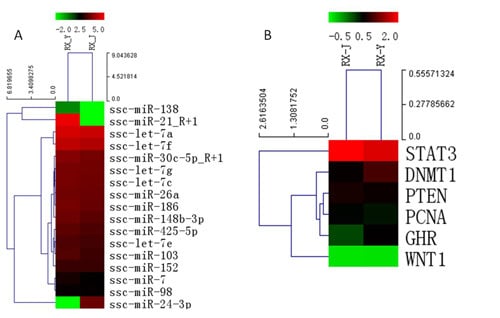
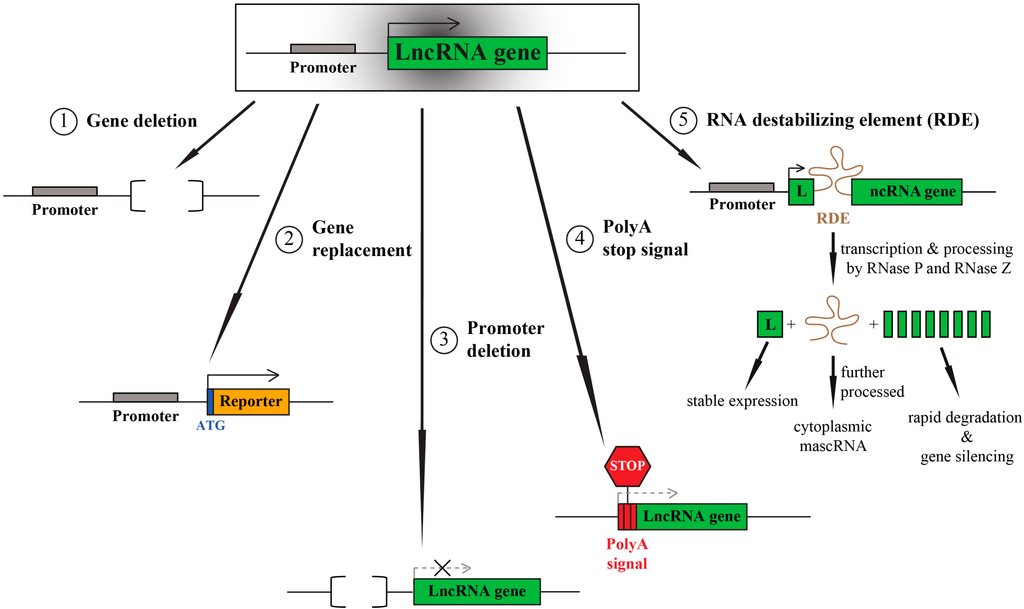
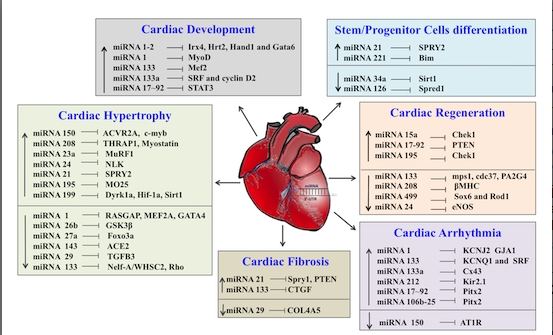
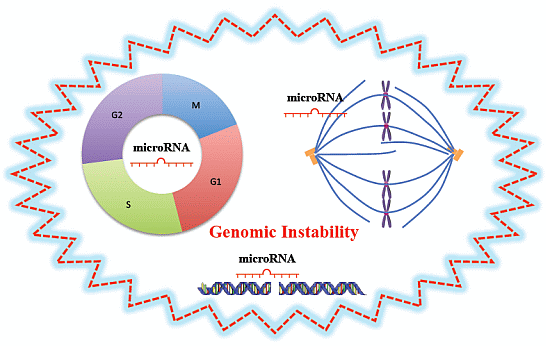
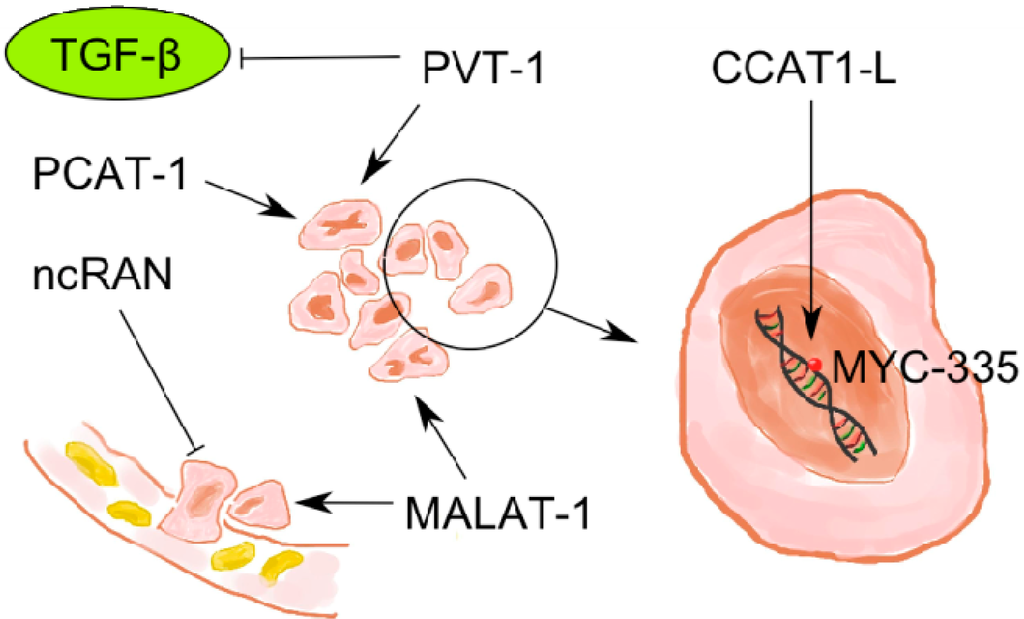
Non-Coding RNAs are currently a hot research topic in many fields of biology, medicine, and chemistry. It is increasingly clear that non-coding RNAs are involved in fundamentally physiological and pathological processes. These processes touch on many important disciplines, from metabolism to cancer. Non-coding RNAs are regulative: they mainly influence biological processes by regulating other (protein-)coding gene expression. By doing this, the cellular properties of development and growth, stem cell regeneration, apoptosis, authophagy, etc., are strictly controlled by non-coding RNAs. This collection is dedicated to summarizing and highlighting the current research concerning the role of non-coding RNAs in regulating the aforementioned functions. The underlying mechanisms of action, the target molecules, the interactor pairs, and the pertinent cellular functions should all be presented. All relevant fields in medicine (with a special focus on metabolism, cancer, and inflammation) are of interest. The classes of non-coding RNAs should include microRNAs, other small non-coding RNAs, and long non-coding RNAs. Original research articles, review articles, and research letters are welcomed.
Dr. Martin Pichler
Collection Editor
Manuscript Submission Information
Manuscripts for the topical collection can be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. All papers will be peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on this website. The topical collection considers regular research articles, short communications and review articles. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page.
Please visit the Instructions for Authors page before submitting a manuscript. The article processing charge (APC) for publication in this open access journal is 2900 CHF (Swiss Francs).
Keywords
- Regulatory RNA
- sRNA
- ncRNA
- lncRNA
- miRNA
- siRNA
- piRNA
- CRISPR RNA
- regulatory small RNA fragments
Related Special Issues
- Regulation by non-coding RNAs 2013 in International Journal of Molecular Sciences (52 articles)
- Non-Coding RNAs 2012 in International Journal of Molecular Sciences (21 articles)
- Non-Coding RNAs in International Journal of Molecular Sciences (12 articles)